Capsule Summary
Eosinophilic esophagitis is associated with increased GATA-3 and T-bet expression in comparison to GERD and inactive EoE; moreover, parallel trends are observed in PPI-REE. Immunohistochemical assessment of the GATA-3+ and T-bet+ cells may help distinguish patients with EoE/PPI-REE from GERD and aid in assessment of treatment response.
Keywords: Eosinophilic esophagitis, proton pump inhibitor-responsive esophageal eosinophilia, GATA-3, T-bet, reflux, pediatric, esophagus
To the Editor
Eosinophilic esophagitis (EoE) is a clinicopathological condition characterized by symptoms of esophageal dysfunction and dense eosinophil infiltration of the esophageal epithelium. The current diagnostic metric requires 15 eosinophils per high-power field (eos/hpf) in at least one mucosal biopsy specimen following 6-8 weeks of treatment with high-dose proton pump inhibitor (PPI).1 While this histologic threshold distinguishes most subjects with EoE several shortcomings exist including: (1) eosinophilia may underestimate the extent of eosinophil activity (2) some patients with gastroesophageal reflux disease (GERD) may have distal esophageal eosinophilia exceeding 15 eos/hpf (3) patients often require more than one biopsy to determine the underlying diagnosis and establish appropriate treatment. Recent evidence suggests that EoE and proton pump inhibitor-responsive esophageal eosinophilia (PPI-REE) have a similar transcriptome2 and are phenotypically indistinguishable.3 Novel histologic biomarkers may further aid in distinguishing causes of esophageal eosinophilia.
Characterizations of mucosal inflammatory responses in EoE and PPI-REE describe a Th2-predominant pattern with overexpression of Th2-associated genes.4 In EoE, IL-5 and IL-13 levels are associated with eotaxin-3 expression which likely drives eosinophil recruitment. In contrast esophageal biopsies from patients with GERD are more commonly associated with a Th1 phenotype defined by increased mRNA expression of IL-1β, IL-8, and IFN-γ.5
T-bet and GATA-3 are transcriptional regulators that drive differentiation of Th0 CD4+ lymphocytes to Th1 and Th2 lineages, respectively. We previously demonstrated the utility of characterizing tissue-specific, immune polarization using immunohistochemistry-based assessments of GATA-3 and T-bet in bladder cancer.6 Given its role in Th2-associated inflammation, we hypothesized that GATA-3 expression would be increased in EoE and PPI-REE and that the ratio of GATA-3/T-bet expression would differentiate these individuals from subjects with GERD.
We performed a retrospective, case control study of children characterized clinically as having EoE (n=24), PPI-REE (n=10), GERD (n=28) and controls (n=32). Subjects diagnosed with EoE were treated with an elimination diet (n=7), swallowed topical steroids (n=12) or a combination of elimination diet and swallowed topical steroids (n=5) and those with PPI-REE were treated with high dose PPI (2 mg/kg/day). All subjects with EoE and PPI-REE demonstrated histologic resolution of esophageal eosinophilia (<15 eos/hpf) after 6-8 weeks of treatment, respectively.
Tissue sections from active and matched post-treatment biopsies were assessed with hematoxylin and eosin and immunohistochemical staining for T-bet and GATA-3 was performed. Slides stained for T-bet and GATA-3 were digitized and staining of the epithelial layer was quantified (See Figure E1). A nuclear algorithm was used to identify T-bet+ and GATA-3+ cells. The number of positive cells was divided by the total area of esophageal epithelium analyzed in order to normalize the number of T-bet+ and GATA-3+ cells/mm2. Polarization of the immune microenvironment was assessed by the G/T ratio (GATA-3+ cells/mm2/T-bet+ cells/mm2). Details regarding the study population, methods for T-bet/GATA-3 staining/quantification, and statistical analysis are detailed in this article’s online repository at jacionline.org.
Table E1 details the demographics and clinical characteristics of the study population. Staining for GATA-3 and T-bet by immunohistochemistry is shown in Figure 1. In comparison to GERD, subjects with active EoE demonstrated increased GATA-3+ cells/mm2 (median 2.81 vs. 8.46 cells/mm2, p<0.0001, AUC 0.88, (0.78, 0.97 95% CI)) and T-bet+ cells/mm2 (median 7.12 vs. 12.01 cells/mm2, p<0.0001, AUC 0.79, (0.66, 0.93 95% CI)) (See Figure 2, panels B, C and F). No statistical differences in GATA-3 and T-bet expression were found between subjects with active EoE and PPI-REE. GATA-3 and T-bet expression was also significantly elevated in active EoE subjects compared to healthy controls. Following treatment, GATA-3 and T-bet expression decreased significantly in EoE subjects (p<0.0001) (See Figure 2, panels C and C). A similar trend was observed for subjects with PPI-REE, such that those with inactive EoE/PPI-REE were indistinguishable from GERD and control subjects based on nuclear transcription factor expression. No significant differences were observed between groups for the G/T ratio (See Figure 2, panel D).
Figure 1. EoE subjects have increased GATA-3 and T-bet expression.
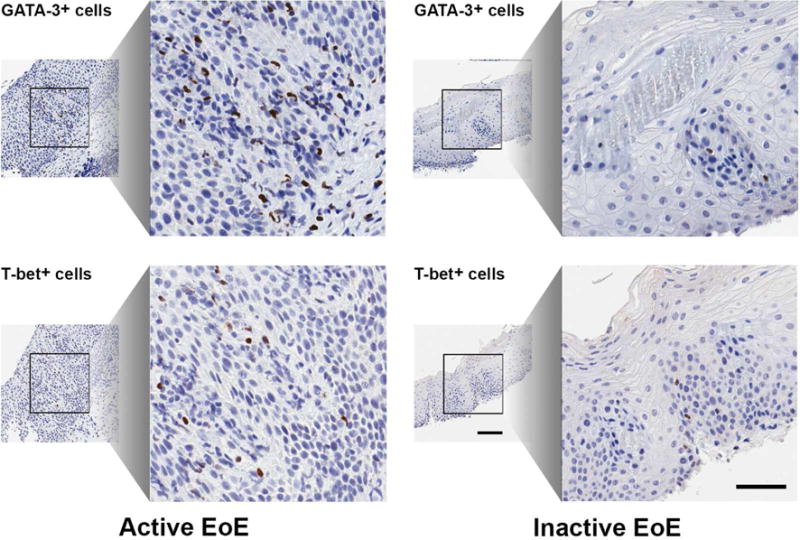
Immunohistochemistry stains for GATA-3 and T-bet in the same subject with active and inactive EoE.
Figure 2. GATA-3+ cells and T-bet+ cells are increased in EoE subjects, correlate with esophageal eosinophilia and decrease in response to treatment.
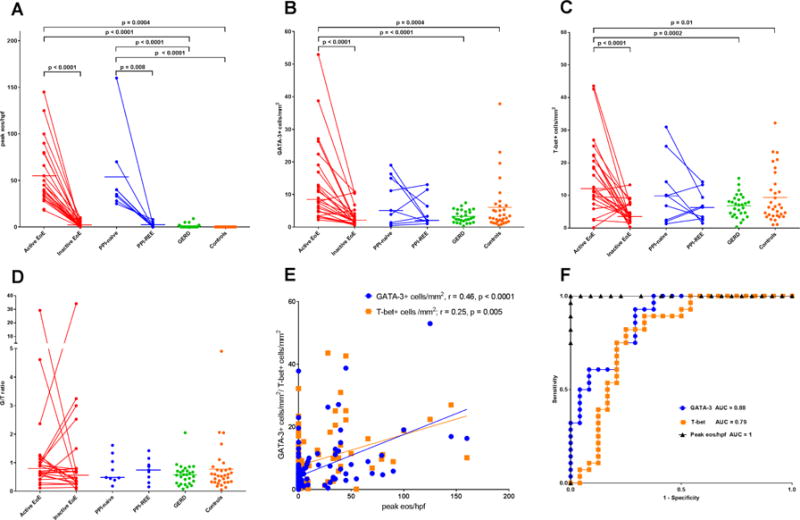
Peak esophageal eosinophil counts are shown in panel A. GATA-3 (panel B) and T-bet (panel C) expression are increased EoE relative to GERD and health controls. Similar trends were noted for PPI-REE subjects but these did not reach statistical significance. The G/T ratio does not distinguish subjects with EoE or PPI-REE (panel D). GATA-3 and T-bet correlate with eos/hpf (panel E). ROC curves for eos/hpf, GATA-3+ cells/mm2, and T-bet+ cells/mm2 suggest GATA-3 and T-bet expression may serve as histologic biomarkers that differentiate active EoE from GERD (panel E).
While EoE has been defined as a type-2 cytokine-mediated disease we observed similar increases in GATA-3+ and T-bet+ cells in subjects with active EoE and PPI-REE. Expression of both markers was associated with active disease and the G/T cellular ratio did not serve as a histologic marker of EoE or PPI-REE. The observed differences were almost identical when the relative percentages of GATA-3+ and T-bet+ cells were compared among groups (See Figure E2), suggesting tissue polarization in addition to cellular infiltration. Our observations corroborate earlier findings5 suggesting that EoE and PPI-REE share a common pathophysiology and that PPI, topical corticosteroids and dietary elimination each result in decreased GATA-3 expression. The significance of increased T-bet expression is unclear but suggests a mixed Th2/Th1 inflammatory infiltrate is associated with active disease. Consistent with this finding, others have demonstrated increased expression of Th1 cytokines (IFN-γ7 and TNF-α8) in EoE.
We acknowledge the limitation that this is a retrospective, single center study of pediatric subjects; therefore, the results cannot be generalized to adults. Second, immunohistochemical assessments were only performed on a single biopsy specimen from each endoscopy; therefore, it is possible that the immune polarization described is confined to areas of maximum eosinophil infiltrate. Although GATA-3 and T-bet expression was similar in EoE and PPI-REE, we did not examine a sufficient number of PPI-REE subjects in order to observe statistical differences between groups or following treatment for PPI-REE subjects. Finally, we did not perform dual staining to ensure the cells positive for GATA-3 and T-bet were CD4+ helper T cells; therefore, these transcription factors may be expressed by other cell types, including innate lymphoid cells, T regulatory cells9 and natural killer cells (See Figure E3). However, these limitations are balanced by a number of strengths including detailed clinical information, automated assessment of GATA-3/T-bet expression, and blinded analysis of tissue samples.
In summary, we observed that EoE subjects have increased GATA-3 and T-bet expression when compared to subjects with GERD and healthy controls. Similar trends were noted for subjects with PPI-REE. Automated assessment of the GATA-3 and T-bet expression may be a useful strategy to distinguish subjects with EoE/PPI-REE and GERD; moreover, these markers may also be useful in assessing treatment response.
Methods
Study Population
Pediatric subjects (ages 1-18 years) were retrospectively identified at the Children’s Hospital of Colorado from 2006-2016. This study was approved by the Colorado Multiple Institutional Review Board (IRB approval number 07-0888).
Case definitions, clinical data, and biospecimen collection
Diagnosis of active EoE was defined by consensus guidelines.1 Subjects with PPI-REE were treated with high-dose PPI for at least 8 weeks and had <15 eos/hpf on repeat endoscopic biopsy. GERD subjects displayed symptoms consistent with reflux as defined by a pediatric gastroenterologist or had an abnormal pH/impedance study. Control subjects had gastrointestinal symptoms necessitating an upper endoscopy with normal histology (0 eos/hpf). Clinical data were collected through retrospective chart review. Asthma, eczema, seasonal allergies and IgE-mediated food allergies were determined by clinical history as documented in the medical record. Tissue analysis was performed using paraffin-embedded esophageal tissue sections from the Children’s Hospital of Colorado. Esophageal eosinophil counts were quantified by a pediatric pathologist. Study personnel were blinded as to case/control status during sample analysis.
Immunohistochemical staining of GATA-3 and T-bet
Tissue sectioning and IHC staining was performed at the Pathology Research Core (Mayo Clinic, Rochester, MN) using the Leica Bond RX stainer (Leica Biosystems, Nussloch, Germany). Five micron FFPE tissue sections were treated with Bond™ Epitope Retrieval 1 (Citrate; Leica Biosystems, Newcastle, UK) for 20 minutes and Protein Block Serum-Free (Dako, Carpinteria, CA) for 5 minutes. GATA-3 (clone L50-823, Biocare Medical, Pacheco, CA), and T-bet (SC-21003, Santa Cruz Biotech, Dallas, TX) were diluted in Antibody Diluent, Background Reducing (Dako, Carpinteria, CA) at 1:400 and 1:150 respectively and applied to tissue for 15 minutes.
The detection system used was Bond™ Polymer Refine Detection System (Leica Biosystems, Newcastle, UK). This system includes the hydrogen peroxidase block, post primary and polymer reagent, DAB, and hematoxylin. Immunostaining visualization was achieved by incubating slides 10 minutes in DAB and DAB buffer (1:19 mixture) from the Bond™ Polymer Refine Detection System. To this point, slides were rinsed between steps with 1X Bond™ Wash Buffer (Leica Biosystems, Newcastle, UK). Slides were counterstained for five minutes using Schmidt hematoxylin and molecular biology grade water (1:1 mixture), followed by several rinses in 1X Bond™ wash buffer and distilled water, this is not the hematoxylin provided with the Refine kit. Once the immunohistochemistry process was completed, slides were removed from the stainer and rinsed in tap water for five minutes. Slides were dehydrated in increasing concentrations of ethyl alcohol and cleared in 3 changes of xylene prior to permanent coverslipping in xylene-based medium.
Image analysis
Tissue sections were digitized (Aperio AT Turbo, Leica Biosystems, Buffalo Grove, IL) and analyzed using Aperio ImageScope software (version 11.2.0.780, Aperio Technologies, Vista, CA). Serial sections of esophageal biopsies where the maxiumum eosinophil focus was located were analyzed for each subject. The submucosal layer was outlined to subtract it from analysis (see Figure E1). Image analysis using a nuclear algorithm was used to quantify the cells staining positive for T-bet and GATA-3. Only nuclei that stained strongly or moderately positive according to the algorithm were considered positive.
Statistical Analysis
Clinical characteristics of the cases and controls were summarized with descriptive statistics. Baseline comparisons of median eos/hpf, GATA-3+ cells/mm2, T-bet+ cells/mm2 and G/T ratios were made with a Mann-Whitney test. Paired comparisons of G/T ratios pre- and post-treatment (EoE and PPI-REE subjects) were performed using a Wilcoxon matched-pairs signed-rank test. Spearman’s rho values were used to assess correlations between eos/hpf and GATA-3+ cells/mm2 or T-bet+ cells/mm2. Statistical comparisons and plots were made with GraphPad Prism (version 7.00 for Windows, GraphPad software, San Diego, California, USA).
Supplementary Material
Acknowledgments
Funded by Mayo Clinic, Phoenix Children’s Hospital Foundation, Don and Kathy Levin Family Foundation (Wright BL, Lee JJ). This work was also supported by U54AI117804 (CEGIR), which is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Disease Research (ORDR), NCATS, and is funded through collaboration between NIAID, NIDDK, NCATS and patient advocacy groups including APFED CURED and EFC, NIH 1K24DK100303 (Furuta GT), and T32DK067009-11 (Nguyen N). This work benefitted from data assembled by the ImmGen consortium.
Declaration of funding: Funded by Mayo Clinic, Phoenix Children’s Hospital Foundation, Don and Kathy Levin Family Foundation (Wright BL, Lee JJ). This work was also supported by U54AI117804 (CEGIR), which is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Disease Research (ORDR), NCATS, and is funded through collaboration between NIAID, NIDDK, NCATS and patient advocacy groups including APFED CURED and EFC, NIH 1K24DK100303 (Furuta GT), and T32DK067009-11 (Nguyen N).
Abbreviations used
- eos/hpf
eosinophils per high-power field
- EoE
eosinophilic esophagitis
- GERD
gastroesophageal reflux disease
- G/T
GATA-3/T-bet
- PPI
proton pump inhibitor
- PPI-REE
proton pump inhibitor responsive esophageal eosinophilia
- SFED
six food elimination diet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. The Journal of allergy and clinical immunology. 2011;128:3–20 e6. doi: 10.1016/j.jaci.2011.02.040. quiz 1-2. [DOI] [PubMed] [Google Scholar]
- 2.Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor-responsive esophageal eosinophilia reveals proton pump inhibitor-reversible allergic inflammation. J Allergy Clin Immunol. 2015;135:187–97. doi: 10.1016/j.jaci.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, Speck O, Woodward K, Gebhart JH, Madanick RD, Levinson S, et al. Clinical and endoscopic characteristics do not reliably differentiate PPI-responsive esophageal eosinophilia and eosinophilic esophagitis in patients undergoing upper endoscopy: a prospective cohort study. Am J Gastroenterol. 2013;108:1854–60. doi: 10.1038/ajg.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. 17 e1–7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald RC, Onwuegbusi BA, Bajaj-Elliott M, Saeed IT, Burnham WR, Farthing MJ. Diversity in the oesophageal phenotypic response to gastro-oesophageal reflux: immunological determinants. Gut. 2002;50:451–9. doi: 10.1136/gut.50.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunez-Nateras R, Castle EP, Protheroe CA, Stanton ML, Ocal TI, Ferrigni EN, et al. Predicting response to bacillus Calmette-Guerin (BCG) in patients with carcinoma in situ of the bladder. Urol Oncol. 2014;32:45 e23–30. doi: 10.1016/j.urolonc.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta SK, Fitzgerald JF, Kondratyuk T, HogenEsch H. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2006;42:22–6. doi: 10.1097/01.mpg.0000188740.38757.d2. [DOI] [PubMed] [Google Scholar]
- 8.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–61. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 9.Yu F, Sharma S, Edwards J, Feigenbaum L, Zhu J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat Immunol. 2015;16:197–206. doi: 10.1038/ni.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


