Abstract
Despite moderate heritability, only one study has identified genomewide significant loci for cannabis-related phenotypes. We conducted meta-analyses of genomewide association study (GWAS) data on 2,080 DSM-IV cannabis dependent cases and 6,435 cannabis exposed controls of European descent. A cluster of correlated single nucleotide polymorphisms (SNPs) in a novel region on chromosome 10 was genomewide significant (lowest p = 1.3E-8). Among the SNPs, rs1409568 showed enrichment for H3K4me1 and H3K427ac marks, suggesting its role as an enhancer in addiction-relevant brain regions, such as the dorsolateral prefrontal cortex and the angular and cingulate gyri. This SNP is also predicted to modify binding scores for several transcription factors. We found modest evidence for replication for rs1409568 in an independent cohort of African-American (896 cases and 1591 controls; p=0.03) but not European-American (781 cases and 1905 controls) participants. The combined meta-analysis (3,757 cases and 9,931 controls) indicated trend-level significance for rs1409568 (p=2.85E-7). No genomewide significant loci emerged for cannabis dependence criterion count (n=8,050). There was also evidence that the minor allele of rs1409568 was associated with a 2.1% increase in right hippocampal volume in an independent sample of 430 European-American college students (fwe-p=.007). The identification and characterization of genomewide significant loci for cannabis dependence is amongst the first steps towards understanding the biological contributions to the etiology of this psychiatric disorder, which appears to be rising in some developed nations.
INTRODUCTION
Cannabis is amongst the most commonly used illicit psychoactive substances in developed nations (1;2). Ten percent of individuals who ever use cannabis meet criteria for lifetime cannabis dependence, which is associated with significant comorbid adverse mental health outcomes (3–5). A recent survey of U.S. adults showed that the past year prevalence of cannabis use disorders has increased from 1.5% to 2.9% in the decade spanning 2002–2012, an increase apparently attributable to a corresponding increase in use during that period of time (6).
About 50–60% of the variance in cannabis use disorders, including DSM-IV dependence, is attributable to the additive effects of genes (i.e., narrow sense heritability)(7). Despite this, only one study to date has successfully identified genomewide significant loci for any cannabis related trait (8). Table 1 provides an overview of six genomewide association studies (GWASs) of cannabis-related phenotypes (9–12), the largest being a recent meta-analysis of GWASs of ever using cannabis, even once during the lifetime (N > 32,000)(13). However, only the recent study by Sherva and colleagues (8) identified genomewide significant loci (three independent regions) for DSM-IV cannabis dependence criterion counts in a sample of European-American (EA) and African-American (AA) descent.
Table 1.
Summary of existing genomewide association studies of cannabis-related phenotypes
| Author, Date | Phenotype | N | Genomewide significant SNPs | Gene-based significance | Heritability |
|---|---|---|---|---|---|
| Agrawal, 2011; PMC3117436 | DSM-IV cannabis dependent | 708 cases, 2346 controls (exposed) | none | - | - |
| Verweij, 2013; PMC3548058 | Cannabis Use | 10,091 | none | none | 6% (p=.28) |
| Agrawal, 2014; PMC3943464 | Factor score of DSM5 criteria | 3,053 | none | C17orf58, BPTF and PPM1D | 21% (p=.13) |
| Minica, 2015; PMC4561059 | Cannabis use Age at initiation |
6,774 | none | none | 25% (p=.002) |
| Sherva, 2016; PMID27028160 | Cannabis dependence symptom count | 14,754 | rs143244591 (chr3:149296148; RP11-206M11.7; p=4.3E-10); rs146091982 (chr10:93900201; SLC35G; p=1.3E-9) rs77378271 (chr8:3215967; CSMD1;p=2.2E-8); |
- | - |
| Stringer, 2016; PMID27023175 | Cannabis use | 32,330 + 5,627 | none | NCAM1, CADM2, SCOC and KCNT2. | 13–20% (p<.001) |
We conducted a meta-analysis of GWAS data on individuals of European descent from five cohorts to identify loci associated with DSM-IV cannabis dependence (N=2,080). We compared individuals who met criteria for DSM-IV cannabis dependence (N=2,080) to controls who did not meet criteria for cannabis dependence but reported having used cannabis, at least once, during their lives (N=6,435). In addition to comprehensive locus (including epigenetic) annotation, we examined whether genomewide significant SNPs were associated with variability in gray matter volume within brain regions (bilateral amygdala, ventral striatum and hippocampus) previously associated with chronic cannabis use and misuse (14;15) among an independent cohort of 430 EA college students. Some prior studies have reported lower gray matter volume in these brain regions, although results are inconclusive. While a majority of studies have attributed such volumetric changes to the effects of chronic cannabis exposure (e.g.,(16)), at least one study has implicated common predisposing influences, such as genetic liability, as the major contributor to the association between casual cannabis use and variability in amygdala volume (17). As this sample of college students included <10 individuals who met criteria for cannabis dependence, we were principally interested in examining whether the top loci that emerged from the GWAS were associated with volumetric differences, whether regional brain volume varied across cannabis users and nonusers and further, whether the effects of top loci on cannabis involvement could be partly attributed to variability in brain volume.
MATERIALS AND METHODS
Samples
Data were drawn from 5 cohorts: (a) a case-control (18) and (b) family GWAS (19;20) component of the Collaborative Study on the Genetics of Alcoholism (COGA; COGA-cc and COGA-f), (c) the Study of Addictions: Genes and Environment (SAGE)(21), (d) the Australian Alcohol (22), Nicotine Addiction Genetics (23), and Childhood Trauma (24) studies (OZALC+) and (e) the Comorbidity and Trauma Study (CATS)(25). Individual studies have been described in detail in related publications and in Supplemental Text. An outline of the samples used in this study is available in Table 2. As the overwhelming majority of the data were on individuals of European-Australian and European-American descent, discovery analyses were restricted to individuals of European descent. All subjects provided informed consent and protocols were approved by the institutional review boards overseeing the individual studies (see Supplemental text).
Table 2.
Sample characteristics of discovery cohorts of European-American and European-Australian (EA) individuals included in meta-analysis (CATS, COGA-cc, COGA-f, OZALC+, SAGE), replication cohort (Yale-Penn) and neuroimaging extension (DNS) samples. Only individuals with a lifetime history of ever using cannabis are included.
| Study | Ncase | Ncontrols | Median age | % Male | % Alcohol dependent | % Nicotine dependent | % Cocaine dependent | % Opioid dependent |
|---|---|---|---|---|---|---|---|---|
| Discovery Samples | ||||||||
| CATS | 799 | 813 | 36 | 57.5 % | 38.8 % | 60.2 % | 24.8 % | 76.1 % |
| COGA-cc | 311 | 593 | 40 | 60.1 % | 79.4 % | 49.0 % | 34.4 % | 13.3 % |
| COGA-f | 368 | 894 | 36 | 50.6 % | 47.0 % | 40.3 % | 13.9 % | 5.7 % |
| OZALC | 357 | 3094 | 43 | 51.7 % | 40.2 % | 50.7 % | 0.4 % | 0.5 % |
| SAGE | 245 | 1041 | 38 | 46.4 % | 55.0 % | 53.4 % | 25.2 % | 9.3 % |
| Replication Samples | ||||||||
| Yale-Penn EA | 781 | 1591 | 38 | 57.7 % | 74.2% | 77.3 % | 78.9 % | 62.0 % |
| Yale-Penn AA | 896 | 1905 | 42 | 54.6 % | 59.9 % | 57.5 % | 75.6 % | 21.2 % |
| Neuroimaging Sample | ||||||||
| DNS | - | - | 19 | 46.7% | 6.3% | 0% | 0% | 0% |
CATS: Comorbidity and Trauma Study; COGA-cc: Case-control component of the Collaborative Study of the Genetics of Alcoholism, COGA-f: family-based component of the Collaborative Study of the Genetics of Alcoholism; OZALC+: Australian alcohol, nicotine and trauma studies; SAGE: Study of Addictions: Genes and Environment; DNS: Duke Neurogenetics Study.
Summary statistics from European ancestry subjects in CATS, COGA-ccGWAS, COGA-fGWAS, OZALC+ and SAGE were combined to form the discovery analysis. Replication analyses were conducted in the Yale-Penn (8) sample which was the major dataset contributing to the prior study by Sherva et al (8). Yale-Penn includes a large number of AA participants; thus, results from both EA and AA subjects were separately examined. Sherva et al also included SAGE data in their discovery cohort and used CATS as a replication sample. In our analyses, only the Yale-Penn component of Sherva et al (8) was used for replication, while SAGE and CATS were part of the discovery cohort.
Genotyping
A variety of Illumina platforms were used to genotype the cohorts (Table S1). Quality control and imputation metrics (26–29) for the individual samples are provided in referenced publications (18;19;21;22;25) and in Table S1.
Phenotype
Cases met criteria for DSM-IV cannabis dependence(30), which included withdrawal (i.e., 3 or more of 7 criteria) in COGA and SAGE but not in CATS or OZALC+. Controls did not meet criteria for cannabis dependence but reported a lifetime history of ever having used cannabis, even once. Follow-up analyses of top loci examined whether excluding those with DSM-IV cannabis abuse or 1–2 dependence criteria modified the results. A natural log-transformed (to account for skewed data) count of DSM-IV dependence criteria (0–6, excluding withdrawal; adding “1” for 0 values) was also analyzed (n=8,050). Finally, the effect of comorbid DSM-IV alcohol, nicotine and opioid dependence was investigated by examining their association with top loci in post hoc analyses.
Statistical analysis
Each sample was analyzed separately using specific analytic protocols that have been validated for that sample (18;22;25;31;32). Prior to meta-analysis, SNPs that did not satisfy quality control standards imposed for the current study were excluded (see Supplemental Text); only SNPs that survived quality control in all 5 samples were included in the meta-analysis. PLINK (v1.07)(33) was used to analyze allele dosage data for SAGE, CATS and COGA-cc. GWAF-GEE (34) was used to analyze the family data for DSM-IV cannabis dependence from COGA-f and OZALC+. Linear mixed models and Merlin-offline(35) were used to analyze criterion counts in COGA-f and OZALC+ respectively. Logistic and linear regressions were used for the diagnosis and count definitions, respectively (see Table S1 for covariates used for each sample). Results were meta-analyzed in METAL (36) using inverse variance weighting procedures and genomic control correction. Gene-based association analyses were conducted using MAGMA (37) with the 1000 Genomes European data (release version 3, May 22, 2014) as the reference panel. Gene boundaries were extended to include a 10kb window at the 3′ and 5′ ends.
Annotation
Top SNPs (p<5E-8) were annotated using a variety of resources that are described in Supplemental Text.
Replication
Replication analyses were conducted in the Yale-Penn study (described in Supplemental Text, related publications (13;38) and Table S1). Cases met criteria for DSM-IV cannabis dependence (NEA=781, NAA=896) and controls (NEA=1,591, NAA=1,905) reported a lifetime history of cannabis use.
Neuroimaging extension
Data on 430 EA college students aged 18–22 years were drawn from the Duke Neurogenetics Study (DNS (39); Supplemental Text). First, we examined the association between genotype (rs1409568, modeled as C-allele carriers vs. T allele homozygotes) and (a) cannabis use (ever used and frequency of use in ever users) and (b) regional brain volume. A Generalized Linear Model in SPM8 was used to test whether genotype predicted regional volume within 6 brain regions (i.e., left and right amygdala, hippocampus, striatum) previously associated with cannabis use and misuse (14;15). Familywise error correction (FWE p < .05) with a 10-voxel extent cluster threshold was applied to each of these 6 anatomical regions of interest (ROIs) derived from the Automated Anatomical Labeling atlas (40) within Wake Forest University Pick Atlas software (41). Additional methodological details are presented in Supplemental Text. Second, we tested whether cannabis use (ever used; frequency of use in ever users) was associated with regional gray matter volume in any of these regions. Third, we examined whether associations between genotype and regional brain volume persisted after controlling for cannabis use. All DNS analyses controlled for sex and age; analyses on regional brain volume also controlled for total intracranial volume (ICV), while analyses including genotype additionally included the first three principal components of ancestry. All non-imaging analyses and group comparisons were conducted using the R (3.1.2) ‘Stats’ package.
RESULTS
Sample characteristics
Samples were relatively similar in age and gender distribution. By ascertainment design, there was considerable overrepresentation of all forms of substance use disorder across the samples (Table 2), with the exception of OZALC+ which included families that were ascertained based on family size rather than substance-related problems.
GWAS results
DSM-IV cannabis dependence
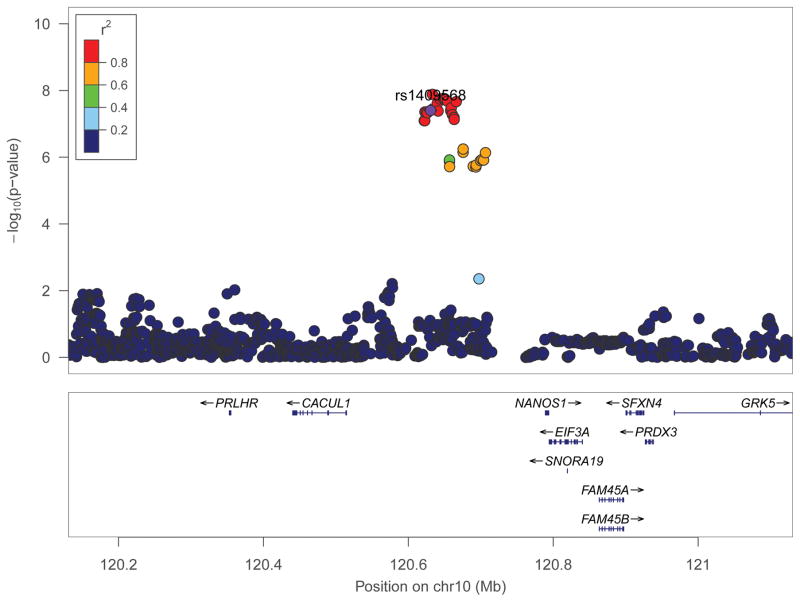
Lambdas for individual studies and meta-analyses were close to 1.0 (Supplemental Table S1; Figure S1A). Genomewide significant loci did not emerge in any individual study. Meta-analysis of summary statistics from the 5 discovery samples (CATS, COGA-cc, SAGE, COGA-f, OZALC) revealed a cluster of genomewide significant SNPs in a region on chromosome 10 (Table 3 for loci at p-value < 10−6; Supplemental S2A for Manhattan plot; full results available upon request), with genomewide significant loci representing a single signal (Figure 1A for regional association plot (42)). The lowest p-value was associated with rs77300175 (p-value = 1.3E-8; Table 3), with stronger contributions from the 3 case-control cohorts (SAGE, CATS, COGA-cc; Supplemental Table S2) than the family-based cohorts (COGA-f and OZALC+).
Table 3.
Association results for SNPs at p-value ≤ 1 × 10−6 in 2,080 cannabis dependent cases and 6,435 cannabis-exposed controls of European-American descent.
| SNP | Chr: position | Function | Effect Allele | Alternate Allele | Meta-analysis
|
Direction of effects** | ||
|---|---|---|---|---|---|---|---|---|
| Effect size (β) | SE | P-value | ||||||
| rs112825709 | 10:120622014 | intergenic | A* | C | 0.50 | 0.09 | 8.04E-08 | +++++ |
| rs151284751 | 10:120622746 | intergenic | A* | C | 0.50 | 0.09 | 8.06E-08 | +++++ |
| rs145575521 | 10:120622907 | intergenic | T | C | −0.51 | 0.09 | 4.39E-08 | − − − − − |
| rs79516280 | 10:120624808 | intergenic | A | G | −0.51 | 0.09 | 4.34E-08 | −−−−− |
| rs75312482 | 10:120626227 | intergenic | T* | C | 0.51 | 0.09 | 4.66E-08 | +++++ |
| rs1409568g | 10:120630785 | intergenic | T | C | −0.50 | 0.09 | 3.95E-08 | −−−−− |
| rs77300175 | 10:120633376 | intergenic | T* | C | 0.53 | 0.09 | 1.30E-08 | +++++ |
| rs7098706g | 10:120639977 | intergenic | T | C | −0.52 | 0.09 | 2.44E-08 | −−−−− |
| rs118006754 | 10:120641184 | intergenic | T* | G | 0.51 | 0.09 | 4.12E-08 | +++++ |
| rs7074123 | 10:120643763 | intergenic | A | C | −0.52 | 0.09 | 1.79E-08 | −−−−− |
| rs7920901 | 10:120648450 | intergenic | T* | C | 0.52 | 0.09 | 1.74E-08 | +++++ |
| rs57602752 | 10:120649972 | intergenic | A | C | −0.52 | 0.09 | 1.88E-08 | −−−−− |
| rs115048844 | 10:120651442 | intergenic | C* | G | 0.52 | 0.09 | 1.86E-08 | +++++ |
| rs1961317 | 10:120654022 | intergenic | T* | C | 0.52 | 0.09 | 2.08E-08 | +++++ |
| rs147702664 | 10:120658617 | intergenic | A | G | −0.57 | 0.10 | 4.07E-08 | −−−−− |
| rs149791363 | 10:120658646 | intergenic | A | C | −0.58 | 0.11 | 3.76E-08 | −−−−− |
| rs150525973 | 10:120659352 | intergenic | T* | C | 0.54 | 0.10 | 3.27E-08 | +++++ |
| rs79277226 | 10:120660716 | intergenic | A* | G | 0.49 | 0.09 | 5.23E-08 | +++++ |
| rs113036365 | 10:120663067 | intergenic | T* | G | 0.48 | 0.09 | 6.37E-08 | +++++ |
| rs60120125 | 10:120663137 | intergenic | T | C | −0.48 | 0.09 | 6.40E-08 | −−−−− |
| rs61538293 | 10:120663338 | intergenic | C | G | −0.49 | 0.09 | 7.34E-08 | −−−−− |
| rs111332403 | 10:120666212 | intergenic | A | G | −0.49 | 0.09 | 2.15E-08 | −−−−− |
| rs12771281 | 10:120675667 | intergenic | C | G | −0.41 | 0.08 | 7.11E-07 | −−−−− |
| rs12413263 | 10:120675738 | intergenic | A | C | −0.40 | 0.08 | 5.67E-07 | −−−−− |
| rs35728709 | 10:120706542 | intergenic | T* | C | 0.41 | 0.08 | 7.30E-07 | +++++ |
indicates that the effect allele is also the minor allele in individuals of European descent.
Order of effect sizes from studies is CATS, COGA-cc, COGA-fGWAS, OZALC+, and SAGE
SNP genotyped in at least one sample. All other SNPs were imputed across samples.
Figure 1.
Regional association plot of chromosome 10 SNPs (centered at rs1409568 ± 500kb) associated with cannabis dependence cases-status (N=2,080) compared with cannabis exposed controls (N=6,435).
Cannabis dependence criterion count
There was no evidence for genomewide significant loci associated with cannabis dependence symptom counts (Supplemental Table S3; Supplemental Figures S1B and S2B). The most promising association was noted for a cluster of SNPs in chromosome 2 (e.g. rs2287641, p=9E-7). The chromosome 10 SNPs were similarly associated, but not at genomewide significant levels (e.g. rs150525973 P = 1.2E-6).
Replication
For the DSM-IV dependence diagnosis, findings were not replicated in Yale-Penn EA participants (Supplemental Table S4); effect sizes were consistently in the same direction, but smaller (e.g. rs1409568: β = −0.072, p=0.6). Consistent with our finding, the T allele of rs1409568 was associated with a reduced likelihood of cannabis dependence among the AA participants from Yale-Penn (β = −0.18, p = 0.028). When results from all datasets, discovery and replication (EA and AA), were meta-analyzed together (Ncase=3,757, Ncontrol=9,931), rs1409568 remained associated with DSM-IV cannabis dependence at a trend level (β = −0.28; p = 2.9E-7). In addition, there was no evidence from our meta-analysis for association between cannabis dependence diagnosis or symptom counts with previously identified loci for cannabis use (i.e., top 10 signals from Stringer et al(13)) and top EA locus from Sherva et al (8) – Supplemental Table S5).
Gene-based association
There was no evidence for enrichment of association within genes for cannabis dependence diagnosis. (Supplemental Table S6); However, for symptom count, MEI1, on chromosome22, was associated at a gene-level (p=2.55E-6; Supplemental Table S7). Several other genes with SNPs of nominal significance clustered in this chromosomal region (Figure S3 for chromosome 22 regional association plot).
Genomic and Epigenomic Annotation
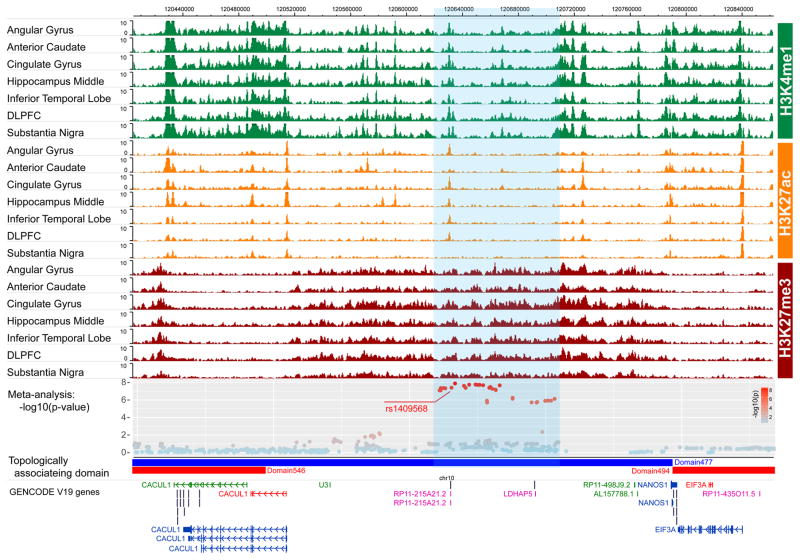
Genomewide significant SNPs on chromosome 10 were not in linkage disequilibrium (r2 ≥ 0.6) with non-synonymous variants in neighboring genes. No significant cis-eQTLs were identified for any chromosome 10 variant in any tissue in GTEx (43) as well as dorsolateral prefrontal cortex (dlPFC) tissue from the Common Mind Consortium data(44). However, there was preliminary evidence that rs1409568 (RegulomeDB score 3a), but not other variants in the region (scores ≥ 5) may have regulatory effects (45). Closer inspection in the Epigenome Browser (46) showed that rs1409568 was accompanied by enhancer-enriched active histone modifications (H3K4me1 and H3K27ac) in a variety of brain tissues (Figure 2). Evidence of an active enhancer was particularly prominent in the dorsolateral prefrontal cortex (dlPFC), angular gyrus, cingulate gyrus and the inferior temporal lobe. There were also enriched H3K4me1 and H3K27ac signals in the middle hippocampus and the substantia nigra, however these signals were not detected at corrected thresholds defined by MACS (q-value cutoff 0.05). All of these regions are strongly implicated in the etiology of addiction (47).
Figure 2.
Epigenetic annotation of rs1409568 on chromosome 10 depicting preliminary in silico evidence for an active enhancer mark.
We determined that rs1409568 was within a chromosome 10 regulatory domain spanning 120,300,000bp – 120,790,000bp that encompassed all of the genome-wide significant SNPs. The regulatory domain included 12 genes, including 3 protein coding genes (PRLHR, CACUL1, NANOS1), 4 pseudogenes (SLC25A18P1, TOMM22P5, RP11-215A21.2, LDHAP5), and 5 non-coding genes (AL356865.1, AL356865.2, U3, RP11-498J9.2, AL15778.1). Seven of 12 genes were expressed in several brain-derived tissues (Supplementary Figure S4). RP11-215A21.1 is the gene closest to rs1409568 (1.3kb from the transcription start site), however, there is no evidence that rs1409568 regulates the expression of any gene within the regulatory domain. The T allele of rs1409568 is conserved within primates, but not between primates and rodents (Supplementary Figure S5).
There was also evidence that rs1409568 altered the binding motif for several transcription factors that are critical during embryogenesis, including those encoded by genes that include homeodomains (e.g., HOXD8, VAX1) and those from the Pit-Oct-Unc (POU) family (e.g., POU4F1, POU4F3, POU6F2: for full list, see Supplemental Table S8). Although predictions were based on common tissue sources, several transcription factors showed brain-related expression (e.g., POU6F2).
We identified 26 CpG probes that corresponded to genes with transcription start sites (TSS) within 1 Mb of rs1409568. Differences in CpG methylation were examined in CT (n=34) and TT (n=313) individuals in tissue from the frontal cortex and cerebellum(48). Only one probe (cg23182539), corresponding to TIA-1 related protein isoform 1 (TIAL1) showed nominal support for change in methylation scores as a function of genotype (Supplemental Table S9), with lower methylation scores in C allele carriers (β = −0.56, p = 0.0017; Wilcoxon p = 0.005). However, methylation change in this gene was not significant after Bonferroni correction (26 probes x 2 regions; pcorrected = 0.00096).
Sensitivity to definition of controls
The chromosome 10 SNPs represented a single signal (Supplemental Figure S6), so follow-up analyses used a representative locus. Controls (N=6,435) included individuals who did not meet criteria for DSM-IV cannabis dependence but may have met criteria for a lifetime history of DSM-IV cannabis abuse or endorsed 1–2 dependence criteria. Exclusion of individuals with abuse (N=1,590) from among the controls yielded similar effect sizes but diminished statistical significance, likely due to the reduced statistical power (rs7098706: b = −0.53, p = 5.90E-7; rs1409568: b = −0.50, p = 1.21E-6). Excluding control individuals with abuse or 1–2 dependence criteria (N=2,152) had a similar effect (e.g., rs7098706 b = −.50, p = 1.85 E-6; rs1409568: b = −0.48, p = 3.95E-6). Thus, heterogeneity within the control population is not responsible for the observed association.
Comorbidity with other substance use disorders
Only nicotine dependence was associated with rs1409568, and in CATS alone (p = .003) – adding nicotine dependence as a covariate to the CATS analysis did not greatly alter the significance of rs1409568 (p = 5.51E-8; Supplemental Table S10). Alcohol dependence was not associated with rs1409568 in any individual study, although the meta-analytic p-value was less than 0.05.
Genotype and brain volumetric variation
In the DNS, 51% of the sample reported lifetime cannabis use, with 12% (n=52), 15% (n=66), 8.8% (n=38) and 15% (n=65) using cannabis 1–2, 3–10, 11–20 and >21 times during their lifetime respectively. Ever using cannabis and the frequency of use within lifetime users were not associated with rs1409568 genotype (C-allele carrier vs TT; only 2 individuals with CC genotype). However, the C allele, which was associated with increased likelihood of cannabis dependence in the meta-analysis, was associated with increased gray matter volume in the right hippocampus (2.13% greater than TT individuals; Cohen’s d = 0.62, maximal voxel p-fwe = 0.007; Bonferroni p-value for 6 a priori regions = .008; Supplemental Figure S7A). This association remained unchanged when cannabis use was included as a covariate in the analysis (Cohen’s d = 0.62, maximal voxel p-fwe = 0.008). Other regions previously associated with cannabis use (i.e., left hippocampus and bilateral amygdala and ventral striatum) showed no relationship with the SNP. Finally, ever having used cannabis was associated with increased volume in a cluster in the left hippocampus (3.18% greater in ever versus never users; Cohen’s d = 0.39, maximal voxel p-fwe = 0.002; Supplemental Figure S7B). No significant volumetric differences were observed for the right hippocampus, where the SNP exerted main effects, nor was the cluster in the left hippocampus in the same region as the cluster in the right hippocampus to which rs1409568 was associated. Lastly, rs1409568 was not associated with hippocampal volume in an independent large meta-analysis (p=0.33; N=12, 516)(49).
DISCUSSION
This study identified a genomewide significant locus on chromosome 10 for cannabis dependence diagnosis in subjects of European descent. To date, only one other (Table 1) study (8) identified genomewide significant loci for cannabis dependence criterion count. The novel locus identified in the present study included a representative SNP, rs1409568, which showed modest evidence for replication in the AA, but not EA, participants from the independent Yale-Penn sample that was part of the only other study with genomewide significant SNPs. The lack of replication in the EA component of Yale-Penn may reflect lower power (i.e., fewer cases than the AA component, or higher minor allele frequency in AA than EA) or ascertainment differences. It is also noteworthy that patterns of LD for the SNPs in Table 3 differ across CEU and ASW populations (based on 1000 Genomes data; Supplemental Figure S8)(50); replication that was noted in the AAs was present in spite of these differences. Nonetheless, associations in the Yale-Penn EA participants were in the same direction as the current meta-analysis.
The genomewide significant chromosome 10 SNPs represent a single LD signal, and are located in a region that is primarily intergenic. However, based on GENCODEv19 (51) annotation, there are multiple genes within the regulatory domain spanning these SNPs. While 5 of these 12 genes are expressed in brain-derived tissues (Supplemental Figure S4), none of the genomewide significant SNPs served as eQTLs for expression of these genes in GTEx, which includes modestly sized samples for a variety of brain tissue, nor in the larger Common Mind Consortium data, which includes 279 dlPFC samples. We found no evidence in the literature for the role of the genes within the regulatory domain in the etiology of addiction-related or other behavioral phenotypes.
One genomewide significant SNP, rs1409568, appears to be located within an active enhancer (52). This finding is consistent with a recent study that reported modest enrichment of H3K27ac marks for a variety of complex traits (e.g., Crohn’s disease)(53). Importantly, there is growing evidence that intergenic genomewide significant loci are disproportionately overrepresented in regulatory regions, such as enhancers (54–56). For example, functional partitioning of SNP-attributable heritability for 11 complex traits found that DNase1 hypersensitivity sites were 1.6- and 5.1-fold enriched in genotyped and imputed data respectively, with enhancers being the most common subcategory, representing 31.7% of total SNP heritability and 9.8-fold enrichment (54).
Importantly, rs1409568 is predicted to bear active enhancer marks in several brain-derived tissues that are critical to addiction, most notably the dlPFC and the cingulate and angular gyri, which play a major role in the development of addictive behaviors, particularly in the regulation of executive control and attentional bias (57). These in-silico findings imply that the C allele is associated with reduced or no binding of several homeodomain-containing (58) developmentally relevant transcription factors, with some difference scores (e.g., POU6F2) being substantial (>8.0). These genes and their products have been variously implicated in embryogenesis and in cell-type specific pathways of differentiation, particularly in visual systems (59–61), but have not been related to behavioral traits thus far.
There was also nominal evidence that rs1409568 genotype was associated with changes in CpG methylation of TIAL1. C allele carriers, on average, had lower methylation scores than T homozygotes. There is no published evidence for a role of the RNA-binding protein encoded by this gene in addictive processes.
In an independent sample, the C allele of rs1409568 was also associated with a modest increase in right hippocampal volume (2.13%) but not with cannabis use itself. The hippocampus has been implicated in addiction (47), including volumetric differences that have been observed in chronic cannabis users (14;15). This, in addition to tentative evidence for the role of rs1409568 as a potential enhancer in the middle hippocampus (Figure 2), indicates that this SNP may regulate neural effects that are central to the development of addictions. The lack of association between cannabis use and genotype is not surprising given the vanishingly low number of problem users (e.g., 12 individuals with cannabis abuse) in the DNS sample.
Cannabis use itself was associated with a modest increase (3.18%) in left (but not right) hippocampal volume. This finding contradicts prior studies that have linked chronic, but not occasional, cannabis use to decreases (not increases) in hippocampal volume. We speculate that the association between cannabis use and increased hippocampal volume may be due to the nature of DNS, which includes casual, non-problem users who are also likely enriched for other factors that might protect against progression to problem use (and against hippocampal deficits). In support of this, we found that cannabis users in DNS were more likely to represent higher socioeconomic status (t=3.70, p<0.001) and even showed modest increases in digit-span performance (t=2.50, p =.013), an index of working memory suggesting that cannabis users in DNS may be characterized by adaptive factors that protect them from progression to problem use. Therefore, if previously documented associations between cannabis use and smaller hippocampal volumes are a consequence of chronic exposure to cannabis, then we would not expect to see these reductions in the DNS.
The minor allele of rs1409568, which was more common in cannabis dependent cases in the meta-analysis, was associated with increased hippocampal volume. This finding is also inconsistent with the hypothesis that liability to heavy cannabis use should relate to decreased hippocampal volume. There are, at least, two plausible explanations for our observation of the opposite association. First, it is possible that the association between rs1409568 and hippocampal volume is independent of its association with cannabis dependence in the meta-analysis. While such a pleiotropic effect adds encouraging evidence favoring a role of rs1409568 in neural regions typically associated with addiction, and augments its functional plausibility, it does not help reconcile the mechanism by which rs1409568 might influence liability to cannabis dependence. Second, the association between rs1409568 and hippocampal volume did not replicate in the large ENIGMA meta-analysis. This raises the possibility that the association is a false positive in DNS and suggests that caution is warranted in its interpretation.
It is also noteworthy that the current study did not replicate previously noted associations for cannabis use (13) or dependence (8). These are not unexpected. For cannabis use, our sample excluded individuals who had never used cannabis, thus limiting our ability to detect loci associated with initiation of cannabis involvement. Our lack of replication of one prior locus identified for cannabis dependence in EAs (rs77378271) might further underscore differences between our European samples and those comprising Yale-Penn. A full meta-analysis of these datasets might yield additional novel loci.
While no single SNP was genomewide significant for the count of DSM criteria, gene-level testing identified MEI1 (meiotic double-stranded break formation protein 1). Relative to other tissues, MEI1 is more robustly expressed in the testes and variants in the gene have been associated with azoospermia due to early and complete meiotic arrest (62). In parallel, there is compelling epidemiological and biological support for the relationship between prolonged/heavy cannabis use and male reproductive health, including fertility. Weekly cannabis use has been associated with a 28–29% reduction in sperm concentration and count (63). The endocannabinoid system actively participates in the regulation of male fertility (64), including by promoting meiosis via CB2 activation (65). Therefore, the possibility of shared genetic pathways to male fertility and heavy cannabis use might provide a plausible alternative to more causal explanations. However, we are not aware of any prior studies that link MEI1 to cannabis use or addiction.
Some limitations are noteworthy. First, despite aggregating across several large datasets, our meta-analytic sample was relatively underpowered to detect small effects and also, for analyses that would allow us to estimate genetic correlations between cannabis dependence and other traits (e.g. cigarettes per day (68) for which genomewide summary statistics are available. Such calculations typically rely on unrelated cases and controls and our study included two samples with complex pedigree structures. Second, we did not have adequate numbers of AA participants for a full examination of loci identified in Sherva et al (8). In EAs, the only SNP associated at genomewide significant levels in Sherva et al was rs77378271 (CSMD1). In the current study, rs77378271 shows some evidence for independent association with cannabis dependence in COGA-cc (p=5.3E-3); however, the meta-analytic p-value was not significant, with indication of heterogeneity across the samples included in the present meta-analysis. We anticipate that additional data on cannabis dependence in both EA and AA participants will be available in the future. Finally, the minor allele frequency for rs1409568 (and related genomewide significant SNPs) was <10% across cases and controls from each sample.
We identified a new genomewide significant locus on chromosome 10 that was associated with vulnerability to cannabis dependence in European ancestry individuals. One of the representative SNPs, rs1409568, showed promising epigenetic evidence and might also contribute to variation in hippocampal volume, which has been related to risk for and resilience to psychiatric disorders, including addictions. Replication, however, was limited to a subset of AA, but not EA, individuals and analyses in the DNS contradicted prior findings for hippocampal volume and did not extend to a broader meta-analysis of hippocampal volume. Therefore, the identification of this chromosome 10 locus should be viewed as preliminary. Future work that aggregates additional cannabis dependent cases and controls, would allow for the detection of smaller effect sizes and a more thorough investigation of comparability of loci across population groups. This is critical, as genomic research into cannabis involvement has lagged behind that of other drugs, despite the pressing public health significance of the problem. Continuing to identify risk factors, both genetic and environmental, that are associated with cannabis dependence is a public health priority, as understanding the genetic etiology of cannabis use disorders can ultimately help to identify individuals who are at greatest risk of the disorders and enhance efforts aimed at prevention and personalizing pharmacotherapy among affected individuals.
Supplementary Material
Acknowledgments
AA acknowledges support from the National Institute on Drug Abuse (NIDA) for the core research pertaining to this study, via K02DA032573 and R01DA023668. CEC received support from the National Science Foundation (DGE-1143954) and the Mr. and Mrs. Spencer T. Olin Fellowship Program. DAAB acknowledges NIMH (T32-GM008151) and NSF (DGE-1143954). JLM acknowledges K01DA037914. LD is supported by an Australian National Health and Medical Research Council (NHMRC) Principal Research Fellowship (#1041472). LJB acknowledges R01DA036583. RB receives additional support from the National Institutes of Health (R01-AG045231; R01-HD083614; U01-AG052564).
COGA: The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); Department of Biomedical and Health Informatics, The Children’s Hospital of Philadelphia; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); J. McClintick, L. Wetherill, X. Xuei, Y. Liu, D. Lai, S. O’Connor, M. Plawecki, S. Lourens (Indiana University); G. Chan (University of Iowa; University of Connecticut); J. Meyers, D. Chorlian, C. Kamarajan, A. Pandey, J. Zhang (SUNY Downstate); J.-C. Wang, M. Kapoor, S. Bertelsen (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon, S. Saccone (Washington University); J. Salvatore, F. Aliev, B. Cho (Virginia Commonwealth University); and Mark Kos (University of Texas Rio Grande Valley). A. Parsian and M. Reilly are the NIAAA Staff Collaborators.
Funding support for GWAS genotyping performed at the Johns Hopkins University Center for Inherited Disease Research was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). GWAS genotyping was also performed at the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine which is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. COGA-f genotypic data are available via dbGaP: phs000763.v1.p1 and COGA-c genotypic data are available via phs000125.v1.p1
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
CATS (dbGaP: phs000277.v1.p1): Funding support for the Comorbidity and Trauma Study (CATS) was provided by the National Institute on Drug Abuse (R01 DA17305); GWAS genotyping services at the Center for Inherited Disease Research (CIDR) at The Johns Hopkins University were supported by the National Institutes of Health [contract N01-HG-65403]. The National Drug and Alcohol Research Centre at the University of New South Wales is supported by funding from the Australian Government under the Substance Misuse Prevention and Service Improvements Grants Fund.
SAGE (dbGaP: phs000092.v1.p1): Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease”(HHSN268200782096C).
OZALC+ (dbGaP: phs000181.v1.p1): Supported by NIH grants AA07535, AA07728, AA13320, AA13321, AA14041, AA11998, AA17688, DA012854, DA019951; by grants from the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498); by grants from the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921); and by the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). GWAS genotyping at CIDR was supported by a grant to the late Richard Todd, PhD, MD, former PI of grant AA13320 and a key contributor to research described in this manuscript. Project 7 data collection was also supported by AA011998_5978.
Yale-Penn: This study was supported by grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535 from the National Institutes of Health (NIH), the Veterans Affairs Connecticut Healthcare Center, and the Philadelphia Veterans Affairs Mental Illness Research, Education and Clinical Center. A portion of the data are available via dbGaP (phs000277.v1.p1).
DNS: The Duke Neurogenetics Study is supported by Duke University and National Institute on Drug Abuse grant DA03369.
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\SAIC-Frederick, Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplements to University of Miami grants DA006227 & DA033684 and to contract N01MH000028. Statistical Methods development grants were made to the University of Geneva (MH090941 & MH101814), the University of Chicago (MH090951, MH090937, MH101820, MH101825), the University of North Carolina - Chapel Hill (MH090936 & MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University St Louis (MH101810), and the University of Pennsylvania (MH101822). The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 09/24/2016.
Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories and the NIMH Human Brain Collection Core. CMC Leadership: Pamela Sklar, Joseph Buxbaum (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Keisuke Hirai, Hiroyoshi Toyoshiba (Takeda Pharmaceuticals Company Limited), Enrico Domenici, Laurent Essioux (F. Hoffman-La Roche Ltd), Lara Mangravite, Mette Peters (Sage Bionetworks), Thomas Lehner, Barbara Lipska (NIMH).
Expression and covariate data for the methylation eQTL analysis was derived from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15745. Genotypes were derived via authorized access to MK, JCW & AG from dbGaP (phs000249.v2.p1). Funding support for the “Brain eQTL (expression data) Study” was provided through the Division of Aging Biology and the Division of Geriatrics and Clinical Gerontology, NIA. The Brain eQTL (expression data) Study includes a genome-wide association study funded as part of the Intramural Research Program, NIA. Funding sources: Z01 AG000949-02 and Z01 AG000015-49.
Footnotes
DISCLOSURE/COI
We disclose that Drs. LJ Bierut, JP Rice, J-C Wang and AM Goate are listed as inventors on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Dr. Kranzler has been a consultant, advisory board member, or CME speaker for Lundbeck, and Indivior. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative (ACTIVE), which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor and Amygdala Neurosciences. Dr. Nurnberger is an investigator for Assurex and a consultant for Janssen.
Reference List
- 1.United Nations Office on Drugs and Crime. World Drug Report 2015. Vienna, Austria: United Nations; 2015. Report No.: Sales No. E.15.XI.6. [Google Scholar]
- 2.Degenhardt L, Ferrari AJ, Calabria B, Hall WD, Norman RE, McGrath J, et al. The global epidemiology and contribution of cannabis use and dependence to the global burden of disease: results from the GBD 2010 study. PLoS ONE. 2013 Oct 24;8(10):e76635. doi: 10.1371/journal.pone.0076635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CY, O’Brien MS, Anthony JC. Who becomes cannabis dependent soon after onset of use? Epidemiological evidence from the United States: 2000–2001. Drug Alcohol Depend. 2005 Jul;79(1):11–22. doi: 10.1016/j.drugalcdep.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Coffey C, Carlin JB, Degenhardt L, Lynskey M, Sanci L, Patton GC. Cannabis dependence in young adults: an Australian population study. Addiction. 2002 Feb;97(2):187–94. doi: 10.1046/j.1360-0443.2002.00029.x. [DOI] [PubMed] [Google Scholar]
- 5.Lynskey MT, Heath AC, Nelson EC, Bucholz KK, Madden PA, Slutske WS, et al. Genetic and environmental contributions to cannabis dependence in a national young adult twin sample. Psychol Med. 2002 Feb;32(2):195–207. doi: 10.1017/s0033291701005062. [DOI] [PubMed] [Google Scholar]
- 6.Hasin DS, Saha TD, Kerridge BT, Goldstein RB, Chou SP, Zhang H, et al. Prevalence of Marijuana Use Disorders in the United States Between 2001–2002 and 2012–2013. JAMA Psychiatry. 2015 Dec 1;72(12):1235–42. doi: 10.1001/jamapsychiatry.2015.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verweij KJ, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG, et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010 Mar;105(3):417–30. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, et al. Genome-wide Association Study of Cannabis Dependence Severity, Novel Risk Variants, and Shared Genetic Risks. JAMA Psychiatry. 2016 Mar;30:10. doi: 10.1001/jamapsychiatry.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, et al. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol. 2011 Jul;16(3):514–8. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal A, Lynskey MT, Bucholz KK, Kapoor M, Almasy L, Dick DM, et al. DSM-5 cannabis use disorder: a phenotypic and genomic perspective. Drug Alcohol Depend. 2014 Jan 1;134:362–9. doi: 10.1016/j.drugalcdep.2013.11.008. Epub;%2013 Nov 16.:362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verweij KJ, Vinkhuyzen AA, Benyamin B, Lynskey MT, Quaye L, Agrawal A, et al. The genetic aetiology of cannabis use initiation: a meta-analysis of genome-wide association studies and a SNP-based heritability estimation. Addict Biol. 2012 Jul 24; doi: 10.1111/j.1369-1600.2012.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minica CC, Dolan CV, Hottenga JJ, Pool R, Fedko IO, Mbarek H, et al. Heritability, SNP- and Gene-Based Analyses of Cannabis Use Initiation and Age at Onset. Behav Genet. 2015 Sep;45(5):503–13. doi: 10.1007/s10519-015-9723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stringer S, Minica CC, Verweij KJ, Mbarek H, Bernard M, Derringer J, et al. Genome-wide association study of lifetime cannabis use based on a large meta-analytic sample of 32 330 subjects from the International Cannabis Consortium. Transl Psychiatry. 2016;6:e769. doi: 10.1038/tp.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batalla A, Bhattacharyya S, Yucel M, Fusar-Poli P, Crippa JA, Nogue S, et al. Structural and functional imaging studies in chronic cannabis users: a systematic review of adolescent and adult findings. PLoS One. 2013;8(2):e55821. doi: 10.1371/journal.pone.0055821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzetti V, Lubman DI, Whittle S, Solowij N, Yucel M. Structural MRI findings in long-term cannabis users: what do we know? Subst Use Misuse. 2010 Sep;45(11):1787–808. doi: 10.3109/10826084.2010.482443. [DOI] [PubMed] [Google Scholar]
- 16.Gilman JM, Kuster JK, Lee S, Lee MJ, Kim BW, Makris N, et al. Cannabis use is quantitatively associated with nucleus accumbens and amygdala abnormalities in young adult recreational users. J Neurosci. 2014 Apr 16;34(16):5529–38. doi: 10.1523/JNEUROSCI.4745-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagliaccio D, Barch DM, Bogdan R, Wood PK, Lynskey MT, Heath AC, et al. Shared Predisposition in the Association Between Cannabis Use and Subcortical Brain Structure. JAMA Psychiatry. 2015 Oct;72(10):994–1001. doi: 10.1001/jamapsychiatry.2015.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, et al. Genome-Wide Association Study of Alcohol Dependence Implicates a Region on Chromosome 11. Alcohol Clin Exp Res. 2010 Mar 1; doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wetherill L, Agrawal A, Kapoor M, Bertelsen S, Bierut LJ, Brooks A, et al. Association of substance dependence phenotypes in the COGA sample. Addict Biol. 2015 May;20(3):617–27. doi: 10.1111/adb.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. 2013 Nov;18(11):1218–24. doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010 Mar 2;107(11):5082–7. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, et al. A Quantitative-Trait Genome-Wide Association Study of Alcoholism Risk in the Community: Findings and Implications. Biol Psychiatry. 2011 Apr 27; doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, et al. Genetic Linkage to Chromosome 22q12 for a Heavy Smoking Quantitative Trait in Two Independent Samples. Am J Hum Genet. 2007 doi: 10.1086/513703. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristjansson S, McCutcheon VV, Agrawal A, Lynskey MT, Conroy E, Statham DJ, et al. The variance shared across forms of childhood trauma is strongly associated with liability for psychiatric and substance use disorders. Brain Behav. 2016 Feb;6(2):e00432. doi: 10.1002/brb3.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson EC, Agrawal A, Heath AC, Bogdan R, Sherva R, Zhang B, et al. Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry. 2015 Aug;4:10. doi: 10.1038/mp.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009 Jun;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010 Dec;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014 Apr;10(4):e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009 Feb;84(2):210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington, DC: American Psychiatric Association; 1994. Revised ed. [Google Scholar]
- 31.Wetherill L, Agrawal A, Kapoor M, Bertelsen S, Bierut LJ, Brooks A, et al. Association of substance dependence phenotypes in the COGA sample. Addict Biol. 2014 May;16:10. doi: 10.1111/adb.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bierut L, Agrawal A, Bucholz K, Doheny KF, Laurie CC, Pugh EW, et al. A Genome-wide Association Study of Alcohol Dependence. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0911109107. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010 Feb 15;26(4):580–1. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007 Nov;81(5):913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010 Sep 1;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015 Apr;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014 Jan;19(1):41–9. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolova YS, Knodt AR, Radtke SR, Hariri AR. Divergent responses of the amygdala and ventral striatum predict stress-related problem drinking in young adults: possible differential markers of affective and impulsive pathways of risk for alcohol use disorder. Mol Psychiatry. 2016 Mar;21(3):348–56. doi: 10.1038/mp.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002 Jan;15(1):273–89. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 41.Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003 Jul;19(3):1233–9. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- 42.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010 Sep 15;26(18):2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013 Jun;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016 Sep 26; doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012 Sep;22(9):1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, et al. The Human Epigenome Browser at Washington University. Nat Methods. 2011 Dec;8(12):989–90. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010 Jan;35(1):217–38. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010 May;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015 Apr 9;520(7546):224–9. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015 Nov 1;31(21):3555–7. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012 Sep;22(9):1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010 Dec 14;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015 Nov;47(11):1228–35. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjalmsson BJ, Xu H, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014 Nov 6;95(5):535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6(10):85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012 Sep 7;337(6099):1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci. 2011 Nov;12(11):652–69. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991 Dec 3;30(48):11357–67. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- 59.Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, O’Connell SM, et al. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000 Dec;28(3):779–92. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- 60.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002 Feb 22;295(5559):1528–32. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 61.McEvilly RJ, Rosenfeld MG. The role of POU domain proteins in the regulation of mammalian pituitary and nervous system development. Prog Nucleic Acid Res Mol Biol. 1999;63:223–55. doi: 10.1016/s0079-6603(08)60724-2. [DOI] [PubMed] [Google Scholar]
- 62.Sato H, Miyamoto T, Yogev L, Namiki M, Koh E, Hayashi H, et al. Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J Hum Genet. 2006;51(6):533–40. doi: 10.1007/s10038-006-0394-5. [DOI] [PubMed] [Google Scholar]
- 63.Gundersen TD, Jorgensen N, Andersson AM, Bang AK, Nordkap L, Skakkebaek NE, et al. Association Between Use of Marijuana and Male Reproductive Hormones and Semen Quality: A Study Among 1,215 Healthy Young Men. Am J Epidemiol. 2015 Sep 15;182(6):473–81. doi: 10.1093/aje/kwv135. [DOI] [PubMed] [Google Scholar]
- 64.Fasano S, Meccariello R, Cobellis G, Chianese R, Cacciola G, Chioccarelli T, et al. The endocannabinoid system: an ancient signaling involved in the control of male fertility. Ann N Y Acad Sci. 2009 Apr;1163:112–24. doi: 10.1111/j.1749-6632.2009.04437.x. [DOI] [PubMed] [Google Scholar]
- 65.Grimaldi P, Orlando P, Di SS, Lolicato F, Petrosino S, Bisogno T, et al. The endocannabinoid system and pivotal role of the CB2 receptor in mouse spermatogenesis. Proc Natl Acad Sci U S A. 2009 Jul 7;106(27):11131–6. doi: 10.1073/pnas.0812789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szutorisz H, Egervari G, Sperry J, Carter JM, Hurd YL. Cross-generational THC exposure alters the developmental sensitivity of ventral and dorsal striatal gene expression in male and female offspring. Neurotoxicol Teratol. 2016 Nov;58:107–14. doi: 10.1016/j.ntt.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, et al. Genome-Wide DNA Methylation Profiling Reveals Epigenetic Changes in the Rat Nucleus Accumbens Associated With Cross-Generational Effects of Adolescent THC Exposure. Neuropsychopharmacology. 2015 Dec;40(13):2993–3005. doi: 10.1038/npp.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010 May;42(5):441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.