Abstract
Aging is an inevitable progressive decline in every physiological function and serves as a primary risk factor for cognitive decline and Alzheimer’s disease. Thus, age-dependent impairments in cognitive function must be understood in association with general aging processes with an integrative approach in a systemic manner. An integrative aging gene network was constructed based on mutual molecular interactions using literature-curated interactome data and separated into functionally distinct modules. To investigate key surrogate biomarkers of the aging brain in the context of the general aging process, co-expression networks were built on post-mortem and Alzheimer’s brain transcriptome data. In both the normal aging brain and the brain affected by Alzheimer’s disease, the immune-related co-expression module was positively correlated with advancing age, whereas the synaptic transmission-related co-expression module was decreased with age. Importantly, the network topology-based analysis indicated that complement system genes were prioritized as a surrogate biomarker in evaluating the process of brain aging. Our public data-centered analysis coupled with experimental validation revealed that the complement system is likely to be a master regulator in initiating and regulating the immune system in the aging brain and could serve as reliable and surrogate biomarkers for the diagnosis of cognitive dysfunction.
Brain ageing: Network map implicates immune-related genes
The body’s immune response to microbes and damaged cells is associated with age-related cognitive decline, a gene network analysis shows. Jisook Moon from CHA University in Seoul, South Korea, and colleagues used published data on gene interactions to construct a network map of molecular determinants behind ageing in the brain. They identified clusters of genes involved in regulating immune responses, in particular those associated with the so-called complement system, which is responsible for activating inflammatory proteins that help clear the body of pathogens and infected host cells. The researchers suggest that complement-related genes may serve as a “master regulator” of brain aging. They propose that measuring expression of these genes could serve as a diagnostic biomarker for the reduced activity of synaptic transmission in brain that leads to cognitive impairment.
Introduction
Aging is defined as an inevitable, progressive loss in physiological functions and it predisposes the aged to diseases and death1. Among the functional declines induced by aging, cognitive impairments are considered the most debilitating aspects of the process2. Consistently, Alzheimer disease (AD) occurs frequently with increasing age, indicating that the age is an obvious risk factor for dysfunction in the brain3. However, the previous studies lack a mechanistic explanation of how aging affects the brain impairments and whether there are certain molecular determinants that serve as a relay between the two processes. Biological relevance of aging to the brain impairments needs to be understood within a systemic framework primarily due to the stochastic nature of it.
Brain aging has been subjected to rigorous data-driven studies4,5. In addition, several databases have provided a manually curated source of aging gene6,7. The accumulation of high-throughput data in public domain facilitates the better understanding of the aging brain in new perspectives. A biological module is an entity consisting of physically connected, functionally coordinated, and/or co-regulated molecular components and interacts with other modules to perform certain actions in living organism8,9. In the case of aging study, module-based approaches are promising in that aging-related molecular changes are hard to discover and frequently caused by combinatorial effects of multiple molecular perturbations rather than an effect of an individual component10. Thus, it seems reasonable to assume that searching for aging-related modules is an efficient strategy to answer the fundamental questions of brain aging using substantial amounts of information.
In our study, we constructed two different networks in two different biological contexts, namely, general aging (non-tissue specific and systemic) and brain aging (tissue-specific): (1) functional association networks of physically connected aging genes and (2) co-expression networks of genes in brain aging. We exploited current biological knowledge of aging to infer the general features of aging in the context of the interactome and construct several aging-related modules that are often associated with various diseases at the module level. Additionally, using transcriptome data of the human brain, we construed co-expressed modules of genes that correlate with expression across increasing aging. To identify the common molecular determinants in regulating general aging processes and specific brain aging, the two different networks were integrated. As a result, two small overlapping clusters were identified and associated with the complement system and the regulation of cytokines. The conserved cluster of genes enriched in the complement system exhibits robust perturbations in the interactome in terms of connectivity significance and tends to locate at the center of the co-expressed modules. Our integrative analysis of brain aging revealed that genes involved in the complement system may constitute a key regulatory module by potentially inducing and governing normal cognitive aging.
Materials and methods
Aging gene selection
We compiled a list of genes related to human aging from two curated databases, Human Ageing Genomic Resources7 and Digital Ageing atlas6. The interactome data were extracted from a previously published literature11. In the study, the aging-related genes that did not form a highly inter-connected sub-network, “the largest connected components” (LCC)12, in the interactome were excluded for analysis to focus more on aging-relevant molecular events.
Functionally distinct modules
The RDAVIDWebService13 was used to annotate the selected aging genes with enrichment analysis of Gene ontology biological processes (GOBP) and KEGG pathways. The significance of enrichment was determined at a false discovery rate of 5%. Next, we constructed a network consisting of the enriched terms connected by their shared genes (combined Jacard and overlap coefficient, >0.375) using the EnrichmentMap cytoscape package14. A clustering algorithm15 was applied to the Enrichment map to identify biological functional units in the context of aging.
Network-based separation between aging-related and disease modules
To understand the relationship between aging and disease in the interactome, disease modules data were extracted from published literature11. We quantified the network-based separation between disease and aging modules. The approach used the shortest distance within same module (dage, ddisease) and between different modules (dage, disease) and quantified the separation between two diseases at the module level:
Negative values of the quantification (Separationage, disease) indicate that the corresponding disease and aging modules are highly likely to topologically and functionally overlap in the context of the interactome.
Co-expression module analysis
Expression data (GSE46706) of post-mortem brain tissue from individuals aged 16–102 were downloaded, processed, and normalized using the oligo package16. We implemented weighted correlation network analysis (WGCNA)17 to identify co-expression modules reflecting aging-specific expression patterns in the brain. Each module was annotated with relevant biological processes. The representative expression of each module was defined as an eigengene (first eigenvector). The same approach was applied to a set of Alzheimer data (GSE48350)18.
Measuring the connectivity significance
To identify context-dependent core modules in brain aging, transcriptome and molecular interaction data were integrated9. Two core modules were derived through the overlay of co-expressed network in the aging hippocampus on the physically connected network in the aging process. Next, we applied an algorithm devised by Barabasi et al.12 for measuring the connectivity significance of and prioritizing two core modules. The ultimate goal of the algorithm is to identify a node that has its most significant interaction with seed genes or set of genes of interest compared with non-seed genes.
As described in ref. 12, the probability that a node with a degree of K in the interactome (N) has Ks links to seed (Sn) is calculated using the hypergeometric distribution. The connectivity significance is calculated as cumulative probability for the higher links than the observed (Ks).
Using the genes identified in our analysis as seed genes, we added the most significant connected genes (according to p-value) to the seed, which was grown through iterative addition to form a connected subnetwork. Then, we created two extended networks starting from the two small, core modules or an individual gene of the two sets. Genes of the two extended sub-networks were compared with each other and the number of overlapped genes was counted for each module by the following:
S(n, individual)
=A set of n genes of an extended module starting from an individual gene
S(n, whole)=A set of n genes of an extended module starting from whole genes.
Excess retention analysis
We investigated whether small sets of genes of interest are close to the hub or periphery in the functional association and co-expression modules using excess retention analysis or “Peeling”. As defined in references to the analysis19,20, the m-core was generated by removing genes with a degree less than m, resulting in a network in which genes with a degree of at least m formed the LCC. A fraction of genes with certain properties () in an m-core (Nm-core) was compared with the fraction of genes with the properties (Ni) in whole genes of a certain network (N).The proportion, which is also excess retention, is expressed as a function of m values ().
Culture of mouse cells
All animal procedures were approved by the Institutional Animal Care and Use Committee in accordance with the Guide for the Care and Use of Laboratory Animals. Mouse hippocampus tissues were dissected from the brain of young (8 weeks, C57BL/6) and old female (25 months). Cells were cultured in basic fibroblast growth factor (20 ng/ml) in serum-free N2 medium. Mouse HT22 (Hippocampus cell line) and BV2 (Microglia cell line) cells were cultured in DMEM media with 10% FBS and penicillin/streptomycin.
Isolation of human placenta-derived stem cells
Human term placentas (≥37 gestational weeks) were obtained from donors with their informed consent according to the Institutional Review Board of the CHA General Hospital, Seoul, Korea. The amniotic membrane isolated from dissected placenta was cultured21.
Quantitative reverse transcription and real- time PCR
Quantitative real-time PCR was performed using the SYBR Green PCR Mix (Bioneer) according to the manufacturer’s protocol. Levels of mRNA were analyzed with relative quantification22.
Immunoblot analysis
Whole cell lysates were normalized using the Bradford reagent (Bio-Rad) and 20–50 μg of the lysate was subjected to 10–15 % SDS-PAGE and transferred to a PVDF membrane (Millipore). Membranes were blocked in TBS-T containing 5% skim milk (Becton Dickinson) and then probed with each antibody (P16, p21, and GAPDH; Santacruz). Immunoreactions were conducted using the ECL Western Blotting system (Millipore).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 for 20 min and then blocked by 10% NGS (normal goat serum). Cells were incubated at 4 °C for overnight with C3 antibody (abcam) diluted in 2% NGS.
RESULTS
Construction of physically connected and functionally distinct modules in aging genes
Aging is a primary factor for developing brain deterioration23. To identify the major drivers of the process, brain aging needs to be understood in the context of general aspects of aging. In addition, aging is attributed to the interplay of multiple molecular components, modulating multiple biological processes, and aging should be elucidated in the context of the interactome11, a network representing all the molecular interactions. In addition to the physical connection, molecular components affected by aging are most likely to form functional networks or modules with similar biological processes. First, we extracted age-related genes from two data repositories6,7, integrated the genes with the interactome data and selected the LCC12 consisting only of the aging genes inter-connected by at least one of molecular interactions. The physically and functionally interconnected sub-network was subjected to functional enrichment analysis using DAVID, a functional annotation tool13. Then, we organized enriched GOBP and KEGG terms into a network in which mutually overlapped terms are connected with each other.
Given that genes with functional similarities tend to be densely connected, the enrichment network was subdivided by a clustering algorithm15 into several clusters of highly inter-connected enriched terms that may capture functionally distinct modules.
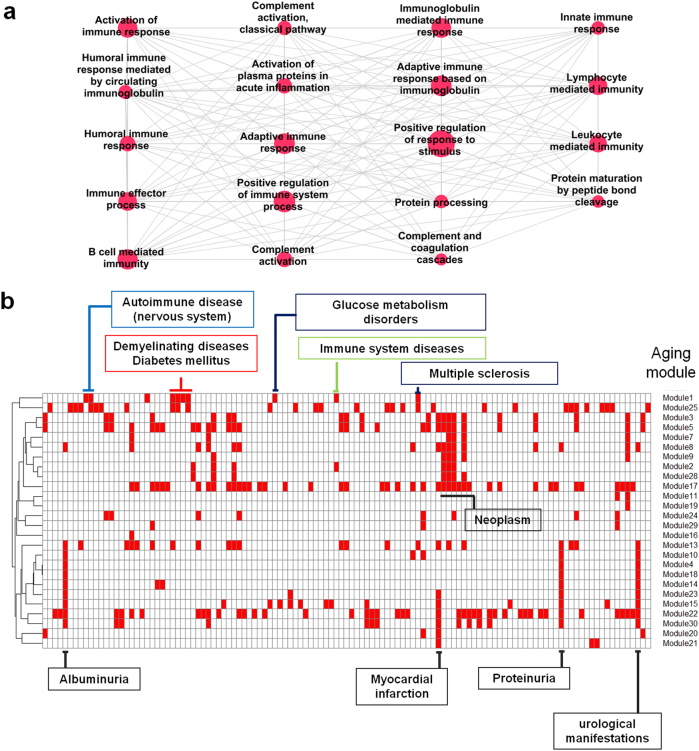
After manual elimination of abundant annotation terms, 30 aging-related functional modules or clustered terms were constructed (Fig. 1, Supplementary Figure S1).The modules were numbered in increasing order from 1 to 30 according to scores of the clustering algorithm15 (Supplementary Table S1); 1 is assigned to the module with the highest score (most densely connected to each other).Next, we assessed whether genes of the clustered terms are significantly interconnected in physical interactions by comparing each of 30 modules with randomly generated networks having the same gene number of the corresponding module and confirmed the highly significant inter-connectivity of them12(Supplementary Figure S2,S3, p < 0.00001), meaning that their components are not randomly distributed in the interactome. As a result, we created functionally and physically distinct modules associated with the aging process. The aging modules were characterized by several biological processes, including immune responses, metabolic processes, cell death, developmental processes, and cancer pathways.
Fig. 1.
Workflow for constructing functional association networks of aging genes. Our analysis was performed on aging genes forming the largest connected component (LCC) in the interactome. The LCC was subjected to gene ontology biological processes (GOBP) and KEGG enrichment analysis. Enriched terms were connected with each other according to their shared genes to generate an enrichment map which was separated into densely connected enriched terms. As a result, 30 aging modules were created. The terms were mapped to the corresponding genes, most of which are physically connected with each other within each aging module
Network-based relationships between aging and disease
In the perspective of graph theory, among the 30 aging modules, the sub-network of clustered terms with the highest score (aging module 1, Fig. 2a) is most similar to the complete graph in which every pair of distinct nodes is connected. Thus, we will focus on the identification of embedded markers for aging in a complex biological system with biological network concepts in the following analysis, despite the fact that important properties, such as the complete graph in terms of graph theory, are not necessarily biologically meaningful.
Fig. 2.
Immune-related aging module and network separations between disease and aging modules. a Aging module 1 with the highest score of MCODE clustering algorithm15 was implicated in immune processes. b Twenty-six aging modules topologically overlapped with several disease modules. Red rectangles indicate topological overlap
In a recent study, Barabasi’s group delineated disease–disease relationships in the context of the module and interactome through the network topology-based approach. Furthermore, they mathematically measured the network-based separations between disease modules and identified the links of otherwise seemingly unrelated diseases. If the values of the measurements are minus, two modules are assumed to be biologically and pathologically related, implying that perturbation in one module is likely to lead to deregulation in the other11. We extracted a list of disease modules manually curated by Barabasi’s group and applied the same strategy to investigate the relationship between diseases and aging at the module level. Specifically, genes of aging module 1 enriched in immune processes are in the neighborhood of modules of autoimmune disease of CNS (79 genes), demyelinating disease (84), multiple sclerosis (65), diabetes (151), immune system disease (377), and glucose metabolism disorders (164), forming unique topological overlaps compared with other modules (Fig. 2a, b). The distinct relevance suggests that perturbation of the aging-related immune module leads to a range of diseases, especially neurodegenerative disease and metabolic disease. In addition, the diseases, which are the first neighbors of module 1, are in the vicinity of each other in the interactome, suggesting high rates of comorbidity (Supplementary Figure S4). More importantly, the disease modules of autoimmune disease of the CNS and glucose metabolism disorders were uniquely related to aging module 1, which consists of genes participating in complement, innate, and adaptive immune responses (Fig. 2a).Neoplasm (454 genes), albuminuria (14), myocardial infarction (10), proteinuria (14), and urological manifestations (14) overlapped with several aging modules, suggesting that the diseases are commonly encountered with age (Fig. 2b).
In terms of identifying reliable and surrogate markers for aging, genes of the term cluster with the highest score (aging module 1) are highly likely to be possible genetic markers for aging. In addition, the genes of aging module 1 could contribute to a major response to aging, given that the aging genes are defined by information regarding gene expression with age across all organs and tissues (general aging).
Thus, it is highly anticipated that genes involved in the immune system are most likely to be molecular determinants for brain aging and dysfunction; however, experimental data related to brain aging such as gene expression are needed to confirm the anticipation and capture the aging-specific information.
Identification of co-expression modules
Thirty aging modules in our analysis are constructed to infer general features of aging processes (general aging) in the context of the interactome. However, for brain aging, additional tissue-specific information is needed. Groups of genes tend to work together and exhibit co-expression patterns to perform certain functions24, highlighting the importance of co-expression module-centered analysis.
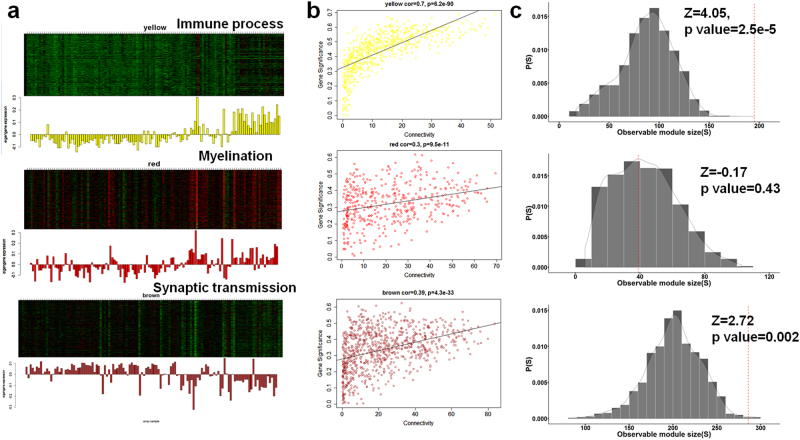
The following are criteria for selecting key sub-networks associated with the aging process of the brain: (1) associated with aging and having similar biological functions; (2) connected to each other in the interactome significantly more than expected; and (3) co-expressed in the aging brain to perform certain functions. To focus principally on brain aging, we performed WGCNA17 on gene expression of post-mortem human brain transcriptome data in the public domain11 and integrated the output of the analysis with that of aging modules. For each brain region, we identified co-expression modules (on average, 13.8 modules per region) and computed the corresponding eigengene (representative expression). To determine the relationship between brain aging and gene expression, we correlated the summarized expression (eigengene) of each module with age information. A module (yellow in Fig. 3a) in the hippocampus exhibited the highest correlation coefficient (0.66) among the detected co-expression modules in 10 brain regions, indicating that the sub-network was the most significantly perturbed module with increasing aging. The module (yellow module, 603 genes) tends to show an age-dependent increase and was enriched in the immune process (Supplementary Figure S5a).In terms of expression profile, a module enriched with myelination and oligodendrocyte differentiation (red module, 447 genes) was positively correlated in expression with the immune-related module(yellow module), whereas the module enriched with synaptic transmission and neurogenesis (brown module, 873 genes) were negatively correlated with the most relevant module, exhibiting an age-dependent decrease in expression (Fig. 3a, SupplementaryFigure S5b, S6).
Fig. 3.
Co-expression network analyses on brain aging. a Three co-expression modules in the aging hippocampus were highly correlated with age in gene expression (positive correlation for genes associated with immune and demyelination; negative correlation for those with synaptic transmission). Each bar graph corresponds to an eigengene (representative expression of each module, left panel). b Linear regressions between intramodular connectivity and gene significance (the extent of correlation with aging). c The observed number of physically connected genes (in the interactome) in each co-expressed module was compared with that of randomly generated modules. From top to bottom, yellow, red, and brown modules were displayed. Yellow module was enriched in immune process, red in myelination and brown in synaptic transmission
In WGCNA17, the intramodular connectivity of certain genes was measured as a sum of correlations with other genes within modules, and genes with higher values correspond to those closer to the hub location in the co-expression network. We performed a linear regression on links between the intramodular connectivity and gene significance (GS, extent of correlation with increasing aging). Co-expressed genes associated with immune process exhibited the highest positive correlation (R > 0.7) between the two measurements. This result indicates that the genes exhibiting a higher correlation with advancing age tend to locate at the hub of the co-expressed network in the context of the aging hippocampus (Fig. 3b).
Given that the molecular components at the hub of a molecular network tend to play an essential role, immune-related genes were also identified as possible molecular determinants for brain aging. Across 10 brain regions, various genes related to immune process were co-expressed and characterized by an age-dependent increase. However, none of the genes exhibited an increased correlation of gene significance with intramodular connectivity compared with the aging hippocampus-specific module. Although the co-expression networks and the interactome differ for defining connectivity between nodes, the co-expressed genes involved in the immune process and synaptic transmission were significantly connected even in the interactome compared with randomly generated modules (Fig. 3c).
Importantly, the relation between the two co-expression modules is similar as that one between immune-related aging genes and several neurodegenerative diseases, such as demyelinating disease in the interactome of aging genes (Fig. 2a, b): the similar biological theme appears in the interactome of general aging and the co-expressed networks of brain aging. Conservation of the relationship indicates that immune-response and synaptic transmission are main components that link general aging process with the aging brain.
At the modular levels in the aging hippocampus, the immune process-enriched module was perturbed the most with advancing age; however, the synaptic transmission-related module was assigned a relatively lower GS compared with the immune module. This finding implies that an increase in the immune process could be an apparent response to advancing age in parallel with a decrease in synaptic transmission and an increase in myelination. Interestingly, genes participating in presynaptic process involved in synaptic transmission (right in Fig. 4; such as CPLX2, STX1B, RIMS1, GRIK5, STX1A, SYN1, DNAJC5, and CADPS2) were anti-correlated in gene expression with genes annotated with complement activation (left in Fig. 4; such as C1QA, C1QB, C1QC, C1R, C1S, and C3). Taken together, aberrant increases in immune response with aging could be considered as a surrogate biomarker for the reduced activity of synaptic transmission in brain.
Fig. 4.
Two co-expression modules in the aging hippocampus. Two representative sets of genes co-expressed with hippocampus aging and associated with immune process (left) and synaptic transmission (right). Two sets are anti-correlated in terms of gene expression. Widths of edges are proportional to the extent of expression correlation
Identifying co-expression modules in AD
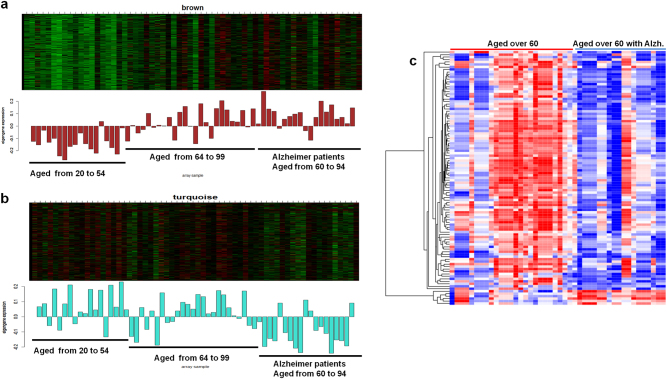
With aging, anti-correlations in gene expression between immune process and synaptic transmission play a prominent role in the brain. Based on the our results, we hypothesized that immune and synaptic transmission processes are possibly correlated with cognitive impairments by aging, and the brain aging-related modules may be more prominent in AD. Thus, we investigated whether the two co-expression modules in the aging hippocampus also appear under AD. Using an expression data set of normal (aged from 20 to 99) and AD hippocampus (60 to 94), the WGCNA was performed. To identify AD-related modules, we correlated the representative expression (eigengene) of each co-expression module with the presence of the disease (AD is coded as 1 and normal as 0). The expression of genes involved in immune process and synaptic transmissions were significantly correlated (p < 0.001) with the presence of AD. The direction of the gene expression (up or down) with age, irrespective of normal or AD, was consistent with the previous co-expression result of the aging hippocampus.
However, AD patients revealed that immune-related genes tend to be upregulated and synaptic transmission-related genes to be downregulated compared with the age-matched normal (Fig. 5a, b). In particular, numerous synaptic transmission genes were significantly downregulated in AD compared with normal, indicating their primary contribution to the disease phenotypes (Fig. 5c, Supplementary Figure S7).The immune-related genes were not significantly, differentially regulated in AD, although the module was primarily characterized by an age-dependent increase in a co-expression pattern.
Fig. 5.
Gene expressions for Alzheimer’s disease. Co-expressed genes were enriched in immune process (a) and synaptic transmission (b). Bar plots correspond to eigengenes. c Differentially expressed genes between AD patients and the age-matched normal were enriched in synaptic transmission. Most of them were downregulated in AD compared with the normal
Instead of directly leading to pathological conditions, increased immune response may consist of main reactions to cognitive impairments and be a clinical indication of cognitive dysfunction induced by decreased synaptic dysfunction. This result further suggests that immune and synaptic transmission processes are likely associated with aging-dependent cognitive impairments, exhibiting a mutually opposite direction in expression.
Identification of brain aging-related metabolic pathways based on gene information
The co-expression and DEG (differentially expressed genes) analysis revealed that decreased activity in synaptic transmission is highly likely to mainly contribute to age-dependent cognitive impairments. Human’s high cognitive function requires neuronal activity coupled with high demands of energy metabolism25. In our analysis, the immune process-enriched module (aging module 1) was in the close neighborhood of the metabolic disorders module (Fig. 2b). Accordingly, we investigated whether there are metabolic pathways relevant to the co-expressed module enriched with synaptic transmission. Gene expression data were used for prediction of relevant metabolites based on the premise that transcripts have dynamic characteristics as metabolites.
Genes of the co-expression module enriched in synaptic transmission (Fig. 3a, brown module) were mapped to metabolites (KEGG compound). The matched compounds in the module were overrepresented in fatty acid biosynthesis and glycolysis and categorized as energy metabolism with topological importance (Supplementary Figure S8). However, genes of the co-expressed module related with the immune process were not enriched with energy metabolism, suggesting that the immune process is not a direct causal link to cognitive dysfunction.
In the case of fatty acid biosynthesis, a series of studies have deciphered relationships between age-related cognitive decline and fatty acid biosynthesis26. According to the literature, metabolic dysfunction in the brain is well reflected in the progressive malfunctioning of glycolysis27.
A recent study demonstrated that the local generation of ATP by glycolytic enzymes is required to meet excess energy demands by syntactic transmission28. This result indicates that perturbation in energy metabolic pathways may be accompanied by aberrant or decreased neuronal activity in the aging brain and that metabolites can be surrogate markers for detecting cognitive dysfunction with genetic markers.
Complement system plays a key role for normal cognitive aging
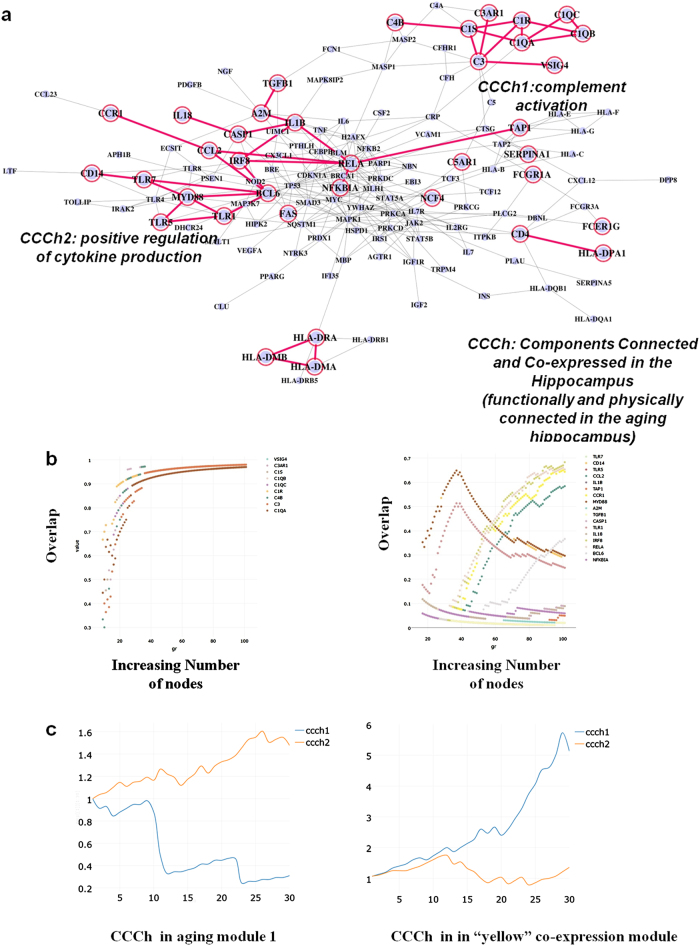
To understand brain aging in the context of the aging process and identify the most relevant genes in age-dependent cognitive impairments, we overlaid genes of the co-expressed module in the aging hippocampus on the 30 aging modules and found that 37 genes were overlapped with aging module 1. Of 37 genes, we identified two overlapped groups of highly connected components whose connectivity were preserved in the interactome (general aging) and co-expression network (brain aging) (Fig. 6a).The two core modules, hereafter called Components Connected and Co-expressed in the aging hippocampus (CCCh) 1 and 2, were enriched in complement system (CCCh1, 9 genes) and positive regulation of cytokines (CCCh2, 17 genes), respectively. After complementing biological evidence of aging with transcriptome data of brain aging, we discovered two small sets of aging genes that are physically connected in the interactome, share similar biological functions and are co-expressed with aging in the hippocampus.
Fig. 6.
Integration of functional association and co-expression networks. a Intersection of the two different networks leads to the identification of two small modules (CCCh1 and CCCh2) connected in the context of general aging and brain aging. CCCh means “Components Connected and Co-expressed in the aging hippocampus”. b Networks starting from each gene or whole genes of CCCh were grown through the iterative addition of the most significant interaction partner until the expanded modules reach100 nodes. The expanded modules starting from individual genes were compared to the expanded module starting from whole genes of CCCh. X axis points to the gene number of expanded modules and Y axis to proportion of overlap between the two expanded modules. Legends in the upper left of the plots correspond to the seed genes (starting individual genes). c Excess retention analysis showed that CCCh1 tends to be located at the periphery of aging module1 (association network or part of interactome) and at the hub of yellow co-expression module. An m-core of x axis means a network consisting of genes with a degree of at least m and is generated by removing the genes with a degree less than m. Y-axis indicates the enrichment of genes with certain properties (in this case, CCCh1 and CCCh2) in each m-core. For example, a set of genes close to or at the hub tend to show an increasing pattern in the analysis but a decrease in genes at the periphery with increasing m values
The two key modules must be prioritized according to biological significance and integrity in the interactome to narrow down the most aging-relevant candidates in the hippocampus whose age-related deterioration may be frequently associated with cognitive problems29.
In WGCNA, the biological significance of each gene can be computed in regard to age by measuring GS. The higher the absolute value of the gene, the more significant correlation to age. We calculated mean values of GS of the two core modules. The GS mean value of CCCh1 (0.554), which was significant compared with randomly generated networks (z = 2.7, p < 0.05), was increased compared with CCCh2 (0.455), which was not significant (z = 0.6, p > 0.1).
A previous research12 devised an algorithm to predict the interaction partners of well-known disease genes and add the most significant connected genes into the existing disease modules based on connectivity significance in an iterative manner. The network-based analysis revealed that the expanded disease modules tend to be robust against perturbations with distinct interaction patterns12. Accordingly, we assumed that the core modules in the aging hippocampus may resemble the network property of disease modules due to the strong relationships between aging and disease.
To measure the integrity of the core modules, we modified the algorithm (Supplementary Figure S9a) by removing N − 1 genes of the two modules and making the remaining one gene as a starting seed gene, thereby constructing N different expanded modules which we compared to the expanded sets of the whole genes of the two (SupplementaryFigure S9b, c, d). If a set of genes has significant links to each other and is robust again perturbation (simulated by removing the N − 1 nodes of a module and making the remaining one gene as a seed set), the genes tend to maintain preferences in connectivity to certain nodes during the expansion process regardless of variations in the set of starting seed genes (starting from one gene (Supplementary Figure S9b) or full genes (Supplementary Figure S9c) of CCCh modules). The integrity was measured as a fraction of shared genes between N different expanded sets (staring from each gene) and the expanded set of the full genes of the two CCChs (Fig. 6b, Supplementary Figure S9d). Genes of two different seed-based expanded modules overlapped in ~100% in 9 genes of CCCh1 but not in CCCh2, suggesting that the genes of CCCh1 have more robust integrity in the interactome compared with CCCh2.
For further confirmation, we removed each gene from the two CCCh and measured the number of iteration steps necessary for the missing link to be recovered and integrated into the modules of N − 1 genes (Supplementary Figure S10). Based on the property of the algorithm, it seems reasonable to assume that if the connectivity significance is high, the missing link is recovered at an early iteration of extension. C1QA, C3, C1R, C1QB, and C1 of complement systems are added according to connectivity significance at iteration step 2 (Supplementary Figure S10a), indicating their robust integrity in the interactome. Conversely, most genes of CCCh2 were not predicted as the significant interaction partners until the expanding modules reached 100 nodes (Supplementary Figure S10b).
Excess retention analysis19, to measure the centrality of molecular components in network (Supplementary Figure S11), revealed that CCCh1 tend to be located at the periphery of aging module 1, but at the hub of the co-expressed module (the aging hippocampus) showing the highest age-relevant expression. Opposite results were obtained for CCCh2 (Fig. 6c). Additionally, we performed one of unsupervised learning, PCA (principal components analysis), using 9 genes of CCCh1 as features to verify the CCCh1 as biomarkers of brain aging.The PCA plot (Supplementary Figure S14) displayed that the older subjects tend to be located in the right side of the plot and the younger subjects in the left side of the plot, indicating that complement genes are good predictors of brain aging.
Taken together, genes of the complement system may be perturbed the most with increasing aging among immune-related aging genes and are highly likely to be surrogate markers for brain aging.
Experimental validation of responses in complement system to stresses
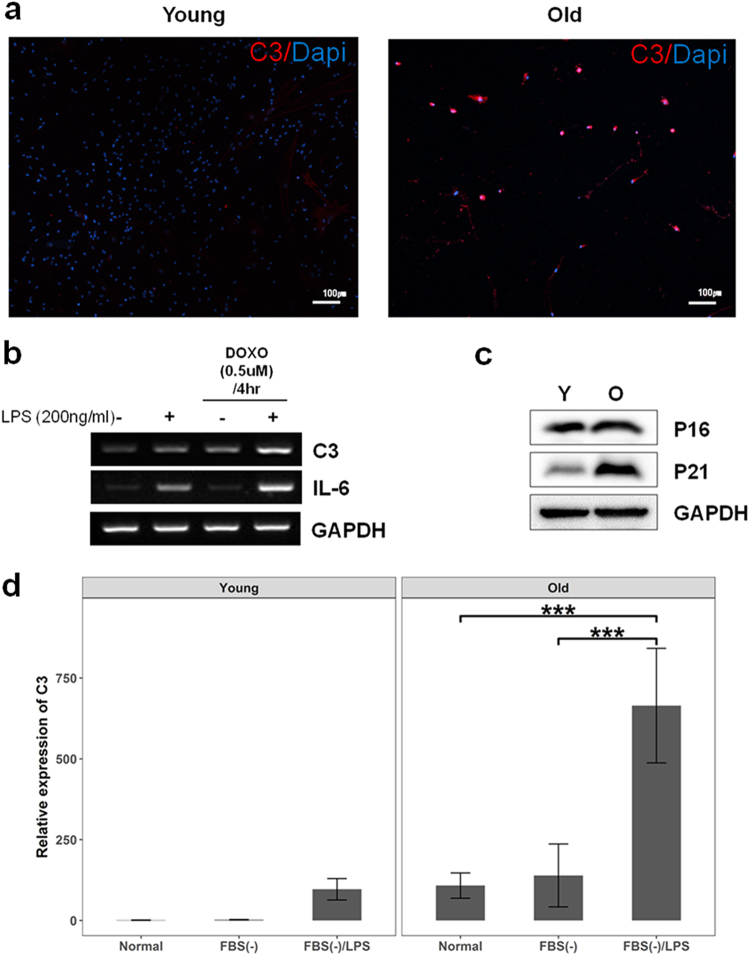
The integration of the aging network with co-expression modules in the aging brain predicted that genes encoding complement system components constitute an important small module in the process, exhibiting time-dependent increased responses to advancing age (Supplementary Figure S12). For experimental validation of the response, we recapitulated environmental stresses, withdrawal of media supplements (fetal bovine serum(FBS)), DOXO and/or lipopolysaccharide (LPS) treatment during cell culture, and expression of C3, a main component of complement system, was quantified through real-time PCR analysis in young and old cells.
In primary hippocampal cells, aged cells are more immune-responsive to C3 than young cells in immunohistochemistry analysis (Fig. 7a). However, because aged primary cells are vulnerable to the treatment for extracting mRNA, HT22, immortalized mouse hippocampal cells, were used for gene expression analysis rather than primary cells. HT22 cells treated with DOXO to recapitulate aged environments displayed increased expression of C3 and IL-6, a pro-inflammatory cytokine, compared with control cells in response to LPS treatment (Fig. 7b).
Fig. 7.
Experimental validation of C3 expression in stem cells and primary cells. a Immunohistochemistry result showed that C3 positive cells in primary hippocampal cells were increased with age (Young: 8 weeks, Old: 25 months). b Cells pre-treated with DOXO displayed increased expression of C3 and IL-6 compared with control cells after LPS treatment. c Aged cells from placental stem cells were determined by increased expression of P21 (O: old, Y: young). dWhen depleted of FBS and treated with LPS, the aged placental stem cells showed dramatic increase compared to the young cells. Interaction effects between cell status (aged or young) and stress treatments (depletion of FBS and LPS treatment) were significant (F2, 24 = 22.96, p < 0.001). Data are expressed as mean ± SD. Scale bars: 100 μm in a,***p < 0.001 for comparison
Next, we investigated C3 expression with age in placental stem cells, which can be relatively easily converted to the aged status. Aging phenotypes of the cells were determined by increased expression of P21 (Fig. 7c).
Two-way ANOVA analysis revealed a significant interaction effect between cell status and stress treatments on C3 expression (F2,24 = 22.96, p < 0.001) with significant main effects of the two variables (F1,24 = 75.81, p < 0.001 for cell status; F2,24 = 46.59, p < 0.001 for stress treatments). Pair-wise comparison using the Benjamini-Hochberg procedure revealed that aged cells depleted of FBS and treated with LPS dramatically increased (p < 0.001) C3 expression as compared with the expression change in young cells (Fig. 7d). This result indicates that C3 responsiveness of young placental stem cells was more robust against external challenges than aged cells. In other words, C3 of aged cells were relatively easily perturbed compared to young cells. Taken together, age may compromise the robustness of the complement system against external challenges.
DISCUSSION
Aging is characterized by a progressive physiological decline in multiple organ systems and the interplay of many processes through diverse molecular interactions. Age-dependent impairments in brain function manifest during the aging process and subsequently lead to dementia. Due to its complex nature, studies of brain aging have been challenged with the task of uncovering its general properties in a systemic manner. In addition, aging-related molecular determinants are likely curated in a broad fashion across a range of databases, impeding the identification of principal aging-related modules probably embedded in diverse biological networks. Thus, the aim of our study is to discover core and reliable biomarkers reflecting normal brain aging based on current biological knowledge in the public domain in concert with network-based approaches.
We performed module-centered analysis on a set of aging genes to better understand the underlying mechanisms of the process, integrating literature-based evidence with interactome data. For efficient analysis, we focused on the core molecular functions of the aging process by selecting aging genes that interact with each other such that the selected genes formed the LCCs12 consisting of only aging genes in the interactome. Compared with randomly generated networks, aging genes defined by our criteria tend to agglomerate in the neighborhood of each other.
To construct functionally isolated modules, LCC of aging genes was subjected to enrichment analysis for GOBP and KEGG, and several enriched terms were generated. The over-represented terms were interconnected to each other by measuring their shared genes to construct an enrichment map (a node as an annotation term) that was subsequently separated into densely connected small networks by a clustering algorithm. Genes involved in each cluster of terms were defined as components of aging modules. Genes of each functionally isolated module are not randomly distributed in the interactome (Supplementary Figure S2, S3).In terms of aging in general, functional association networks consisting of physically connected aging genes were constructed using literature-curated molecular interaction and annotation data.
At the module level, the extent of separation between aging and disease modules was measured11. At the modular level, aging module 1, which was enriched in innate, adaptive and complement immune responses, was topologically overlapped with the module associated with autoimmune disease of the nervous system. The remaining two aging modules involved in the immune process (13 and 25) were not overlapped (Supplementary Figure S13). In addition, the genes associated with demyelinating autoimmune disease were within close proximity to aging module 1.This finding indicates that the module most likely plays an important role in mediating immune-related nervous diseases in the context of aging.
We used literature-curated aging genes and interactome data to construct aging modules. However, the result derived from the literature-curated evidence is highly likely to be biased toward most well-studied molecules and molecular interactions. Furthermore, aging genes were curated based on information of multiple organs, probably lacking tissue specific interpretations of the aging brain24. Accordingly, to improve tissue-specificity and focus more on brain aging, the transcriptome profile of post-mortem human brain was analyzed and integrated with the aging modules. Given that molecular components tend to behave similarly in the gene expression to perform biological functions under certain circumstances, we constructed co-expression modules and correlated the expression profile of each module with age information across 10 brain regions. Among the 10 regions, a set of genes co-expressed in the aging hippocampus was most positively correlated with increasing aging and enriched in immune process, exhibiting an age-dependent increase in immune activity. Most importantly, the co-expressed genes involved in immune reaction exhibited topological importance at a network perspective. The higher correlation with increasing age, the closer the genes are located to hubs in the correlation network (r > 0.7). The network properties may render the immune-related genes more robust to perturbation in the context of brain aging and aberrational changes in immune system or dysfunction induced by the aging could be molecular determinants for cognitive impairments.
Interestingly, genes involved in synaptic transmission and myelination were negatively and positively correlated with the immune related genes in expression, respectively, indicating that the immune-related genes play a transcriptional regulatory role in the brain aging. In age-matched (>60) comparisons of AD patients and a control group for gene expression, genes involved in the immune process and synaptic transmission showed AD-specific patterns. Immune-related genes tend to be upregulated in AD compared with normal brain, but the differences were not statistically significant. However, most of the differentially expressed genes in AD were downregulated and related to synaptic transmission in AD, which may reflect the pathological phenotypes. Similar to brain aging, an anti-correlation between immune and synaptic processes may underlie the mechanisms of age-dependent brain impairments. Increased immune reactions would constitute a main response of the brain to increasing age, and the altered responses could trigger aberrational expression changes in genes of synaptic transmission, which plays a potential causal role in inducing brain impairments30. This result is consistent with a recent report demonstrating that microglia and complement induce and mediate synapse loss in AD and indicating the close association between immune responses and cognitive impairments18,31. Also, in an earlier study of brain aging, genes involved in inflammation were upregulated and those in synaptic function downregulated32, but the study only considered changes in gene expression.
Next, given that aging phenotypes are reflective of interplays between multiple molecules, we attempted to infer metabolic pathways relevant to the aging brain exclusively based on expression data.
As shown in normal brain aging and AD, expression changes in synaptic transmission are clear manifestations of an age-dependent decline in brain functionality. Thus, it is reasonable to ask to what metabolic pathways pathological changes in synaptic transmission are closely linked. Using KEGG ortholog data, genes of the module involved in synaptic transmission were converted to the most likely relevant metabolites. The metabolites were statistically overrepresented in fatty acid biosynthesis and glycolysis, which are associated with brain dysfunction. Although the conversion between different molecular levels is conducted based only on information of the functional similarity among different species (ortholog genes), the co-expressed genes, which are the closest to phenotypes of the aging brain, could predict distinct enzymatic activity in functional decline.
In total, 37 genes of aging module 1 overlapped with the co-expressed module enriched in immune process. Then, we explored connectivity of 37 genes in the interactome and identified the two small groups of inter-connected components (preserved connectivity in the physical interaction and co-expressed interaction): the complement system and positive regulation of cytokine production. It is widely accepted that genes should be functionally and physically connected in a cooperative manner and exhibit correlated expression to complete certain tasks. The integrative strategy results in two small composite modules by overlaying physically interacted networks with a molecular profile of transcriptome data, adding dynamic and aged brain-specific information to the aging modules with static characteristics. Although, in general, co-expressed networks have transient interactions under certain conditions, an inter-connectivity of the composite modules was conserved in the two different networks. The two sub-graphs of genes embedded in the integrated networks satisfied the following properties: the genes of networks are (1) physically connected in the interactome; (2) classified with similar gene ontology terms (mainly associated with immune processes); (3) work together under the aging hippocampus at the transcription level and exhibit a distinct co-expression pattern; and (4) are strongly correlated with the aging process. Hence, they can be considered “active modules” that are associated with brain aging9.
In addition, individual gene significance (GS) (correlation of gene expression with aging) and significant connectivity was measured to prioritize the two core gene sets. Given GS values, genes of complement system are likely to clearly reflect the aging hippocampus better than those involved in positive regulation of cytokine production. We then hypothesized that core and active modules are robust to perturbation and maintain interaction patterns under harsh conditions causing loss of molecular interactions. The robust integrity of the two genes sets was measured by computing connectivity.
For calculating significant connectivity, we applied a module expansion method that is devised to predict and add other molecular determinants to the existing disease modules in each iterative expansion step according to the significance of connectivity in the interactome. The analysis started from “seed genes” of interest: a subnetwork was grown from the two gene sets. Another subnetwork was grown from each gene of the two sets. The number of overlapping genes between the different expanded subnetworks implicates the extent of integrity. CCCh1 (associated with complement system) genes exhibit more robust integrity in the interactome, as determined by the result that each gene was extended to include similar genes as the expanded sets starting from all genes of the complement set during the module extension process but not in that of cytokine regulation. Therefore, the module-based analysis, which exhibits greater predictive power than single gene-based analysis9, leads to the identification of valuable biomarkers for the brain aging, particularly the aging hippocampus. Cognitive impairments during aging may be induced by aberrant expression of genes associated with synaptic transmission, as noted in AD microarray data. However, immune-related genes were not differentially expressed but demonstrated a co-expression pattern in AD. In addition, the expression of immune response correlates with synaptic transmission genes and are most likely associated with brain aging based on molecular interaction information. Among the immune-related genes, complement-related genes are a network hotspot that may be a master regulator in brain aging. As a “gatekeeper”, these genes should be robust against internal and external challenges to protect the flow of molecular aberrations during aging to other components in the brain.
Electronic supplementary material
Acknowledgements
This work was supported by Bio & Medical Technology Development Program through the National Research Foundation funded by the Ministry of Science, ICT & Future Planning (NRF-2017M3A9B4025699, NRF-2017M3A9B4025709, and MSIP; NRF-2015R1C1A2A01055144) of South Korea.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s12276-018-0057-6.
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deary IJ, et al. Age-associated cognitive decline. Br. Med. Bull. 2009;92:135–152. doi: 10.1093/bmb/ldp033. [DOI] [PubMed] [Google Scholar]
- 3.Hebert LE, et al. Age-specific incidence of Alzheimer’s disease in a community population. JAMA. 1995;273:1354–1359. doi: 10.1001/jama.1995.03520410048025. [DOI] [PubMed] [Google Scholar]
- 4.Berchtold NC, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl Acad. Sci. USA. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue H, et al. A modular network model of aging. Mol. Syst. Biol. 2007;3:147. doi: 10.1038/msb4100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig T, et al. The Digital Ageing Atlas: integrating the diversity of age-related changes into a unified resource. Nucleic Acids Res. 2015;43:D873–878. doi: 10.1093/nar/gku843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacutu R, et al. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho DY, Kim YA, Przytycka TM. Chapter 5: network biology approach to complex diseases. PLoS Comput. Biol. 2012;8:e1002820. doi: 10.1371/journal.pcbi.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitra K, Carvunis AR, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park C, et al. Systematic identification of an integrative network module during senescence from time-series gene expression. BMC Syst. Biol. 2017;11:36. doi: 10.1186/s12918-017-0417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menche J, et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghiassian SD, Menche J, Barabasi AL. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput. Biol. 2015;11:e1004120. doi: 10.1371/journal.pcbi.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fresno C, Fernandez EA. RDAVIDWebService: a versatile R interface to DAVID. Bioinformatics. 2013;29:2810–2811. doi: 10.1093/bioinformatics/btt487. [DOI] [PubMed] [Google Scholar]
- 14.Lotia S, Montojo J, Dong Y, Bader GD, Pico AR. Cytoscape app store. Bioinformatics. 2013;29:1350–1351. doi: 10.1093/bioinformatics/btt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cribbs DH, et al. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J. Neuroinflamm. 2012;9:179. doi: 10.1186/1742-2094-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han HW, Ohn JH, Moon J, Kim JH. Yin and Yang of disease genes and death genes between reciprocally scale-free biological networks. Nucleic Acids Res. 2013;41:9209–9217. doi: 10.1093/nar/gkt683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuchty S, Almaas E. Peeling the yeast protein network. Proteomics. 2005;5:444–449. doi: 10.1002/pmic.200400962. [DOI] [PubMed] [Google Scholar]
- 21.Kim KS, et al. Long-term immunomodulatory effect of amniotic stem cells in an Alzheimer’s disease model. Neurobiol. Aging. 2013;34:2408–2420. doi: 10.1016/j.neurobiolaging.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat. Rev. Neurosci. 2006;7:278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magistretti PJ, Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron. 2015;86:883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 26.Solfrizzi V, et al. Dietary fatty acids in dementia and predementia syndromes: epidemiological evidence and possible underlying mechanisms. Ageing Res. Rev. 2010;9:184–199. doi: 10.1016/j.arr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Vlassenko AG, Raichle ME. Brain aerobic glycolysis functions and Alzheimer’s disease. Clin. Transl. Imaging. 2015;3:27–37. doi: 10.1007/s40336-014-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang S, et al. Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron. 2016;90:278–291. doi: 10.1016/j.neuron.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Driscoll I, et al. The aging hippocampus: cognitive, biochemical and structural findings. Cereb. Cortex. 2003;13:1344–1351. doi: 10.1093/cercor/bhg081. [DOI] [PubMed] [Google Scholar]
- 30.Berchtold NC, et al. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol. Aging. 2013;34:1653–1661. doi: 10.1016/j.neurobiolaging.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.