To the Editor
Chronic rhinosinusitis (CRS) pathophysiology has yet to be clearly defined, largely because the diagnosis itself likely represents a heterogeneous syndrome rather than a distinct clinical entity. Several investigators have shown that CRS with (CRSwNP) and without polyps (CRSsNP) can both be linked with either Th1-, Th2, or Th17-associated inflammatory signatures1, 2, and there likewise appears to be a substantial geographic predisposition to select types of inflammatory burden3. Recent attempts to identify inflammatory endotypes have presented a more nuanced approach to the classification of CRS, with Tomassen et al. recently using surgically-obtained tissue to identify potential CRS endotypes based entirely on inflammatory biomarkers4. While such studies have preferentially used tissue-based assessment of inflammatory mediators, this approach may have some logistical limitations as it is inherently invasive and subject to variations in site-specific protein expression throughout the sinonasal cavity5, 6. Minimally invasive approaches to disease endotyping have been reported for asthma and other respiratory diseases, typically employing analysis of sputum, mucus, or epithelial brushings7–9. The aim of the current study was to define CRS endotypes in a U.S. population based entirely on analysis of mucus collected via a minimally invasive approach. Going one step further, we sought to additionally define the clinical relevance of putative endotypes by assessing phenotypic characteristics, metrics of disease severity, quality of life, and surgery outcomes in these populations.
90 CRS patients undergoing endoscopic sinus surgery were prospectively enrolled in the study (Table E1). Mucus was collected using an absorbent polyurethane sponge and we subsequently assayed 18 different mucus inflammatory mediators that reflect Th1/Th2/Th17-associated inflammation. Since a priori selection of biological variables risks biasing data based on preconceived hypotheses, we attempted to include all cytokines that could be practically analyzed using a single platform, only excluding growth factors and most chemokines. Association between immunologic variables was assessed using principal component analysis with a five factor solution explaining 71.3% of the data variance (Figure E1A). The first component (PC1) included IL-1β, IL-6, IL-8, TNF-α, and eotaxin, while the second was composed of IL-5, IL-9, and IL-13 (Table E2). The remaining factors were composed of IL-2, IL-4, IL-12, and IL-21 (PC3), IFN-γ and IL-10 (PC4), and IL-3 and IL-17A (PC5). Hierarchical cluster analysis was then performed using the factor scores calculated for each patient (Figure 1A). This suggested an optimum of 5 or 6 clusters, with mathematical modeling and visual evaluation of the dendogram validating a 6 cluster model (Figure E1B). The mean factor scores for each of the 6 clusters are shown in Figure 1B. Clusters were compared against each other and all were significantly different (p<0.001) on all of the factors, except for factor 4. Individual cytokines were all significantly different in at least one cluster (Table E3).
Figure 1. Identification of inflammatory endotypes using cluster analysis of CRS mucus.
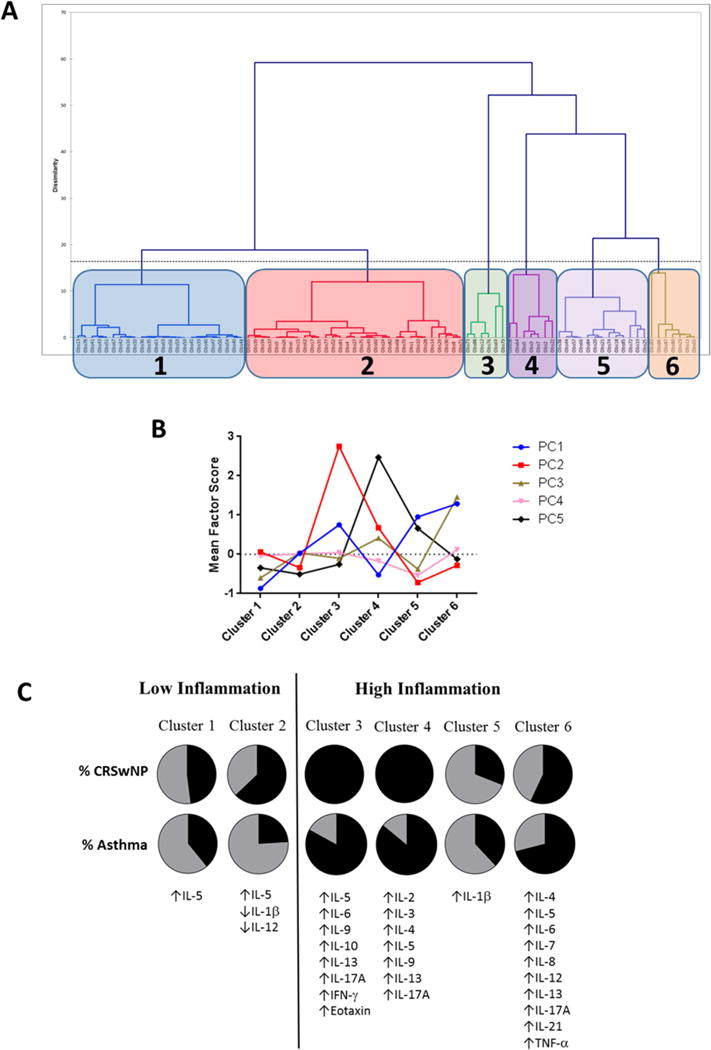
(A) Dendogram representing hierarchical cluster analysis of CRS patients based on principal component analysis of 18 mucus-derived biological variables. (B) Mean factor scores for each of the 6 CRS Clusters. (C) Asthma/polyp prevalence and differences in mucus cytokine levels among each of the 6 CRS clusters, compared to healthy controls. CRSwNP, chronic rhinosinusitis with nasal polyps; PC, principal component.
When compared to a control population, cluster 1 and cluster 2 were defined by low levels of most cytokines and were largely indistinguishable from healthy non-CRS patients (Figure 1C). Cluster 3 and Cluster 4 both carried a predominantly Th2 signature, with Cluster 3 defined by high levels of IL-5, IL-6, IL-9, IL-10, IL-13, IL-17A, IFN-γ, and eotaxin. Cluster 4 also had elevated levels of IL-5, IL-9 and IL-13, though these were lower than in Cluster 3. Cluster 4 additionally was distinguished by elevated IL-2, IL-3, and IL-4, and was noted to have higher levels of IL-17A than Cluster 3. Cluster 5 was characterized by very high levels of IL-1β, while Cluster 6 had elevated IL-4, IL-5, IL-6, IL-7, IL-8, IL-12, IL-13, IL-17A, IL-21, and TNF-α. Cluster 5 was the only cluster in which IL-5 levels were not elevated compared to controls. Similar to recently characterized clusters in a European population4, our results suggest that Th2-associated inflammation is present in a majority of CRS patients, but dominant in only a small minority (Clusters 3 and 4). One in five patients (Cluster 5) presented with a Th2-low, pro-inflammatory signature. Interestingly, more than half of patients (Clusters 1 and 2) were characterized by disease with low overall inflammatory burden, that did not correspond to a distinctly Th1-, Th2- or Th17-associated signature. Collectively, the inflammatory endotypes outlined in this study confirm that CRS is a heterogeneous inflammatory disease, and suggest that endotypes likely do not adhere to strict T-helper cell features.
Endotypes were then compared against phenotypic characteristics of the study population (Table 1). Cluster 3 and Cluster 4 were both composed exclusively of CRSwNP patients and had the highest levels of comorbid asthma, at 83% and 86%, respectively. These clusters were also characterized by tissue eosinophilia, worse objective measures of disease severity (SNOT-22, CT, SIT scores) and a high incidence of prior endoscopic sinus surgery. Cluster 1 and Cluster 2 were characterized by mild disease, with low CT scores and good olfactory function, and a low incidence of prior surgery. These groups were heterogeneous, with each composed of a fairly equal number of CRSwNP patients (48% and 63%, respectively), and a majority were non-asthmatic. Cluster 5 was composed primarily of CRSsNP patients (69%) and had a low incidence of comorbid asthma (38%). This cluster was characterized by mild-to-moderate disease, but had a fairly high incidence of prior endoscopic sinus surgery (69%). All patients in Cluster 6 had prior endoscopic sinus surgery, but were heterogeneous in terms of polyp status.
Table 1. Differences in clinical and phenotypic characteristics among CRS endotypes.
Post-hoc analysis of clinical and phenotypic characteristics were examined for each of the 6 identified CRS endotypes. Values are presented as either the mean +/− standard deviation or median with interquartile range, depending on the normalcy of the data. Differences between groups were assessed using the Kruskal-Wallis test or Chi-square analysis. BOLD, p < 0.05. AERD, aspirin-exacerbated respiratory disease; AFRS, allergic fungal rhinosinusitis; BMI, body mass index; CT, computed tomography; NCS, nasal corticosteroid; LTR, anti-leukotriene; SIT, smell identification test; SNOT-22, sinonasal outcome test-22.
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | Cluster 6 | p-value | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| No. | 31 | 24 | 6 | 7 | 13 | 7 | |
|
| |||||||
| Age (years) | 49.5 +/− 12.9 | 44.3 +/− 9.9 | 55.7 +/− 17.6 | 47.3 +/− 13.6 | 52.7 +/− 17.0 | 49.3 +/− 10.5 | 0.33 |
| Sex, no. (% female) | 19 (61) | 8 (33) | 2 (33) | 1 (14) | 5 (38) | 5 (71) | 0.08 |
| Race, no. (% white) | 30 (97) | 20 (83) | 6 (100) | 5 (71) | 13 (100) | 7 (100) | 0.08 |
| Current smoker, no. (%) | 4 (13) | 2 (8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0.51 |
| BMI (kg/m2) | 30.7 +/− 5.2 | 29.8 +/− 7.6 | 28.4 +/− 6.4 | 30.9 +/− 2.9 | 28.3 +/− 7.7 | 27.2 +/− 4.6 | 0.60 |
| Nasal polyps, no. (%) | 15 (48) | 15 (63) | 6 (100) | 7 (100) | 4 (31) | 4 (57) | 0.01 |
| Asthma, no. (%) | 12 (39) | 8 (24) | 5 (83) | 6 (86) | 5 (38) | 5 (71) | 0.03 |
| Allergic Rhinitis, no. (%) | 16 (52) | 17 (71) | 3 (50) | 1 (14) | 8 (62) | 6 (86) | 0.31 |
| AERD, no. (%) | 4 (13) | 3 (13) | 2 (33) | 3 (43) | 0 (0) | 0 (0) | 0.07 |
| AFRS, no. (%) | 1 (3) | 5 (21) | 1 (17) | 2 (29) | 1 (8) | 0 (0) | 0.19 |
| Taking NCS, no. (%) | 26 (84) | 20 (83) | 4 (67) | 6 (86) | 9 (69) | 6 (86) | 0.79 |
| Taking LTR, no. (%) | 5 (16) | 10 (42) | 4 (33) | 3 (43) | 0 (0) | 3 (43) | 0.04 |
| SNOT-22 score | 45.0 +/− 18.4 | 47.7 +/− 11.5 | 51.3 +/− 36.3 | 59.0 +/− 17.3 | 42.0 +/− 21.3 | 47.2 +/− 25.9 | 0.80 |
| CT score | 14.5 (11.0–19.0) | 15.0 (11.5–20.8) | 22.0 (19.3–23.3) | 18.0 (17.5–23.5) | 16.0 (11.0–17.8) | 13.0 (10.0–15.5) | 0.02 |
| SIT score | −7.0 (−24.5–3.0) | −4.0 (−21.0–3.0) | −29.0 (−31.0–24.0) | −16.0 (−26.6–0.8) | −2.5 (−19.8–1.0) | −7.0 (−17.5–2.0) | 0.03 |
| Prior surgery, no. (%) | 9 (29) | 8 (33) | 4 (66) | 5 (71) | 9 (69) | 7 (100) | 0.002 |
| # of prior surgeries | 0.0 (0.0–1.0) | 0.0 (0.0–1.0) | 1.5 (0.0–3.8) | 2.0 (1.0–3.0) | 1.0 (0.0–2.0) | 1.0 (1.0–1.0) | 0.004 |
| Tissue eosinophils/HPF | 19.0 (1.0–50.0) | 52.5 (15.0–100.0) | 86.5 (41.8–213.8) | 50.0 (37.8–100.0) | 5.0 (0.5–43.5) | 35.0 (1.0–100.0) | 0.01 |
Immunologic and phenotypic characteristics of Clusters 3–6 were collectively suggestive of more severe disease. We explored this further by separating subjects into 2 groups, the first composed of Clusters 1 and 2, and the second composed of Clusters 3–6. This distinction was based on differences in overall inflammatory burden (Figure 1C), and corresponded to differences in clinical characteristics and further analysis of the dendogram (Figure E2A). Subjects in Clusters 3–6 had a greater likelihood of prior endoscopic sinus surgery than those in Clusters 1 and 2 (75.8% vs. 37.8%; p = 0.001) (Figure E2B) and had a greater number of prior surgeries (median, 1.0 vs. 0.0, p < 0.0001) (Figure E2C). Since this association was largely based on retrospective data, we then analyzed postoperative changes in SNOT-22 scores for patients in each collective group. Postoperative SNOT-22 scores improved in Clusters 1 and 2, and this benefit was maintained one year after surgery. In contrast, postoperative SNOT-22 scores in Clusters 3–6 improved initially, but then returned almost to baseline at one year after surgery (Figure E2D). At greater than 6 months follow-up this corresponded to a significantly greater % improvement in postoperative SNOT-22 score for patients in clusters 1 and 2, compared to those in clusters 3–6 (mean 52.0% ± 11.0 vs. −44.2% ± 50.2%, respectively; p=0.04) (Figure E2E, F). Collectively, these data suggest that cluster analysis of mucus biomarkers can both define CRS inflammatory endotypes and potentially predict postoperative outcomes and need for revision surgery.
We recognize that there are some limitations to the current study that warrant discussion. First, the study only enrolled patients receiving endoscopic sinus surgery and may fail to capture a representative population of CRS patients. It is likely that enrolling only surgical patients could bias the study toward those with greater disease severity. Second, though statistically adequate, the total number of subjects in the study was somewhat small (n = 90), resulting in some clusters with fewer than 10 patients. Validation of the disease clusters identified in this study will most certainly require larger prospective cohorts that potentially incorporate patients treated both medically and surgically.
Our study is the first to report inflammatory CRS endotypes in a North American population based on cluster analysis of biological variables, and the first to utilize minimally invasive collection of sinonasal mucus for this purpose. Similar to tissue-based results from a European population4, we identified diverse inflammatory endotypes that differ substantially with respect to phenotype and disease behavior. In order to further define the clinical relevance of clusters identified in the current study, we analyzed post hoc inter-cluster differences among common measures of CRS disease severity and quality of life. The ease by which mucus can be collected potentially allows for longitudinal studies that assess the stability of inflammatory signatures over time, as well as impacts of therapeutic interventions.
Supplementary Material
CAPSULE SUMMARY.
CRS endotypes with prognostic potential can be identified via cluster analysis of mucus cytokine levels. These clusters correspond to distinct inflammatory signatures and differ based on phenotypic characteristics, disease severity, and surgical outcomes.
Acknowledgments
This project was supported by NIH RO3 DC014809 (J.H.T.), L30 AI113795 (J.H.T.), and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
ABBREVIATIONS
- AERD
aspirin-exacerbated respiratory disease
- AFRS
allergic fungal rhinosinusitis
- CRSsNP
chronic rhinosinusitis without nasal polyps
- CRSwNP
chronic rhinosinusitis with nasal polyps
- SIT
smell identification test
- SNOT-22
22 item sinonasal outcome test
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: No relevant disclosures
Conflicts of interest: None
References
- 1.Tan BK, Klingler AI, Poposki JA, Stevens WW, Peters AT, Suh LA, et al. Heterogeneous inflammatory patterns in chronic rhinosinusitis without nasal polyps in Chicago, Illinois. J Allergy Clin Immunol. 2017;139:699–703e7. doi: 10.1016/j.jaci.2016.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens WW, Ocampo CJ, Berdnikovs S, Sakashita M, Mahdavinia M, Suh L, et al. Cytokines in Chronic Rhinosinusitis. Role in Eosinophilia and Aspirin-exacerbated Respiratory Disease. Am J Respir Crit Care Med. 2015;192:682–94. doi: 10.1164/rccm.201412-2278OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang BF, Cao PP, Long XB, Zhang XH, Xu K, Cui YH, et al. Distinct mucosal immunopathologic profiles in atopic and nonatopic chronic rhinosinusitis without nasal polyps in Central China. Int Forum Allergy Rhinol. 2016;6:1013–9. doi: 10.1002/alr.21799. [DOI] [PubMed] [Google Scholar]
- 4.Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137:1449–56e4. doi: 10.1016/j.jaci.2015.12.1324. [DOI] [PubMed] [Google Scholar]
- 5.Weibman AR, Huang JH, Stevens WW, Suh LA, Price CPE, Lidder AK, et al. A prospective analysis evaluating tissue biopsy location and its clinical relevance in chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol. 2017;7:1058–64. doi: 10.1002/alr.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt MP, Soler ZM, Kao SY, Metson R, Stankovic KM. Topographic gene expression in the sinonasal cavity of patients with chronic sinusitis with polyps. Otolaryngol Head Neck Surg. 2011;145:171–5. doi: 10.1177/0194599811402030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinks TS, Brown T, Lau LC, Rupani H, Barber C, Elliott S, et al. Multidimensional endotyping in patients with severe asthma reveals inflammatory heterogeneity in matrix metalloproteinases and chitinase 3-like protein 1. J Allergy Clin Immunol. 2016;138:61–75. doi: 10.1016/j.jaci.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133:388–94. doi: 10.1016/j.jaci.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo CS, Pavlidis S, Loza M, Baribaud F, Rowe A, Pandis I, et al. A Transcriptome-driven Analysis of Epithelial Brushings and Bronchial Biopsies to Define Asthma Phenotypes in U-BIOPRED. Am J Respir Crit Care Med. 2017;195:443–55. doi: 10.1164/rccm.201512-2452OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


