Summary
In rice, amylose content (AC) is controlled by a single dominant Waxy gene. We used CRISPR/Cas9 to introduce a loss-of-function mutation into the Waxy gene in two widely-cultivated elite japonica varieties. Our results show that mutations in the Waxy gene reduce AC and convert the rice into glutinous ones without affecting other desirable agronomic traits, offering an effective and easy strategy to improve glutinosity in elite varieties. Importantly, we successfully removed the transgenes from the progeny. Our study provides an example of generating improved crops with potential for commercialization, by editing a gene of interest directly in elite crop varieties.
Yield and quality traits are two of the most important targets for rice breeders. Recent technical advances in high-throughput sequencing have accelerated the mapping and identification of many Quantitative Trait Loci (QTLs) making it possible to use tools such as CRISPR/Cas9 to perform precise gene editing for plant improvement. In the last few years, a number of reports have described the use of gene editing to study yield-related QTLs, such as Gn1a, IPA1, GS3 and DEP1, and explore their function in different varieties (Li et al. 2016). Food security considerations make yield an obvious target for rice improvement. However, economic growth and improved living standards in many rice-majority consuming countries are shifting public attention towards quality characteristics such as flavor, cooking, appearance and nutrition, which are strongly linked to starch physical properties. Starch is the major component in rice grain endosperm and it is a mix of two glucans: amylose and amylopectin. Amylose is primarily a linear polysaccharide while amylopectin is highly branched. The amylose content (AC) refers to the percentage of starch instead of the percentage of total weight in the grain, and is arguably the most important quality indicator in rice, especially for cooking and eating quality (Juliano 1998). Based on AC values, rice is commercially classified into five groups: waxy (0–5%), very low (5%–12%), low (12%–20%), intermediate (20%–25%), and high (25%–33%) AC (Juliano 1992). In general, cooking of high AC varieties results in dry, firm and well separated rice grains, becoming hard after cooling; Intermediate AC varieties is soft but not sticky after cooking; Low and very low AC (5%–20%) varieties, have a soft and sticky texture after cooking; Finally, waxy rice, also called glutinous rice, is especially sticky when cooked (Juliano 1998). The quality of some of the hybrid rice grown in China, especially the indica hybrids, is considered poor owing to their high AC that makes them hard and dry when cooked. Therefore, a major objective for breeders is to improve grain quality by decreasing AC in rice, mostly in the indica hybrids.
The Waxy (Wx) gene (LOC_Os06g04200) of rice which encodes a granule-bound starch synthase (GBSS), also known as Waxy protein, is responsible for the synthesis of amylose in endosperm (Wang et al. 1995). There are two major Wx alleles in cultivated rice, Wxa and Wxb, with indica cultivars mostly containing the Wxa allele while most japonica cultivars contain the Wxb allele (Wang et al. 1995). The main difference between the alleles is a G/T polymorphism that results in differential splicing of the gene affecting mRNA stability (Wang et al. 1995; Larkin and Park 2003). As a result, the Wxa allele produces 10-fold higher mRNA and protein levels than Wxb (Isshiki et al. 1998). Waxy expression levels are positively correlated with AC, making it feasible to alter the AC by manipulating the Waxy gene and a number of approaches have been attempted including conventional breeding and genetic manipulation. Ma et al (2015) showed that CRISPR/Cas9-generated mutations in the Waxy gene led to reduced AC in a japonica cultivar of rice, Taichung 65 (Ma et al. 2015). Transgenic rice lines containing Waxy antisense constructs also produced seeds with reduced amylose levels and improved quality; and the decreased AC was also observed in hybrids obtained with the transgenic lines (Terada et al. 2000; Liu et al. 2003; Liu et al. 2005). In contrast, introduction of the Wxa cDNA into Waxy null-mutant Japonica rice lines increased the AC by 6%–11% (Itoh et al. 2003). Traditional breeding methods are effective but time-consuming and laborious, therefore we used CRISPR/Cas9-mediated gene editing to introduce a loss-of-function mutation into the Waxy gene to reduce AC in two widely-cultivated elite japonica varieties, Xiushui134 (XS134) and Wuyunjing 7 (9522). Our results show that mutations in the Waxy gene produce decreased AC in rice offering an effective strategy of generating improved crops in elite varieties without affecting other desirable agronomic traits.
We sequenced the waxy gene XS134 and 9522 and confirmed that both varieties contain the typical Wxb allele (Figure S1). We designed a CRISPR/Cas9 construct targeting the first exon of the Waxy gene with the expectation to produce a null mutation (Figure 1A). The 20-nt target sequence for the sgRNA was carefully chosen to avoid off-target effects using the web-based tool CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). Embryogenic calli from the elite rice varieties XS134 and9522 were co-cultivated with Agrobacterium tumefaciens carrying binary vectors with the CRISPR/Cas9 cassette and transgenic T0 plants analyzed to detect the presence of mutations in the target regions using Sanger sequencing. A very high mutagenesis efficiency was observed with 82.76% of T0 transformants carrying mutations in XS134 plants and 86.96% in 9522 (Table S1). It has been reported that CRISPR/Cas9-induced mutations mainly take place in transformed calli cells and homozygous mutations in the target genes can be readily found in T0 plants (Zhang et al. 2014). In our case we found one homozygous T0 plant in XS134 (4.17%) and 3 in 9522 (15%) (Table S1). Most of the mutation types detected in the target sequence produced frameshifts in the coding region giving rise to non-functional proteins (Figure 1B). To test for possible off-target effects, we identified the locus in the rice genome with the highest probability for such effects based on the sequence of the sgRNA used in our study. No mutations were found in any of the 46 T0 plants analyzed for both varieties by sanger sequencing (Table S2) suggesting that off-target effects can be avoided or reduced by careful selection of the target site.
Figure 1. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties.
(A) Schematic diagram of the targeted site in the Waxy gene. Black arrows indicate the start codon, targeted site and stop codon. The numbers in brackets indicate the distance to the start codon (ATG). The sequence of the targeted site is shown with the protospacer adjacent motif (PAM) sequences labeled in blue color. (B) Examples of mutations at the Waxy locus in CRISPR-waxy T0 generation plants of two rice varieties, 9522 and XS134. The targeted sequence is highlighted in red and the PAM sequences are in blue. Mutations are marked in green color. (C) Phenotypes of CRISPR-waxy mutants and their corresponding WTs.
A number of T1 plants containing CRISPR/Cas9-induced homozygous frameshift mutations in the Waxy gene (hereupon referred to as “CRISPR-waxy” mutants) were selected for phenotypic analysis. All plants were grown under natural field conditions in the Shanghai region, China (30°N, 121°E) during the normal rice-growing season from mid-May to mid-October. For both varieties, the mutated waxy plants were indistinguishable from WT controls in plant height (Figures 1C, S2A), grain number per panicle (Figure S2B), panicle number per plant (Figure S2C), yield per plot (Figure S2D), seed width (Figure S3A, C, F and H), seed length (Figure S3B, D, G and I) and 1,000 grains weight (Figure S3E, J). These results confirm our expectation that mutation of the Waxy gene does not affect important agronomic traits in rice, an essential pre-requisite to achieve our aim of crop improvement without compromising agronomic characteristics in elite varieties.
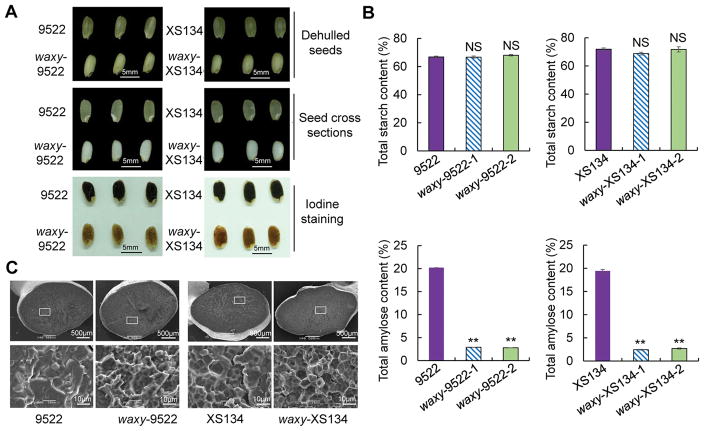
Grains of CRISPR/Cas9 mutated plants had a ‘waxy’ appearance being white and fully opaque in contrast with the typical ‘non-waxy’ translucent appearance of 9522 and XS134 seeds (Figure 2A). Cross-sections of seeds also revealed a milky white and opaque appearance in the CRISPR-waxy mutants, compared to translucent endosperm of WT seeds (Figure 2A). The two starch components in rice, amylose and amylopectin, have different iodine binding capacity. Amylose has a high binding capacity and turns deep blue when stained with iodine, while amylopectin has much lower iodine-binding capacity, turning red-brown upon staining. As expected, endosperms in cross-sections of 9522 and XS134 WT seeds turned dark blue when stained with an iodine solution while CRISPR-waxy endosperms turned red-brown (Figure 2A), revealing that CRISPR-waxy endosperms had a lower amylose/amylopectin ratio than WT. Quantification of the amylose content confirmed the staining results with CRISPR-waxy seeds showing a significant reduction in AC compared to XS134 and 9522 WT controls (Figure 2B; Table S3). Total starch content was unchanged in CRISPR-waxy and control plants (Figure 2B; Table S3). The low AC in the CRISPR-waxy seeds qualify then as waxy or glutinous rice, implying that we successfully transformed two non-glutinous varieties, XS134 and 9522 into new glutinous varieties by using CRISPR/Cas9 to edit the Waxy gene. The Waxy gene has been reported to affect the gel consistency (GC) and gelatinization temperature (GT) of rice (Liu et al. 2003; Tian et al. 2009). Our analysis showed a 2-fold increase in GC and a marked reduction in GT for CRISPR-waxy seeds compared to their respective WT (Table S3). Scanning electron microscopy revealed clear differences in the starch structure of CRISPR-waxy and WT seeds with WT showing sharp edges while CRISPR-waxy seeds were more irregular (Figure 2C). Finally we addressed the issue of social acceptance of genetically modified foods by removing the transgenes from CRISPR/Cas9-edited waxy rice lines by self-pollination in the T2 generation (Table S4; Figure S4). To identify transgene-free lines we analyzed individuals by PCR using specific primers for the U6-sgRNA, 35S promoter and Nos terminator sequences. Two out of 48 waxy-XS134 and 4 out of 20 waxy-9522 T2 plants were transgene-free (Table S4).
Figure 2. Grain phenotypes, total starch and total amylose content and scanning electron micrographs of endosperms in mature seeds of waxy mutants and corresponding WT lines.
(A) Grain phenotypes of CRISPR-waxy mutants and their corresponding WTs. Upper row, phenotype of the dehulled seeds. Middle row, endosperm phenotypes in seed cross-sections. Bottom row, iodine-staining of endosperm in cross sections of seeds. (B) Total starch and total amylose content in CRISPR-waxy mutants and their corresponding WTs. Data is presented as means ± sd. n=4; **P < 0.01, two-tailed, two-sample t-test; NS: no significant. (C) Scanning electron micrographs of endosperms in CRISPR-waxy mutants and their corresponding WTs.
Glutinous rice (waxy rice) is popular in many Asian countries. Compared with non-glutinous rice, waxy rice provides unique characteristics for numerous food and non-food applications, such as brewing. In this study, we used CRISPR/Cas9 to mutate the Waxy gene and converted two elite non-glutinous rice varieties into glutinous varieties without changing any of the important agronomic traits that had been laboriously introduced over long and costly breeding programs. Therefore, the newly created variety is already elite and can be immediately used without the process of trait introgression by repeated backcrossing to parental lines. In addition, the ability to remove the transgene cassettes in the T2 generation allowed us to produce non-GM lines containing the desired mutation with surgical precision, thus helping to circumvent GM-related regulations and public controversy.
Aside from conventional elite varieties, CRISPR/Cas9 editing of the Waxy gene can also be used in the production of hybrid rice. The Waxy gene has a dosage effect in the triploid rice endosperm (Kumar and Khush 1986), making it possible to manipulate AC in hybrid rice by editing the Waxy gene in sterile or restorer lines. Waxy mutations in sterile lines would give rise to a Wx/wx/wx endosperm genotype; while mutations in restorer lines, would result in a Wx/Wx/wx genotype allowing the production of hybrid rice with different AC.
In summary, we have used CRISPR/Cas9 to mutate the Waxy gene in two elite cultivated rice lines, XS134 and 9522 and developed new rice lines with lower amylose content while maintaining all the desired agronomic traits and successfully removed the transgenes from the progeny to produce transgene-free lines. Our study provides an example of generating improved crops with potential for commercialization, by editing genes of interests directly in elite crop varieties.
MATERIALS AND METHODS
Plant materials and growth conditions
XS134 is an elite japonica rice variety with high yield and good quality, which is mainly planted in Jiangsu Province, Zhejiang Province and Shanghai area, China (http://ricedata.cn). 9522 is also an elite japonica rice variety with high yield, high disease resistance and good quality, which is cultivated widely in the south of Jiangsu Province, China (http://ricedata.cn). The CRISPR/Cas9 vector targeting the Waxy gene was constructed as previously described (Zhang et al. 2014). The CRISPR/Cas9 cassette was transferred into 9522 and Xiushui 134 (XS134) callus by Agrobacterium tumefaciens-mediated transformation using the strain EHA105 (Hiei et al. 1997). Transgenic rice lines were grown in paddy fields in Shanghai, China, during normal rice-growing seasons. Mature seeds were harvested and dried for germination. T1 and T2 seeds were collected following the same procedure. Sequences of the primers used for vector construct and detection are listed in Table S5.
Phenotype and genotype assays
Plant height, grain number per panicle, and panicle number per plot weremeasured in the paddy fields. For plot experiment, the planting density was 20 plants in 65.5cm×101.5cm area and he plot yields determined when the seeds were harvested and dry. Seeds were collected for each plant and weighted after drying at 37°C for two weeks. 1,000 grains weight, grain length and width were measured using an SC-A grain analysis system (Wseen company, China). PCR amplification was carried out using primer pairs flanking the sgRNA target site and putative off-target designated locus (Table S5). The PCR products were sequenced by sanger method.
Grain phenotype and iodine-staining of endosperm
The hull of rice seeds was removed to observe the external appearance of the grain in both 9522 and XS134 varieties. Grains were cut through the center to expose the endosperm. 0.2% iodine reagent was dropped on the endosperm surface and photographs taken after 3–5 minutes.
Determination of total starch and amylose content
Total starch content was determined according to the Megazyme Total Starch assay procedure (Megazyme Intenational Ireland, www.megazyme.com). D-Glucose stains with GOPOD (Glucose Oxidase Plus Peroxidase and 4-aminoantipyrine) reagent were determined at 510nm. Amylose content was measured following the procedure described in GB/T 15683-2008/ISO 6647-1: 2007. The amylase-iodine blue color was determined at 720 nm.
Evaluation of grain GC and GT
Gel consistence (GC) was evaluated according to Cagampang et al. (Cagampang et al. 1973). Quartic measurements were performed for each sample. Gelatinization temperature (GT) was indirectly estimated via the alkali digestion test (Little et al. 1958). Six whole-grain and same size, milled rice samples were placed in small plastic boxes containing 2 mL 1.7% potassium hydroxide (KOH) and incubated at 30°C in an oven. After 23 hours, grain appearance and disintegration were visually rated based on a standard numerical scale.
Scanning electron microscopy of starch granules
Rice grains were dried in an oven at 42°C for 2 days and cooled in a desiccator. Cross-sections of the samples were manually snapped and sputter-coated with gold palladium on copper studs. Magnifications of 50× and 2000× were used to observe endosperm and starch granule morphology.
Supplementary Material
Acknowledgments
We thank Huangwei Chu from Shanghai Academy of Agriculture Sciences for providing 9522 and Xiushui 134 seeds. This work was supported by the Chinese Academy of Sciences and by US NIH Grants R01GM070795 and R01GM059138 (to J.-K.Z.). H. Z. gratefully acknowledges the support of the International Postdoctoral Exchange Fellowship Program of China under grant 20140029.
Footnotes
AUTHORS CONTRIBUTIONS
J.-K.Z. and H.Z. designed research and supervised the study; J.Z. and H.Z. performed research; H.Z., J.Z., J.R.B., and J.-K.Z. analyzed data; and H.Z., J.R.B., J.-K.Z. and J.Z. wrote the paper.
References
- Cagampang GB, Perez CM, Juliano BO. A gel consistency test for eating quality of rice. J Sci Food Agric. 1973;24(12):1589–1594. doi: 10.1002/jsfa.2740241214. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T, Kubo T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol. 1997;35(1–2):205–218. [PubMed] [Google Scholar]
- Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 1998;15(1):133–138. doi: 10.1046/j.1365-313x.1998.00189.x. [DOI] [PubMed] [Google Scholar]
- Itoh K, Ozaki H, Okada K, Hori H, Takeda Y, Mitsui T. Introduction of Wx transgene into rice wx mutants leads to both high-and low-amylose rice. Plant Cell Physio. 2003;44(5):473–480. doi: 10.1093/pcp/pcg068. [DOI] [PubMed] [Google Scholar]
- Juliano B. Structure, chemistry, and function of the rice grain and its fractions. Cereal Foods World. 1992;37:772–772. [Google Scholar]
- Juliano B. Varietal impact on rice quality. Cereal Foods World. 1998;43:207–222. [Google Scholar]
- Kumar I, Khush G. Gene dosage effects of amylose content in rice endosperm. J Genet. 1986;61(6):559–568. [Google Scholar]
- Larkin PD, Park WD. Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.) Mol Breeding. 2003;12(4):335–339. [Google Scholar]
- Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 System. Front Plant Sci. 2016;7(12217):377. doi: 10.3389/fpls.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RR, HILDER GB, Dawson EH. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 1958;35(2):111–126. [Google Scholar]
- Liu Q, Wang Z, Chen X, Cai X, Tang S, Yu H, Zhang J, Hong M, Gu M. Stable inheritance of the antisense Waxy gene in transgenic rice with reduced amylose level and improved quality. Transgenic Research. 2003;12(1):71–82. doi: 10.1023/a:1022148824018. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yu H, Chen X, Cai X, Tang S, Wang Z, Gu M. Field performance of transgenic indica hybrid rice with improved cooking and eating quality by down-regulation of Wx gene expression. Mol Breed. 2005;16(3):199–208. [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Terada R, Nakajima M, Isshiki M, Okagaki RJ, Wessler SR, Shimamoto K. Antisense Waxy genes with highly active promoters effectively suppress Waxy gene expression in transgenic rice. Plant Cell Physiol. 2000;41(7):881–888. doi: 10.1093/pcp/pcd008. [DOI] [PubMed] [Google Scholar]
- Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA. 2009;106(51):21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Zheng FQ, Shen GZ, Gao JP, Snustad DP, Li MG, Zhang JL, Hong MM. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995;7(4):613–622. doi: 10.1046/j.1365-313x.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12(6):797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.