Abstract
While testicular nuclear receptor 4 (TR4) may promote prostate cancer (PCa) metastasis, its roles in the clear cell renal cell carcinoma (ccRCC) remains unclear. Here we found a higher expression of TR4 in ccRCC tumors from patients with distant metastases than those from metastasis-free patients, suggesting TR4 may play positive roles in the ccRCC metastasis. Results from in vitro ccRCC cell lines also confirmed TR4’s positive roles in promoting ccRCC cell invasion/migration via altering the microRNA (miR-32-5p)/TR4/HGF/Met/MMP2-MMP9 signaling. Mechanism dissection revealed that miR-32-5p could suppress TR4 protein expression levels via direct binding to the 3'UTR of TR4 mRNA, and TR4 might then alter the HGF/Met signaling at the transcriptional regulation via direct binding to the TR4-response-elements (TR4RE) on the HGF promoter. Then the in vitro data also demonstrated the efficacy of Sunitinib, a currently used drug to treat ccRCC, could be increased after targeting this newly identified miR-32-5p/TR4/HGF/Met signaling. The preclinical study using the in vivo mouse model with xenografted ccRCC cells confirmed the in vitro cell lines data. Together, these findings suggest that TR4 is a key player to promote ccRCC metastasis and targeting this miR-32-5p/TR4/HGF/Met signaling with small molecules including TR4-shRNA or miR-32-5p may help to develop a new therapy to better suppress the ccRCC metastasis.
Keywords: nuclear receptor, TR4, renal cell carcinoma, metastasis, microRNA, HGF/Met
Introduction
Renal cell carcinoma (RCC) is the most lethal urological tumor, constituting 2%–3% of adult malignancies in the United States1. Clear cell RCC (ccRCC) is the most common histologic subtype and accounts for 70% of RCC2. Although a higher incidence of small renal masses is being detected, approximately one-third of patients still present with or will develop metastatic lesions during the course of the disease3 and the detailed mechanism(s) of metastasis remain unclear.
Testicular nuclear receptor 4 (TR4), also known as NR2C2 (nuclear receptor subfamily 2, group C, member 2), is a transcriptional factor and a member of the nuclear receptor family4–6. Earlier studies from TR4 knockout (TR4KO) mice revealed that TR4 might play important roles in fertility, bone formation, neural development and metabolism7–11. Recent studies also found TR4 might play a negative role to suppress prostate cancer (PCa) initiation, yet played a positive role to promote PCa metastasis12–14, which is opposite the androgen receptor that promotes PCa initiation and suppresses PCa metastases15. These contrasting results prompted us to investigate TR4 potential impacts on the ccRCC progression.
The microRNAs (miRNAs) are a class of short (19-22 nucleotides), noncoding RNA sequences that can negatively regulate gene expression via interacting with the 3' untranslated region (3'UTR) of target mRNAs16 or positively regulate gene expression via interacting with the 5' promoter region17. Studies have documented the role of miRNAs in cellular proliferation and invasion in a variety of tumors18–20 including RCC 21–23. Whether TR4 may also function in the network of miRNAs to influence the ccRCC progression, however, remains unclear.
Here we report that TR4 might play a positive role in promoting ccRCC metastasis via up-regulating the HGF/Met signaling. We also demonstrated miR-32-5p might suppress ccRCC metastasis via suppressing the TR4/HGF/Met signaling.
Materials and Methods
Cell culture
ACHN, OSRC-2 and SW839 cell lines were purchased from the American Type Culture Collection (Rockville, MD) in December of 2009. After cells were received, the cell lines were frozen in liquid N2 after the first 3 passages with 50 ampules of cell stock. After an ampule is thawed, cells are used for the designed experiments within 15 passages. All cell lines were cultured in Dulbecco's Modified Eagle's Medium (Invitrogen, Grand Island, NY, USA) supplemented with 10% FBS (v/v), penicillin (25 units/ml), streptomycin (25 ug/ml), 1% L-glutamine, and 10% fetal bovine serum (FBS). All cell lines were cultured in a 5% (v/v) CO2 humidified incubator at 37°C.
Cell invasion assay
The invasion capability of RCC cells was determined using the Matrigel-coated Transwell assay. Briefly, cells were harvested and seeded at 5 × 104 cells/well with serum-free media into the upper chambers coated with Matrigel (Trivigan, #3500-096-K). Then 750 μl 10% FCS media was placed in the lower chambers for 24 hours incubation at 37°C in 5% (v/v) CO2 incubator. The cells invaded into the matrigel were fixed by 4% paraformaldehyde and stained with 0.1% (w/v) crystal violet. The invaded cells were counted in ten randomly chosen microscope fields (100×) in each experiment for quantification. Each sample was run in triplicate and in multiple experiments.
3D invasion assay
The 3D invasion assay was as previously described24. Briefly, 1 ×104 cells were suspended in 200 ml media containing 1% Matrigel and then (Trivigen, #3500-096-K) and then plated into the previously prepared collagen/Matrigel mixture coated plates. Media was replenished every 3 days for 2 weeks and then spheres and protrusions numbers were recorded under microscope.
Wound healing assay
We seeded cells into 6-well tissue culture plates at a density that after 24 hours of growth, they should reach ~70–80% confluence as a monolayer. We then gently and slowly scratched the monolayer with a sterile 200 μl pipette tip across the center of the well. After scratching, we gently washed the well twice with media to remove the detached cells and replenished the well with fresh media. We then incubated the cells for an additional 24 hours, washed the cells twice with 1x PBS, and then took photos. The percentage of decreased wound width was compared between the control group and experimental group.
RNA extraction and qRT-PCR analysis
For RNA extraction, total RNAs were isolated using Trizol reagent (Invitrogen). 1 μg of total RNA was subjected to reverse transcription using Superscript III transcriptase (Invitrogen). Quantitative real-time PCR (qRT-PCR) was conducted using a Bio-Rad CFX96 system with SYBR green to determine the mRNA expression level of a gene of interest. Expression levels were normalized to the expression of GAPDH RNA. The miRNAs were isolated using PureLink® miRNA kit. Briefly, 50 ng small RNAs were processed for poly A addition by adding 1 unit of polymerase with 1 mM ATP in 1xRT buffer at 37°C for 10 minutes in 10 μl volume, heat inactivated at 95°C for 2 minutes. We then added 50 mmol anchor primer to 12.5 μl, and incubated at 65°C for 5 minutes. For the last step of cDNA synthesis, we added 2 μl 5x RT buffer, 2 μl 10 mM dNTP, 1 μl reverse transcriptase to total 20μl, and incubated at 42°C for 1 hour. Quantitative real-time PCR (qT-PCR) expression levels were normalized to the expression of 5s RNA and/or U6.
Immunoblot Analysis
Cells were lysed in RIPA buffer and proteins (30 μg) were separated on 8–10% SDS/PAGE gel and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking membranes, they were incubated with appropriate dilutions of specific primary antibodies including TR4 (ab109301, Abcam, Cambridge, MA, U.S.A.), HGF (AP1724b, One World Lab, San Diego, CA,U.S.A.), Bi-Phospho-MET/HGFR (Y1234/Y1235) (AM1000a, One World Lab.), Met (sc-10, Santa Cruz Biotechnology, Dallas, Texas, U.S.A.), MMP9 (A2095, One World Lab) and MMP2 (sc-13594, Santa Cruz Biotechnology) the blots were incubated with HRP-conjugated secondary antibodies and visualized using the ECL system (Thermo Fisher Scientific, Rochester, NY, USA).
Luciferase Assay
The human promoter region of HGF was constructed into pGL3-basic vector (Promega, Madison, WI, USA). Cells were plated in 24-well plates and the cDNA were transfected using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. The pRL-TK was used as the internal control. 959 bp fragments of TR4 3'UTR containing wild type or mutant miRNA-responsive elements were cloned into the psiCHECK™-2 vector construct (Promega) downstream of the Renilla luciferase ORF. Luciferase activity was measured by Dual-Luciferase Assay (Promega) according to the manufacturer’s manual.
Chromatin Immunoprecipitation Assay (ChIP)
Cell lysates were precleared sequentially with normal rabbit IgG (sc-2027, Santa Cruz Biotechnology) and protein A-agarose. Anti-TR4 antibody (ab109301, Abcam) 2.0 μg was added to the cell lysates and incubated at 4 °C overnight. For the negative control, IgG was used in the reaction. Specific primer sets was designed to amplify a target sequence within human HGF promoter and PCR products were identified by agarose gel electrophoresis.
Renal capsule implantation
OSRC-2 cells expressing Luciferase and expressing pLKO.1-shTR4, pLKO.1-scramble, or pLVTHM-miR-32-5p (2X106) were injected into the left renal capsule of 6-week-old male athymic nude mice (NCI) (n = 8 mice per group). We checked the body weights every week and used the non-invasive in vivo imaging system (IVIS) every 2 weeks to monitor the tumor/metastasis growth. After 8 weeks, mice were sacrificed, and tumors were excised. Studies on animals were conducted with approval from the Animal Research Ethics Committee of the University Of Rochester Medical Center.
H&E and immunohistochemical (IHC) staining
Tissues were fixed in 10% (v/v) formaldehyde in PBS, embedded in paraffin, and cut into 5 μm sections and used for H&E staining and IHC staining with specific primary antibodies against TR4, and HGF (AP1724b, One World Lab). To enhance antigen exposure, the slides were treated with 1× EDTA at 98°C for 10 minutes for antigen retrieval. The slides were incubated with endogenous peroxidase blocking solution, and then were incubated with the primary antibody at 4°C overnight. After rinsing with Tris-buffered saline, the slides were incubated for 45 minutes with biotin-conjugated secondary antibody, washed, and then incubated with enzyme conjugate horseradish peroxidase (HRP)-streptavidin. Freshly prepared DAB (Zymed, South San Francisco, CA) was used as the substrate to detect HRP. Finally, slides were counter-stained with hematoxylin and mounted with aqueous mounting media. Positive cells were calculated as the number of immunopositive cells × 100% divided by total number of cells/field in 10 random fields at 400× magnification.
Statistical Analysis
Data were expressed as mean±SEM from at least 3 independent experiments. Statistical analyses involved paired t-test with SPSS 17.0 (SPSS Inc., Chicago, IL). P <0.05 was considered statistically significant.
Results
Higher expression of TR4 is correlated with ccRCC metastasis
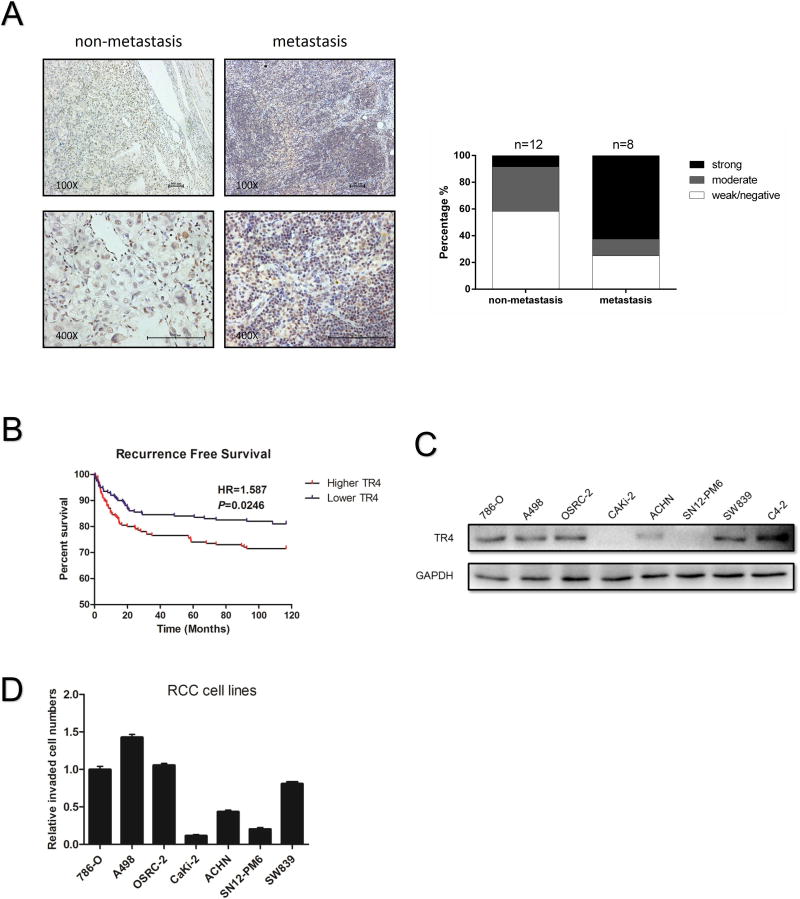
The higher TR4 expression has been found in PCa patients with a higher Gleason score12, 13. The linkage of TR4 expression in the different stages of ccRCC, however, remains unclear. We first applied the IHC staining with TR4 antibody to examine the TR4 expression in human clinical ccRCC samples and found higher TR4 expression in ccRCC from patients with metastases (Supplementary Table 1 shows the Clinical parameters for the ccRCC patients) than those from metastasis-free patients (Fig. 1A), suggesting TR4 may play positive roles to promote the ccRCC metastasis. Results from database analysis in the TCGA (TCGA website: http://cancergenome.nih.gov/) of 606 primary ccRCC samples also revealed that patients with higher TR4 expression had significantly lower recurrence-free survival (HR=1.587) than patients with lower TR4 expression (Fig. 1B). Furthermore, we found that patients with higher TR4 expression had lower overall survival (HR=1.397) than patients with lower TR4 expression (Fig. S2A), but with less significance.
Figure 1. The correlation between TR4 expression and ccRCC metastasis.
(A), IHC staining results investigating TR4 levels in tumor tissues from ccRCC patients with distant metastases versus tumor tissues from metastasis-free patients. Clinical specimens were obtained from Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China. IHC staining was performed using TR4 antibody (1:300). Left panels show imaging and quantification is shown on right. Magnification, 100× (upper) and 400× (lower). (B), Recurrence-free survival curves of ccRCC patients analyzed according to the TR4 expression (data downloaded from TCGA website). (C-D), The correlation between TR4 expression using western blot assay (C) and ccRCC cell lines invasive capacity (D) as determined by Matrigel-coated Transwell assay. Data presented as mean±SEM.
To confirm these clinical results in the in vitro multiple ccRCC cell lines, we first assayed the TR4 expression in various ccRCC cells and results using Transwell assay revealed the higher TR4 expression with higher invasion capacities in 786-O, A498, OSRC-2,,and SW839 cells compared to other ccRCC cells including Caki-2, ACHN, and SN12-PM6 (Fig. 1C-D) with C4-2 cells as a positive control.
Together, results from Fig. 1A-D and Supplementary Fig. S2A suggest that TR4 may play a positive role to promote the ccRCC metastasis.
TR4 promotes RCC cells invasion and migration in vitro
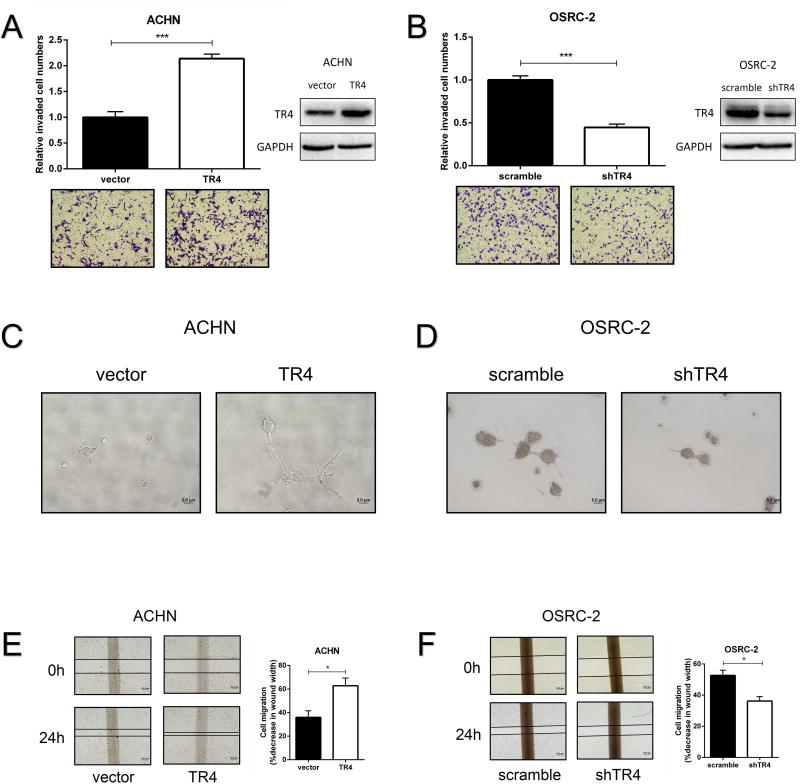
We then applied the Matrigel-coated Transwell invasion assay25 to confirm our initial findings via altering the TR4 expression using a lentiviral system in both VHL-wild type ACHN and VHL-mutant OSRC-2 cells. The results revealed that adding TR4-cDNA led to significantly increase the invasion capacity of the ACHN cells that have lower endogenous TR4 (Fig. 2A), and knocking-down TR4 could significantly decrease the invasion capacity of the OSRC-2 cells that have higher endogenous TR4 (Fig. 2B), Similar results were obtained when we replaced OSRC-2 cells with the ccRCC SW839 cells (Supplementary Fig. S3A).
Figure 2. The effect of TR4 on invasion and migration in ccRCC cell lines.
(A-B), Invasion ability of TR4 overexpressed ACHN cells (A) and TR4 knocked-down OSRC-2 cells (B) were compared to their control vector or scramble cells using Matrigel-coated Transwell assay. Quantification of invaded cell numbers in upper panels, images in lower panels and insets are WB of viral infections. (C-D), Invasion ability of TR4 overexpressed ACHN cells (C) and TR4 knocked-down OSRC-2 cells (D) were compared to their control vector or scramble cells using 3D invasion assay. (E-F), Migration ability of TR4 overexpressed ACHN cells (E) and TR4 knocked-down OSRC-2 cells (F) were compared to their control vector or scramble cells using wound healing assay. The percentage of decreased wound width was compared between the control group and experimental group. Each sample was run in triplicate and in multiple experiments. Quantitations at right. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05 and ***P < 0.005.
We also replaced the Matrigel-coated Transwell invasion assay with the 3D invasion assay to confirm the above findings and results revealed that adding TR4-cDNA increased the cell invasion in ACHN cells (Fig. 2C) and knocking-down TR4 with TR4-shRNA decreased the cell invasion in OSRC-2 cells (Fig. 2D). Finally, results from wound healing assays also indicated that adding TR4-cDNA increased the cell migration in ACHN cells (Fig. 2E), and knocking-down TR4 decreased the cell migration in OSRC-2 and SW839 cells (Fig. 2F and Supplementary Fig. S3B, respectively).
Together, results from Fig. 2A-F and Supplementary Fig. S3A-B demonstrate that TR4 plays positive roles to promote ccRCC cells invasion and migration independent of VHL status.
Mechanism dissection of how TR4 enhances ccRCC cell invasion: via altering the HGF/Met/MMP2-MMP9 signaling
Since early studies indicated TR4 might function via altering the CCL2/CCR2 and TGFβR2/p-SMAD3 signals to promote the metastasis in PCa cells12, 13, we then analyzed the expression of CCL2, TGFβR2 and p-SMAD3 after manipulating the TR4 expression in ACHN and OSRC-2 cells. The results revealed that altering TR4 (either adding TR4-cDNA or knocking-down with shTR4) led to little impact on these three proteins expression (Supplementary Fig. S4A).
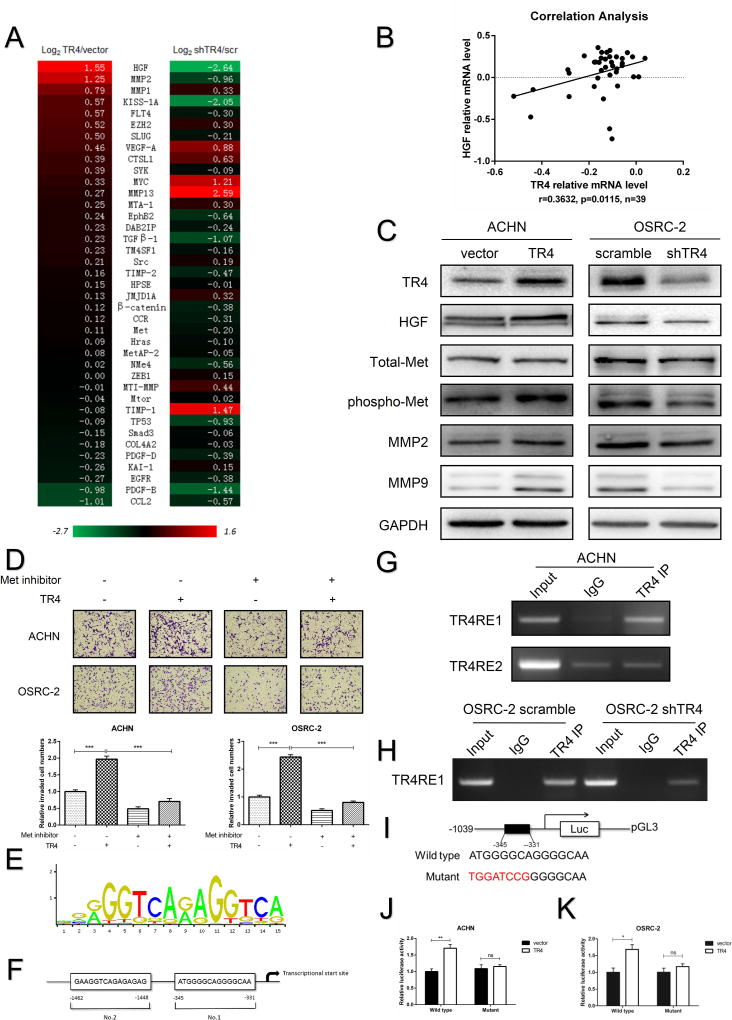
We hen analyzed many reported RCC metastasis-associated genes mRNA expressions and found some were altered after manipulating the TR4 expression. Among these genes, we found HGF mRNA expression was significantly altered (Fig. 3A), and results from the NCBI GEO DataSets (GEO GSE29609) analysis of the ccRCC sample array also revealed a significant positive correlation (r=0.3632, p=0.0115) between the TR4 expression and HGF expression in 39 ccRCC specimens (Fig. 3B).
Figure 3. HGF/Met signaling plays a role in TR4-mediated invasion and migration in vitro.
(A), Metastasis associated genes screened in TR4 knocked-down ACHN cells versus their scramble control cells and TR4 overexpressed ACHN versus their vector control cells using qRT-PCR assay. (B), Correlation analysis of TR4 and HGF using Pearson correlation coefficient with the data downloaded from GEO DataSets websites. (C), Immunoblot analysis of HGF, phospho-Met, total-Met, MMP2 and MMP9 changes via overexpressing TR4 in ACHN cells (left) and knocking down TR4 in OSRC-2 cells (right). (D), Rescue assay using ACHN and OSRC-2 cells treated with/without Met inhibitor showed partially reversed TR4-induced cell invasion ability. Upper panels show representative images and quantifications are shown in the lower panels. (E), TR4RE motif sequences. (F), Two putative TR4REs on the HGF promoter as predicted by JASPAR. (G), ChIP assay results of the HGF promoter TR4REs in ACHN cells. (H), ChIP assay results in OSRC-2 cells showed knocking down TR4 decreased the physical binding between TR4 and TR4RE1. (I), The wild type and mutant pGL3-HGF promoter reporter constructs. (J-K), Luciferase activity after transfection of wild-type or mutant HGF promoter reporter construct in the overexpressed TR4 ACHN (J) and OSRC-2 (K) cells compared to the control cells. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.005, and ns=not significant.
We then confirmed the above results at the protein level showing adding TR4-cDNA led to increase the expression of HGF and phospho-Met with unchanged total-Met expression in ACHN cells (Fig. 3C). In contrast, knocking-down TR4 with TR4-shRNA led to significantly decreased HGF and phospho-Met with unchanged total-Met expression in OSRC-2 cells (Fig. 3C).
As early studies indicated that HGF/Met signaling might promote cancer cell migration and invasion via elevating MMP2/MMP9 expression26, 27, we also assayed these potential downstream signals, and results revealed that adding TR4-cDNA led to increase the expression of MMP2 and MMP9 in ACHN cells and knocking-down TR4 with TR4-shRNA suppressed the expression of MMP2 and MMP9 in OSRC-2 cells (Fig. 3C). Similar results were also obtained in SW839 cells showing knocking-down TR4 suppressed HGF/Met/MMP2-MMP9 signaling (Supplementary Fig. S4B).
Finally, using interruption approaches with Met kinase inhibitor SU11274 (5 μmol/L) for the cell invasion assay, we found Met kinase inhibitor could reverse/block the TR4-enhanced ccRCC cells invasion in ACHN and OSRC-2 cells (Fig. 3D).
Together, results from Fig. 3A-D and Supplementary Fig. S4A-B suggest that the TR4 may function via altering HGF/Met/MMP2-MMP9 signaling to promote the ccRCC cells invasion.
Mechanism dissection of how TR4 up-regulates HGF expression: via transcriptional regulation
To dissect the mechanism how TR4 could modulate the expression of HGF at the transcriptional level, we first applied the JASPER database to search for potential TR4 response elements (TR4REs) on the 2kb HGF 5' promoter region (Fig. 3E), and found two putative TR4REs (-1462~-1448; -345~-331) (Fig. 3F). Results from the chromatin immunoprecipitation (ChIP) assay revealed that TR4 could bind to the TR4RE located 331-345 bp from the transcription start site of HGF in ACHN cells (Fig. 3G). We also found that TR4 could bind to this region in the OSCR-2 cells and knocking-down TR4 could decrease this binding (Fig. 3H).
We then constructed the HGF promoter luciferase reporter by inserting a 1 kb 5' promoter region of HGF containing either wild-type or mutated TR4RE into the pGL3 luciferase backbone (Fig. 3I), and results revealed that adding TR4-cDNA significantly increased luciferase activity in ACHN and OSRC-2 cells with wild type and not mutated TR4RE (Fig. 3J-K).
Together, results from Fig. 3E-K suggest that TR4 can increase HGF expression at the transcriptional level via binding to the TR4RE located on its 5' promoter region.
Targeting the TR4 with miR-32-5p
To search for the small molecules that can target this newly identified TR4/HGF/Met/MMP2-MMP9 signaling, we focused on the microRNAs (miRNAs) as recent studies indicated that miRNAs might function as key regulators to alter the cancer progression28, and some selective miRNAs have been linked to the ccRCC cell invasion21–23, 29. Results from multiple databases (TargetScan, miRDB and microRNA.org) analyses also revealed some potential candidate miRNAs that might suppress TR4 expression via targeting its 3'UTR.
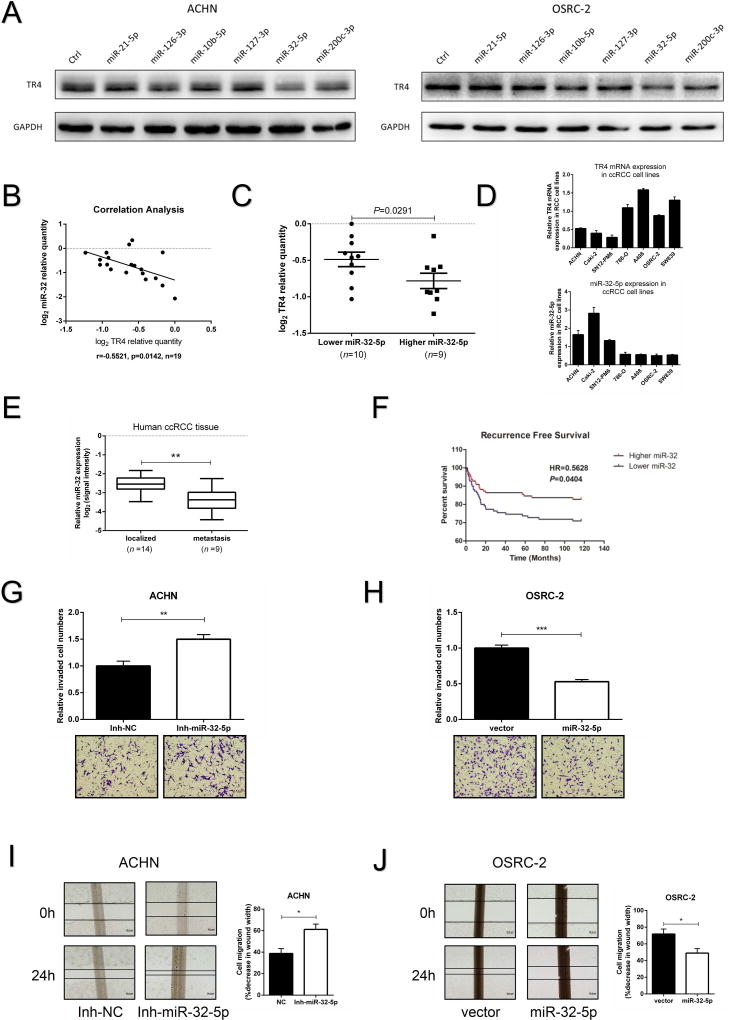
We then focused on 6 miRNAs whose expression were down-regulated in patients with distant metastasis based on the data from Wotschofsky Z’, s microarray28, and results from these miRNAs effects on TR4 expression found altering the miR-32-5p led to decrease TR4 expression in both ccRCC cell lines (Fig. 4A). Importantly, treating with mir-32-5p inhibitor (5 nmol/L, MIN0000090, Qiagen) resulted in an increase of TR4 mRNA in ACHN cells and a decreased TR4 mRNA expression after adding the miR-32-5p in OSRC-2 and SW839 cells (Supplementary Fig. S5A-C).
Figure 4. Expression of miR-32-5p’s in ccRCC tissue samples and effects on invasion and migration in vitro.
(A), Some screened miRNAs that could target TR4 are shown using immunoblot analysis in ACHN (left) and OSRC-2 (right) cells. (B), Correlation analysis of miR-32-5p and TR4 mRNA levels using Pearson correlation coefficient in 19 ccRCC samples. (C), The 19 ccRCC samples were divided into 2 groups according to their miR-32-5p expression and ccRCC tissues with lower miR-32-5p expression displayed higher TR4 level, and vice versa. (D), The negative correlation tendency of miR-32-5p (lower panel) and TR4 (upper panel) in ccRCC cell lines. (E), The expression of miR-32-5p in ccRCC tissue samples from patients with distant metastases compared to tissues from metastasis-free patients (data from GEO DataSets website). (F), Recurrence-free survival curves of ccRCC patients analyzed according to the miR-32 expression data from TCGA website, (top curve, Higher miR-32 and bottom curve, Lower miR-32). (G,H), Invasion ability of miR-32-5p inhibitor treated ACHN cells (G), and miR-32-5p overexpressed OSRC-2 cells (H) were compared to their control cells using Matrigel-coated Transwell assays. (I-J), Migration ability of miR-32-5p inhibitor treated ACHN cells(I), and miR-32-5p overexpressed OSRC-2 cells (J) were compared to their control cells using wound healing assays. The percentage of decreased wound widths were compared between the control group and experimental group. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05, **P < 0.01, and ***P < 0.005.
Results from a clinical sample survey also revealed a significant negative correlation (r=−0.5521, p=0.0142), using the Pearson correlation coefficient, between the expression of miR-32-5p and TR4 in 19 ccRCC patient specimens (Fig. 4B), with lower miR-32-5p expression linked to the higher TR4 mRNA expression, and vice versa (Fig. 4C). Similar results also occurred showing a negative correlation between miR-32-5p and TR4 expression in multiple ccRCC cell lines (Fig. 4D).
A further analysis of Wotschofsky Z’s miRNA microarray raw data in GEO DataSets (GEO GSE37989) indicated that miR-32-5p was down-regulated in ccRCC tissue samples from patients with distant metastases compared to those from metastasis-free patients (Fig. 4E). Results from a database from TCGA (TCGA website: http://cancergenome.nih.gov/) that included miRNA expression data from 326 primary ccRCC samples also revealed that patients with a higher miR-32 expression had significantly higher recurrence-free survivals (HR=0.5628) than patients with lower miR-32 expression (Fig. 4F). Furthermore, we also found that patients with higher miR-32 expression had higher overall survival (HR=0.8254) than patients with lower miR-32 expression (See Fig. S2B), but with less significant results.
We also applied the Matrigel-coated Transwell invasion assay to confirm the consequence of miR-32-5p expression, and results revealed that knocking-down miR-32-5p significantly promoted ACHN cells invasion (Fig. 4G) and treating with miR-32-5p inhibitor via lentiviral system markedly suppressed ccRCC cells invasion in OSRC-2 (Fig. 4H) and SW839 cells (Supplementary Fig. S5D). Wound healing assays also indicated that knocking-down miR-32-5p promoted cell invasion in ACHN cells (Fig. 4I) and adding miR-32-5p decreased cell migration in OSRC-2 (Fig. 4J) and SW839 cells (Supplementary Fig. S5E).
Together, results from Fig. 4A-J and Supplementary Figs. S2B & S5A-E demonstrate that miR-32-5p is linked to the ccRCC metastasis and is the upstream of TR4 that can suppress the TR4 expression to inhibit the ccRCC cell invasion and migration.
The miR-32-5p functions through altering the TR4/HGF/Met/MMP2-MMP9 signaling to suppress the ccRCC metastasis
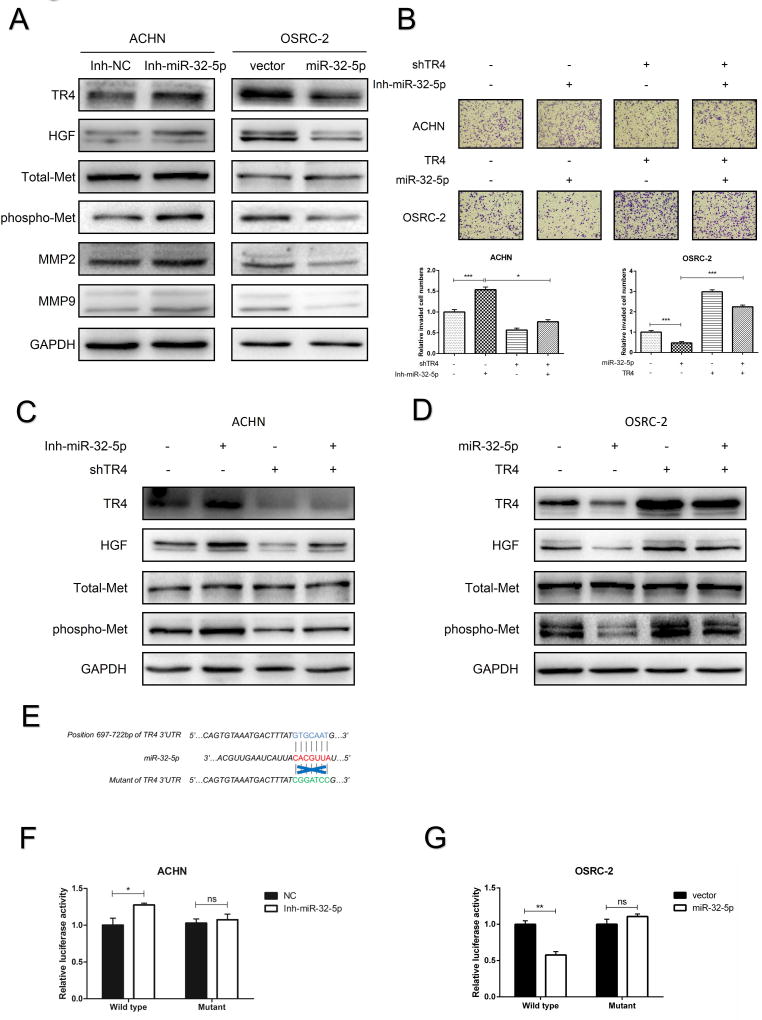
To further link the miR-32-5p-suppressed TR4 to the TR4/ HGF/Met/MMP2-MMP9 signaling for altering the ccRCC metastasis, we first confirmed that knocking-down miR-32-5p increased the expression of TR4, HGF, and phosphorylated-Met (phospho-Met) expression with little impact on the total-Met expression in ACHN cells (Fig. 5A) and adding miR-32-5p decreased the expression of TR4, HGF, and phosphorylated-Met in OSRC-2 (Fig. 5A) and SW839 cells (Supplementary Fig. S6A). As expected, altering the miR-32-5p expression also led to regulate the gene expression of MMP2 and MMP9, the downstream targets of the TR4/HGF/Met axis (Fig. 5A).
Figure 5. Mechanism dissection how miR-32-5p suppresses ccRCC invasion and migration.
(A), The protein levels of TR4, HGF, Total-Met, phospho-Met, MMP2 and MMP9 were determined by immunoblot analysis after using the miR-32-5p inhibitor in ACHN cells (left) and infection with miR-32-5p in OSRC-2 cells (right). (B), The invasion abilities of ACHN cells (upper panels. +/− shTR4 and +/− miR-32-5p inhibitor) and OSRC-2 cells (middle panels +/− TR4 and +/− miR-32-5p) were determined by Matrigel-coated Transwell assay. Quantitations are on lower panels. (C-D), The protein levels of TR4, HGF, Total-Met and phospho-Met were determined using immunoblot analysis of ACHN cells (c, +/− shTR4 and +/− miR-32-5p inhibitor) and OSRC-2 cells (d, +/− TR4 and +/− mir-32-5p). (E), Sequence alignment of the TR4 3'UTR with potential wild type versus mutant miR-32-5p targeting sites. (F-G), Luciferase reporter activity after transfection of wild type TR4 3’UTR reporter construct in the ACHN (F) and OSRC-2 (G) cells treated with inhibitor of miR-32-5p or overexpressing miR-32-5p, respectively, vs control cells. Data presented as mean±SEM. P<0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.005, ns=not significant.
Importantly, using the rescue assay, we found knocking-down TR4 could partially block the cell invasion induced by miR-32-5p inhibitor in ACHN cells and adding TR4-cDNA could partially reverse the miR-32-5p-suppressed cell invasion in OSRC-2 cells (Fig. 5B). Knocking down TR4 could also partially block the HGF and phospho-Met expression induced by miR-32-5p inhibitor in ACHN cells (Fig. 5C) and adding TR4 could partially revers the miR-32-5p-suppressed HGF and phospho-Met expression in OSRC-2 cells, with little influence on the total Met expression in both cells (Fig. 5D).
Together, results from Fig. 5A-D and Supplementary Fig. S6A suggest that miR-32-5p suppressed ccRCC cells invasion via altering the TR4/, HGF/Met/MMP2-MMP9 signaling.
Mechanism dissection of how miR-32-5p decreases TR4 expression: via 3'UTR regulation
To further dissect the mechanism how miR-32-5p can regulate TR4 expression, we focused on the 3’UTR regulation, as early studies indicated that miRNAs could negatively regulate gene expression via interacting with the 3'UTR of target mRNAs16 or positively regulate gene expression via interacting with the 5' promoter region17. We first identified one predicted miRNA-responsive-element that matched the seed sequence of miR-32-5p in the 3'UTR of TR4 gene (Fig. 5E). We then inserted a 959bp fragment from the TR4 3'UTR that contains the predicted miR-32-5p target site (or its mutation) into a dual-luciferase reporter backbone psiCHECK™-2 (Fig. 5E). As expected, the luciferase assay results revealed that treating wit iR-32-5p inhibitor significantly increased luciferase activity in the ACHN cells (Fig. 5F), and adding miR-32-5p markedly decreased luciferase activity in OSRC-2 cells transfected with wild type TR4 3'UTR, but not the mutant TR4 3'UTR (Fig. 5G). Similar results were also obtained when we replaced OSRC-2 with SW839 cells (Supplementary Fig. S6B).
Together, results from Fig. 5E-G and Fig. S6B suggest that miR-32-5p can directly and specifically regulate TR4 expression through binding to its 3'UTR.
Preclinical study using in vivo mouse model to confirm TR4 and miR-32-5p roles in the ccRCC metastasis
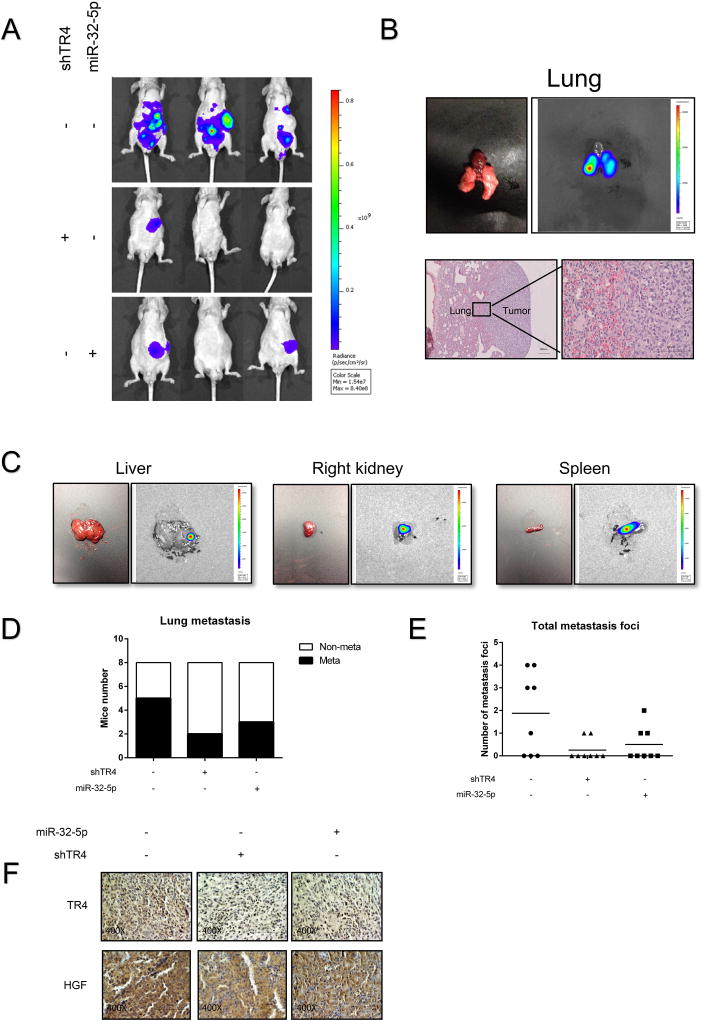
To confirm all above in vitro cell lines data in the in vivo mouse model, we prepared orthotopic xenografts of OSRC-2 cells with luciferase expression and with/without shTR4 or miR-32-5p. After 8 weeks, a dramatic decrease of metastatic luciferase signal using the In Vivo Imaging Systems (IVIS) was found in mice injected with the OSCR-2-Luc-shTR4 cells and OSCR-2-Luc-miR-32-5p cells group compared to the OSCR-2-Luc control group (Fig. 6A). Similar reduced metastatic foci in lung, spleen, liver, and right kidney using H&E staining or IVIS were also found in OSCR-2-Luc-shTR4 group and OSCR-2-Luc-miR-32-5p group compared to the OSCR-2-Luc control group (Fig. 6B-C). The OSCR-2-Luc control group also had relative higher rates of lung metastasis and more metastasis foci than the OSCR-2-Luc -shTR4 group and OSCR-2-Luc-miR-32-5p group (Fig. 6D-E). Importantly, our IHC staining from these ccRCC xenografts also demonstrated that OSCR-2-Luc-miR-32-5p tumors had a lower TR4 expression compared to tumors from the OSCR-2-Luc control group and the tumors from the OSCR-2-Luc miR-32-5p group. Also the OSCR-2-Luc-shTR4 group had a lower HGF expression compared to those from the OSCR-2-Luc control group, which was consistent with our in vitro findings (Fig. 6F).
Figure 6. In vivo mice studies confirmed the role of TR4 and miR-32-5p in ccRCC metastasis.
(A), IVIS imaging was used to determine the metastasis in mice injected with OSRC-2-Luc-shTR4 cells and OSC-2-Luc-miR=32-5p cells compared to the OSRC-2-Luc control cells. Here are the representative image. (B), Representative bioluminescent images of lung metastasis (upper panel); HE staining confirm the tumor tissue in lung (lower panel). (C), Representative bioluminescent images of metastatic foci in liver, right kidney and spleen. (D-E), Quantification of the lung metastases (D) and total metastatic foci (E) in the 3 groups of mice. (F), Representative images of IHC staining for TR4 and HGF in the 3 groups of mice.
Together, results from preclinical studies using in vivo mouse model (Fig. 6A-F) demonstrate that targeting this newly identified miR-32-5p/TR4/HGF/Met signaling with small molecules including the TR4-shRNA or miR-32-5p can suppress ccRCC metastasis.
Sunitinib suppresses ccRCC metastasis via altering the miR-32-5p-TR4 signal
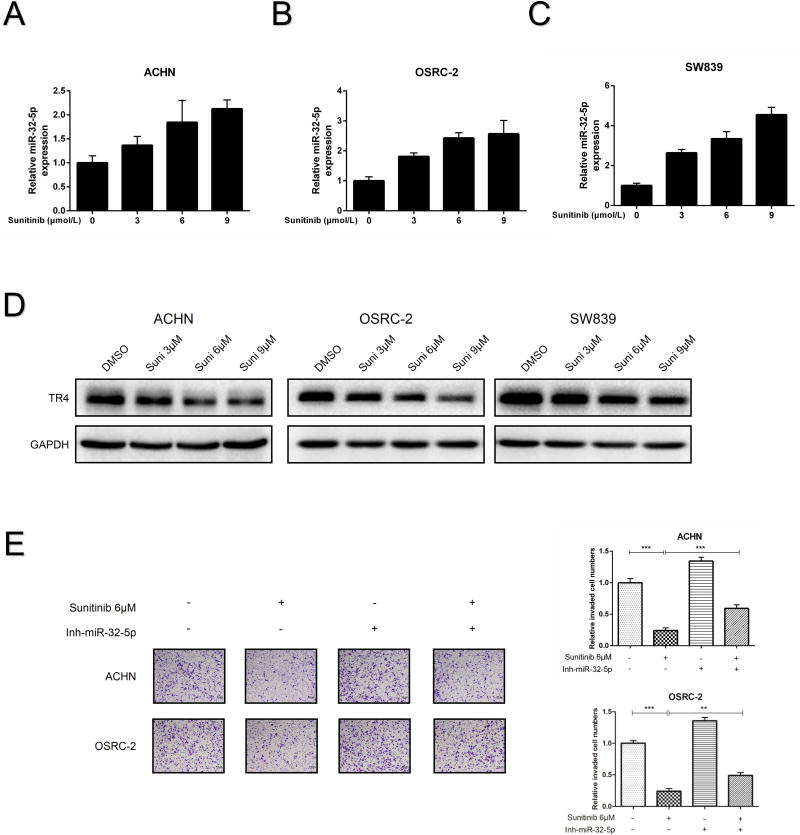
Interestingly, we found this newly identified miR-32-5p/TR4/HGF/Met signaling might also impact the current Sunitinib-chemotherapy to suppress the ccRCC. We first found that treating with Sunitinib could increase miR-32-5p expression (Fig. 7A-C) and decrease the TR4 protein (Fig. 7D) in a dose-dependent manner in ACHN, OSRC-2 and SW839 cells. Importantly, suppressing miR-32-5p with the miR-32-5p inhibitor led to partially reverse the Sunitinib-suppressed ccRCC cell invasion in both ACHN and OSRC-2 cells (Fig. 7E), suggesting that targeting this miR-32-5p/TR4/HGF/Met signaling pathway can also alter the efficacy of Sunitinib-suppressed ccRCC cell invasion.
Figure 7. Sunitinib suppressed ccRCC metastasis via inducing miR-32-5p and suppressing TR4.
(A-C), The qRT-PCR shows miR-32-5p expression increases in a dose dependent manner after the Sunitinib treatment in ACHN (A), OSRC-2 (B) and SW839 (C) cells. (D), The immunoblot analysis shows TR4 expression decreases in a dose dependent manner after the Sunitinib treatment in ACHN (left), OSRC-2 (middle) and SW839 (right) cells. (E), The invasion abilities of ACHN (upper) and OSRC-2 (lower) cells were determined using Transwell assay after Sunitinib treatment (vs. control) and after treatment with miR-32-5p inhibitor (vs. control). Quantitation is at right. Data presented as mean±SEM. P <0.05 was considered statistically significant. **P < 0.01, ***P < 0.005
Discussion
ccRCC is a highly metastatic tumor that may resist most chemotherapy and radiotherapy, and anti-VEGF targeted therapy remains as the first line therapy for metastatic ccRCC even though it may only last for 6–15 months30. Therefore, a new therapy to better suppress ccRCC metastasis is much needed to extend patients survival. We provided the first preclinical evidence showing targeting the newly identified miR-32-5p/TR4/HGF/Met signaling with small molecules including miR-32-5p or TR4-shRNA could further suppress the ccRCC metastasis. This is clinically significant and may help us to develop a new therapy to better suppress ccRCC metastasis.
Interestingly, mechanism dissection found that TR4 might play a positive role to promote RCC metastasis via altering the HGF/Met/MMP2-MMP9 signaling, which is opposite to the early studies showing TR4 might play negative roles to suppress PCa metastasis via altering CCL2/CCR signaling12. The precise reasons why TR4 utilized different signaling pathways to influence these two urological tumors’ metastasis is not clear.
Inappropriate activation of HGF/Met signaling31 may function via autocrine and paracrine manners to impact the tumor growth, angiogenesis, and metastasis in various types of cancers including ccRCC32, 33. While our results from ccRCC cell lines suggest that the HGF/Met signaling may function through autocrine signaling to promote ccRCC cell invasion/migration, it is possible that upon activation via phosphorylation or gene amplification of Met, the HGF/Met signaling can also function through a paracrine axis involving cancer-associated fibroblasts and endothelium34, 35 to influence the ccRCC metastasis. Our results confirmed that HGF-induced phosphorylation of Met and not amplification of Met to mediate the TR4-induced ccRCC metastasis.
Early studies identified some compounds, including polyunsaturated fatty acids and the thiazolidinedione36, 37, might function as TR4 ligands/activators. We found here that miRNAs might also be able to alter the TR4 expression, which might have the potential to be developed as a new therapy to better suppress the ccRCC. A growing body of evidence indicates that the miRNAs might play a vital role in altering gene expression and used as diagnostic and therapeutic agents in clinical trials38. Our finding that miR-32-5p might function as an metastasis suppressor in ccRCC via directly targeting the 3'UTR of TR4 to suppress the TR4-HGF/Met signaling is interesting since earlier studies suggested miR-32 could also function as an oncogene to promote PCa39 and colorectal carcinoma40 progression.
In summary, TR4 may up-regulate HGF/Met signaling to promote ccRCC metastasis and miR-32-5p may target TR4/HGF/Met signaling to suppress ccRCC metastasis. Using small molecules including TR4-shRNA or miR-32-5p may help to potentially develop new therapies to better suppress the ccRCC metastasis.
Supplementary Material
Supplementary Figure 2. (A), Overall survival curves of ccRCC patients analyzed according to the TR4 expression (data downloaded from TCGA website). (B), Overall-free survival curves of ccRCC patients analyzed according to the miR-32 expression (data downloaded from TCGA website).
Supplementary Figure 3. The effect of TR4 on invasion and migration in ccRCC SW839 cell line. (A), Invasion ability of TR4 knocked-down SW839 cells were compared to the control cells using Matrigel-coated Transwell assay. Quantification of invaded cell numbers in upper panels, images in lower panels and insets are WB of viral infections. (B), Migration ability of TR4 knocked-down SW839 cells were compared to the control scramble cells using wound healing assay. Quantification at right. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05 and ***P < 0.005.
Supplementary Figure 4. TR4 regulates HGF/Met signaling in SW839 cell line. (A), Immunoblot analysis of CCL2, TGFβR2 and P-SMAD3 changes via overexpressing TR4 in ACHN cells (left) and knocking down TR4 in OSRC-2 cells (right). (B), Immunoblot analysis of HGF, phospho-Met, total-Met, MMP2 and MMP9 changes via knocking down TR4 in SW839 cells.
Supplementary Figure 5. The effect of miR-32-5p on TR4 mRNA expression and invasion and migration in SW839 cell line. (A-C), TR4 mRNA level changes analyzed using qRT-PCR after treating ACHN cells with miR-32-5p inhibitor (A) and overexpressing miR-32-5p in OSRC-2 (B) and SW839 (C) cells. (D), Invasion ability of miR-32-5p overexpressed SW839 cells were compared to the control cells using Matrigel-coated Transwell assay. (E), Migration ability of miR-32-5p overexpressed SW839 cells were compared to the control cells using wound healing assays. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05 and ***P < 0.005.
Supplementary Figure 6. Mechanism dissection how miR-32-5p suppresses ccRCC invasion and migration in SW839 cell line. (A), The protein levels of TR4, HGF, Total-Met, phospho-Met, MMP2 and MMP9 were determined using immunoblot analysis after infection with miR-32-5p in SW839 cells versus vector control. (B), Luciferase reporter activity after transfection of wild type or mutant TR4 3'UTR reporter construct in SW839 overexpressing miR-32-5p cells vs control cells. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. **P < 0.01, ns not significant.
Supplementary Figure 7. Western blots quantification. (A), quantification for Fig 3C. (B), quantification for Fig 5A. (C), quantification for Fig 5C. (D), quantification for Fig 5D. (E), quantification for Fig 7D. (F), quantification for Fig 1C.
Supplementary Table 1. Clinical parameters for ccRCC patients.
Novelty & Impact Statements.
We first demonstrated TR4’s positive roles in promoting ccRCC metastasis via altering the HGF/Met signaling with clinical and preclinical studies using in vitro multiple cells lines and in vivo mouse model. We then dissected the molecular mechanisms showing TR4 could function via modulating the miR-32-5p expression to alter the HGF/Met signaling. Finally, we demonstrated that targeting this newly identified signaling with small molecules including miR-32-5p or TR4-shRNA can further suppress ccRCC metastasis, as well as alter the Sunitinib-chemotherapy efficacy to suppress the ccRCC metastasis.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81702508), NIH grants (CA155477 and CA156700), George Whipple Professorship Endowment and Taiwan Department of Health Clinical Trial, Research Center of Excellence (DOH99-TD-B-111-004 to China Medical University, Taichung, Taiwan) and China 973 Program (2012CB518305). We thank Karen Wolf for help preparing the manuscript.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Comperat E, Camparo P. Histological classification of malignant renal tumours at a time of major diagnostic and therapeutic changes. Diagnostic and interventional imaging. 2012;93:221–31. doi: 10.1016/j.diii.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Stadler WM. Targeted agents for the treatment of advanced renal cell carcinoma. Cancer. 2005;104:2323–33. doi: 10.1002/cncr.21453. [DOI] [PubMed] [Google Scholar]
- 4.Chang C, Da Silva SL, Ideta R, Lee Y, Yeh S, Burbach JP. Human and rat TR4 orphan receptors specify a subclass of the steroid receptor superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:6040–4. doi: 10.1073/pnas.91.13.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YF, Lee HJ, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol. 2002;81:291–308. doi: 10.1016/s0960-0760(02)00118-8. [DOI] [PubMed] [Google Scholar]
- 6.Ding XF, Yu SC, Chen BD, Lin SJ, Chang C, Li GH. Recent advances in the study of testicular nuclear receptor 4. J Zhejiang Univ Sci B. 2013;14:171–7. doi: 10.1631/jzus.B1200357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu X, Lee YF, Liu NC, Chen YT, Kim E, Shyr CR, Chang C. Targeted inactivation of testicular nuclear orphan receptor 4 delays and disrupts late meiotic prophase and subsequent meiotic divisions of spermatogenesis. Mol Cell Biol. 2004;24:5887–99. doi: 10.1128/MCB.24.13.5887-5899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SJ, Ho HC, Lee YF, Liu NC, Liu S, Li G, Shyr CR, Chang C. Reduced osteoblast activity in the mice lacking TR4 nuclear receptor leads to osteoporosis. Reprod Biol Endocrinol. 2012;10:43. doi: 10.1186/1477-7827-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YT, Collins LL, Uno H, Chou SM, Meshul CK, Chang SS, Chang C. Abnormal cerebellar cytoarchitecture and impaired inhibitory signaling in adult mice lacking TR4 orphan nuclear receptor. Brain Res. 2007;1168:72–82. doi: 10.1016/j.brainres.2007.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins LL, Lee YF, Heinlein CA, Liu NC, Chen YT, Shyr CR, Meshul CK, Uno H, Platt KA, Chang C. Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15058–63. doi: 10.1073/pnas.0405700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi H, Kim SJ, Park SS, Chang C, Kim E. TR4 activates FATP1 gene expression to promote lipid accumulation in 3T3-L1 adipocytes. FEBS Lett. 2011;585:2763–7. doi: 10.1016/j.febslet.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Ding X, Yang DR, Lee SO, Chen YL, Xia L, Lin SJ, Yu S, Niu YJ, Li G, Chang C. TR4 nuclear receptor promotes prostate cancer metastasis via upregulation of CCL2/CCR2 signaling. Int J Cancer. 2015;136:955–64. doi: 10.1002/ijc.29049. [DOI] [PubMed] [Google Scholar]
- 13.Qiu X, Zhu J, Sun Y, Fan K, Yang DR, Li G, Yang G, Chang C. TR4 nuclear receptor increases prostate cancer invasion via decreasing the miR-373-3p expression to alter TGFbetaR2/p-Smad3 signals. Oncotarget. 2015 doi: 10.18632/oncotarget.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Yu S, Ding X, Jing C, Xia L, Wang M, Matro E, Rehman F, Niu Y, Li G, Chang C. The role of testicular nuclear receptor 4 in chemo-resistance of docetaxel in castration-resistant prostate cancer. Cancer Gene Ther. 2014;21:411–5. doi: 10.1038/cgt.2014.41. [DOI] [PubMed] [Google Scholar]
- 15.Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2014;33:3225–34. doi: 10.1038/onc.2013.274. [DOI] [PubMed] [Google Scholar]
- 16.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen J, Zhang Y, Lai P, Fan X, Zhou X, Lin J, Li M, et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res. 2014;74:3031–42. doi: 10.1158/0008-5472.CAN-13-2193. [DOI] [PubMed] [Google Scholar]
- 19.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, Wu J, Li M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–79. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoni E, van der Horst G, van de Merbel AF, Chen L, Rane JK, Pelger RC, Collins AT, Visakorpi T, Snaar-Jagalska BE, Maitland NJ, van der Pluijm G. miR-25 Modulates Invasiveness and Dissemination of Human Prostate Cancer Cells via Regulation of alphav- and alpha6-Integrin Expression. Cancer Res. 2015;75:2326–36. doi: 10.1158/0008-5472.CAN-14-2155. [DOI] [PubMed] [Google Scholar]
- 21.Chen XY, Wang XG, Ruan AM, Han WW, Zhao Y, Lu X, Xiao P, Shi HC, Wang R, Chen L, Chen SY, Du QS, et al. miR-141 Is a Key Regulator of Renal Cell Carcinoma Proliferation and Metastasis by Controlling EphA2 Expression. Clinical Cancer Research. 2014;20:2617–30. doi: 10.1158/1078-0432.CCR-13-3224. [DOI] [PubMed] [Google Scholar]
- 22.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, Yamamura S, Ueno K, Zaman MS, Singh K, Chang I, Deng G, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–21. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamasaki T, Seki N, Yoshino H, Itesako T, Hidaka H, Yamada Y, Tatarano S, Yonezawa T, Kinoshita T, Nakagawa M, Enokida H. MicroRNA-218 inhibits cell migration and invasion in renal cell carcinoma through targeting caveolin-2 involved in focal adhesion pathway. J Urol. 2013;190:1059–68. doi: 10.1016/j.juro.2013.02.089. [DOI] [PubMed] [Google Scholar]
- 24.Xiang B, Muthuswamy SK. Using three-dimensional acinar structures for molecular and cell biological assays. Methods Enzymol. 2006;406:692–701. doi: 10.1016/S0076-6879(06)06054-X. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Zhang D, Zhang LL, Zhu GD, Sun Y, Wu KJ, Wang XY, He DL. PrLZ Expression Is Associated With the Progression of Prostate Cancer LNCaP Cells. Mol Carcinogen. 2009;48:432–40. doi: 10.1002/mc.20481. [DOI] [PubMed] [Google Scholar]
- 26.Baek JH, Birchmeier C, Zenke M, Hieronymus T. The HGF Receptor/Met Tyrosine Kinase Is a Key Regulator of Dendritic Cell Migration in Skin Immunity. Journal of Immunology. 2012;189:1699–707. doi: 10.4049/jimmunol.1200729. [DOI] [PubMed] [Google Scholar]
- 27.Torres KE, Zhu QS, Bill K, Lopez G, Ghadimi MP, Xie XB, Young ED, Liu JH, Nguyen T, Bolshakov S, Belousov R, Wang SZ, et al. Activated MET Is a Molecular Prognosticator and Potential Therapeutic Target for Malignant Peripheral Nerve Sheath Tumors. Clinical Cancer Research. 2011;17:3943–55. doi: 10.1158/1078-0432.CCR-11-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 29.Wotschofsky Z, Liep J, Meyer HA, Jung M, Wagner I, Disch AC, Schaser KD, Melcher I, Kilic E, Busch J, Weikert S, Miller K, et al. Identification of Metastamirs as Metastasis-associated MicroRNAs in Clear Cell Renal Cell Carcinomas. Int J Biol Sci. 2012;8:1363–74. doi: 10.7150/ijbs.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 31.Gherardi E, Sharpe M, Lane K, Sirulnik A, Stoker M. Hepatocyte growth factor/scatter factor (HGF/SF), the c-met receptor and the behaviour of epithelial cells. Symp Soc Exp Biol. 1993;47:163–81. [PubMed] [Google Scholar]
- 32.Horie S, Aruga S, Kawamata H, Okui N, Kakizoe T, Kitamura T. Biological role of HGF/MET pathway in renal cell carcinoma. J Urol. 1999;161:990–7. [PubMed] [Google Scholar]
- 33.Ciamporcero E, Miles KM, Adelaiye R, Ramakrishnan S, Shen L, Ku S, Pizzimenti S, Sennino B, Barrera G, Pili R. Combination strategy targeting VEGF and HGF/c-met in human renal cell carcinoma models. Mol Cancer Ther. 2015;14:101–10. doi: 10.1158/1535-7163.MCT-14-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia CC, Wang TT, Liu W, Fu BS, Hua X, Wang GY, Li TJ, Li X, Wu XY, Tai Y, Zhou J, Chen GH, et al. Cancer-Associated Fibroblasts from Hepatocellular Carcinoma Promote Malignant Cell Proliferation by HGF Secretion. Plos One. 2013;8 doi: 10.1371/journal.pone.0063243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueki T, Fujimoto J, Suzuki T, Yamamoto H, Okamoto E. Expression of hepatocyte growth factor and its receptor c-met proto-oncogene in hepatocellular carcinoma. Hepatology. 1997;25:862–6. doi: 10.1002/hep.510250413. [DOI] [PubMed] [Google Scholar]
- 36.Xie S, Lee YF, Kim E, Chen LM, Ni J, Fang LY, Liu S, Lin SJ, Abe J, Berk B, Ho FM, Chang C. TR4 nuclear receptor functions as a fatty acid sensor to modulate CD36 expression and foam cell formation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13353–8. doi: 10.1073/pnas.0905724106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim E, Liu NC, Yu IC, Lin HY, Lee YF, Sparks JD, Chen LM, Chang C. Metformin inhibits nuclear receptor TR4-mediated hepatic stearoyl-CoA desaturase 1 gene expression with altered insulin sensitivity. Diabetes. 2011;60:1493–503. doi: 10.2337/db10-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 39.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Janne OA, Seppala J, Lahdesmaki H, Tammela TL, Visakorpi T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31:4460–71. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 40.Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, Tan W, Wei G, Zhou Y. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12:30. doi: 10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 2. (A), Overall survival curves of ccRCC patients analyzed according to the TR4 expression (data downloaded from TCGA website). (B), Overall-free survival curves of ccRCC patients analyzed according to the miR-32 expression (data downloaded from TCGA website).
Supplementary Figure 3. The effect of TR4 on invasion and migration in ccRCC SW839 cell line. (A), Invasion ability of TR4 knocked-down SW839 cells were compared to the control cells using Matrigel-coated Transwell assay. Quantification of invaded cell numbers in upper panels, images in lower panels and insets are WB of viral infections. (B), Migration ability of TR4 knocked-down SW839 cells were compared to the control scramble cells using wound healing assay. Quantification at right. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05 and ***P < 0.005.
Supplementary Figure 4. TR4 regulates HGF/Met signaling in SW839 cell line. (A), Immunoblot analysis of CCL2, TGFβR2 and P-SMAD3 changes via overexpressing TR4 in ACHN cells (left) and knocking down TR4 in OSRC-2 cells (right). (B), Immunoblot analysis of HGF, phospho-Met, total-Met, MMP2 and MMP9 changes via knocking down TR4 in SW839 cells.
Supplementary Figure 5. The effect of miR-32-5p on TR4 mRNA expression and invasion and migration in SW839 cell line. (A-C), TR4 mRNA level changes analyzed using qRT-PCR after treating ACHN cells with miR-32-5p inhibitor (A) and overexpressing miR-32-5p in OSRC-2 (B) and SW839 (C) cells. (D), Invasion ability of miR-32-5p overexpressed SW839 cells were compared to the control cells using Matrigel-coated Transwell assay. (E), Migration ability of miR-32-5p overexpressed SW839 cells were compared to the control cells using wound healing assays. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. *P < 0.05 and ***P < 0.005.
Supplementary Figure 6. Mechanism dissection how miR-32-5p suppresses ccRCC invasion and migration in SW839 cell line. (A), The protein levels of TR4, HGF, Total-Met, phospho-Met, MMP2 and MMP9 were determined using immunoblot analysis after infection with miR-32-5p in SW839 cells versus vector control. (B), Luciferase reporter activity after transfection of wild type or mutant TR4 3'UTR reporter construct in SW839 overexpressing miR-32-5p cells vs control cells. Each sample was run in triplicate and in multiple experiments. Data presented as mean±SEM. P <0.05 was considered statistically significant. **P < 0.01, ns not significant.
Supplementary Figure 7. Western blots quantification. (A), quantification for Fig 3C. (B), quantification for Fig 5A. (C), quantification for Fig 5C. (D), quantification for Fig 5D. (E), quantification for Fig 7D. (F), quantification for Fig 1C.
Supplementary Table 1. Clinical parameters for ccRCC patients.