Abstract
Delineating the lineage of neural cells that captures the progressive steps in their specification is fundamental to understanding brain development, organization, and function. Since the earliest days of embryology, lineage questions have been addressed with methods of increasing specificity, capacity, and resolution. Yet, a full realization of individual cell lineages remains challenging for complex systems. A recent explosion of technical advances in genome-editing and single-cell sequencing has enabled lineage analysis in an unprecedented scale, speed, and depth across different species. In this review, we discuss the application of available as well as future genetic labeling techniques for tracking neural lineages in vivo in the mammalian nervous system.
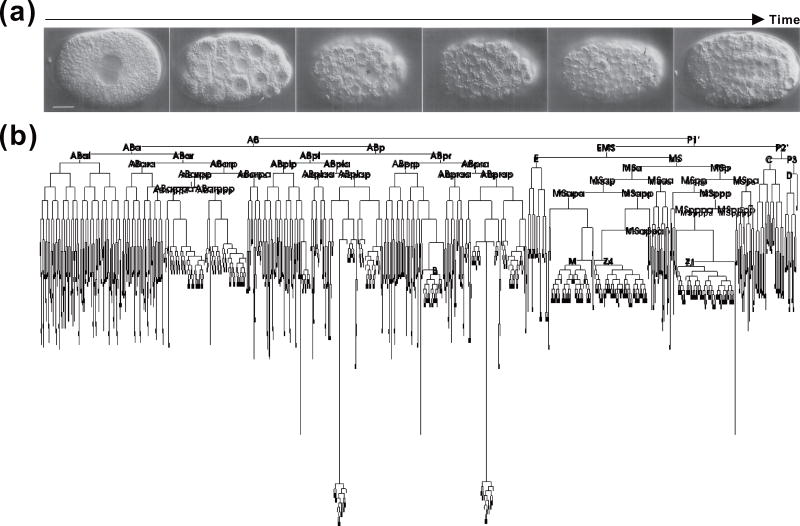
The lineage of a particular cell refers to the history of cellular divisions that sequentially tracks its mother cell up to the original ancestor. Resolving cell lineage relationships of a tissue or organism is important because it provides direct information on how particular cells progressively acquire their final identities. It also helps in the understanding of how gene mutations and environmental factors can perturb the process and result in malformed cells. One of the most successful examples of cell lineage analysis is the full description of the cell lineage tree of C. elegans (Figure 1) [1–3]. Taking advantage of the organism’s transparency, John Sulston and others used Normarski differential interference contrast (DIC) microscopy and traced the complete genealogy of all cells in a nematode by direct visualization and manual drawings. This landmark achievement not only immediately informed the comprehensive lineage relationships between cells, but also served as a fundamental framework instructing subsequent experimental design and data interpretation.
Figure 1. Lineage analysis of C. elegans by direct visualization.
(a) DIC images of the progressively developing nematode embryos (adapted based on Ref. 1). Scale bar: 10 µm. (b) A complete lineage tree of the C. elegans (adapted based on Wikipedia).
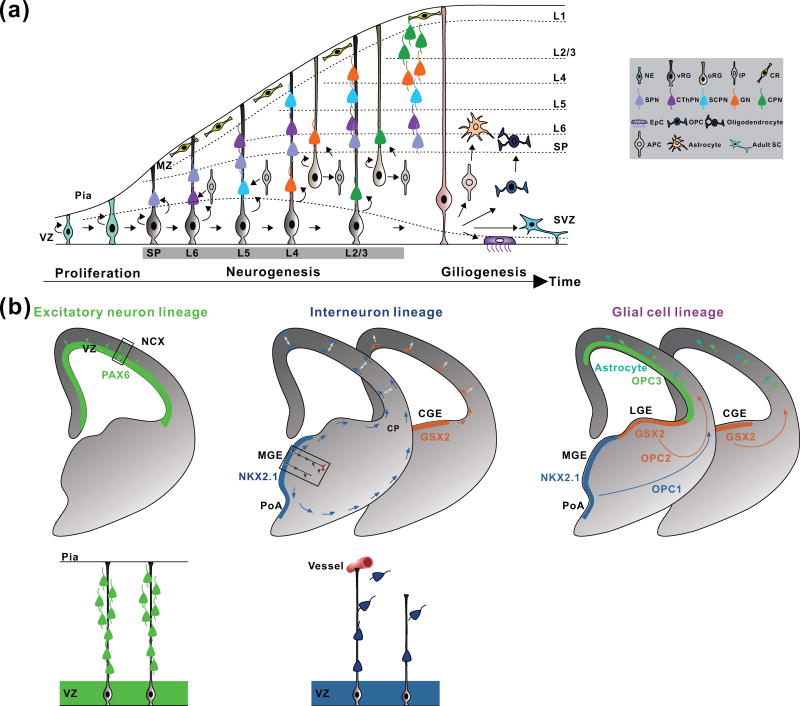
The mammalian nervous system such as the cerebral cortex contains a large number of neural cells that are highly diverse with complex progenitor cell origins (Figure 2). Commanding a complete census of all neural cell types in different brain regions as well as their developmental origins is a prerequisite to understanding their development, organization, and function. Since the ground-breaking work of Ramón y Cajal, neural cells have been increasingly defined; yet, our understanding of neural cell diversity, especially the origin of cells, remains limited. A fundamental source of neural cell diversity is likely rooted in the process of genesis during development. Moreover, the developmental history and lineage relationship may influence the structural and functional organization of neural networks [4–15]. Therefore, it is important to systematically delineate the lineage history and relationship of neural cells in the mammalian nervous system.
Figure 2. Neural lineages in the mammalian cerebral cortex.
(a) A schematic representation of the progressive development of the cerebral cortex. As time progresses, neuroepithelial cells (NEs) proliferate and give rise to radial glial progenitor cells in the ventricular zone (vRGs), which account for the major neural progenitor cells in the developing cortex. vRGs initially undergo symmetric division to amplify the progenitor pools and then undergo asymmetric division to produce distinct cortical neurons (SPNs, subplate neurons; CThPN, cortico-thalamic projection neurons; SCPN, subcerebral projection neurons; GNs, granular neurons; CPN, callosal projection neurons) in a temporally progressive manner via intermediate progenitor cells (IPs) and outer subventricular zone radial glial progenitor cells (oRGs). New-born neurons migrate radially in a birth date-dependent inside-out fashion to constitute the future cortex. Towards the end of neurogenesis, a fraction of RGs proceeds to gliogenesis, generating astrocytes, oligodendrocytes, ependymal cells (EpCs), and adult subventricular zone stem cells (adult SCs). CR, Cajal-Retzius cell; MZ, marginal zone; OPC, oligodendrocyte precursor cell; APC, astrocyte precursor cell; (b) The progenitor cell origins of excitatory principal neurons, inhibitory interneurons, and glial cells in the cerebral cortex. Excitatory neurons are generated by PAX6-expressing RGs in the dorsal telencephalon, migrate radially, and form ontogenetic radial units in the cortex. Inhibitory interneurons are produced by NKX2.1-and GSX2-expressing RGs that often contact periventricular blood vessels in the ventral telencephalon, including the medial ganglionic eminence (MGE), preoptic area (PoA), and caudal ganglionic eminence (CGE), migrate tangentially, and frequently form ontogenetic clusters in the cortex. Oligodendrocytes arise from both the dorsal and ventral telencephalon in different waves, whereas astrocytes are mostly generated by progenitors in the dorsal telencephalon. LGE, lateral ganglionic eminence; CP, cortical plate.
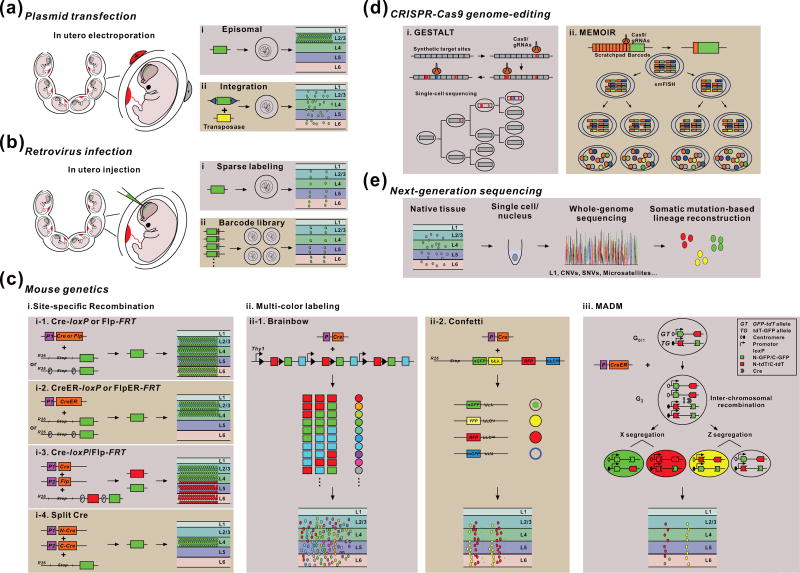
In complex, non-transparent species, mosaic marking of cells is essential for tracking cells of interest. This was initially fulfilled by the development and application of various vital dyes or tracers. While informative, the non-selective nature of these dyes and tracers in labeling cells limits their use. With the development of genetically encoded enzymatic (e.g. β-galactosidase, β-gal) and fluorescent (e.g. green fluorescence protein, GFP) cellular markers, it became theoretically possible to selectively label any ancestor or progenitor cells and directly trace their lineage progression and progeny output. Below we discuss the major genetic labeling technologies that allow lineage analysis in the mammalian nervous system in vivo (Figure 3).
Figure 3. Genetic labeling techniques for in vivo neural lineage tracing in the mammalian nervous system.
(a) Plasmid transfection, such as in utero electroporation, introduces the marker gene into progenitor cells either episomally (i) or integrated into the genome via a transposase (ii) for progeny labeling and lineage tracing. (b) Sparse retrovirus infection introduces the maker gene without (i) or with (ii) the barcodes into the genome of dividing progenitor cells for progeny labeling and lineage tracing. (c) Mouse genetics approaches for engineering the marker gene into the genome of progenitor cells via site-specific recombination (i), multi-color labeling (ii), or MADM (iii) for progeny labeling and lineage tracing. (d) CRISPR-Cas9-based genome-editing such as GESTALT (i) or MEMOIR (ii) introduces traceable genomic edits that are progressively and stably accumulated over cell division for lineage reconstruction. (e) Next-generation single cell whole-genome sequencing allows the detection of somatic mutations such as L1 retrotransposon, CNVs, SNVs, and Microsatellites for lineage reconstruction.
Plasmid transfection for lineage tracing
Conceptually, using genetic markers for lineage tracing is rooted in the classic cell transplantation and chimeric embryo experiments that rely on native genetic differences [16,17]. It is worth noting that cell lineage under transplantation condition can be different from that in native condition due to the changes in cellular context.
The emergence of recombinant DNA technology enabled the introduction of exogenous genetic markers into animals for cell labeling and lineage tracing. In utero electroporation has emerged as an effective technique for transfecting one or more enzymatic or fluorescent protein coding plasmids into neural progenitors in vivo (Figure 3a) [18,19]. By adjusting the direction of electrical pulses and the operation time, it is possible to achieve a degree of spatial (i.e. location) and temporal control of DNA transfection. Target cell specificity can also be implemented by using specific promoters or mouse lines to drive marker gene expression [20].
While the labeling density can be adjusted, in utero electroporation of plasmid often transfects a large number of cells and thereby is not suitable for lineage analysis at the individual progenitor cell (i.e. clonal) level. Another limitation is the nonpermanent nature of marker gene expression. Upon transfection, the marker gene plasmid is carried episomally and passed on to the subsequent daughter cells passively at the completion of cell division. It becomes progressively diluted in progeny at each round of division. As a result, the late-born progeny of a lineage may not be reliably labeled due to the low abundance of the marker gene plasmid. Therefore, plasmid transfection techniques usually do not label the entire lineage.
The transposon system offers a solution to plasmid loss due to cell divisions. A class II transposable element (i.e. DNA sequence) can jump in the genome through a cut-and-paste mechanism in the presence of a transposase protein such as TOL2, Sleeping Beauty, or Piggyback [21]. Thus, it offers a strategy to integrate the exogenous marker gene into the genome of transfected cells. The transposon system is an attractive tool for transgenesis and lineage tracing, especially in non-genetically tractable animals, such as ferrets and primates. Of note, however, random genomic integration can lead to disruptions of endogenous genes at or near the integration site (i.e. mutagenesis) that might confound experimental results. On the other hand, the rate of mutagenesis is reduced by a limited round of transposition events due to the transient expression of the transposase and the heterozygous state of most mutations.
Retrovirus infection for lineage tracing
Retroviruses can be considered as transposable elements and thereby act as an agent of gene transfection to deliver an exogenous marker gene for permanent lineage tracing (Figure 3b) [22,23]. While some retroviruses (e.g. Lentiviruses) infect both dividing and non-dividing cells [24], other retroviruses (e.g. Maloney murine leukemia virus) infect only dividing cells and subsequently allow genomic integration of exogenous DNA into one of the two daughter cells [25,26]. The integrated marker gene is then stably inherited by all the descendants of the labeled cells. The infection time and rate (i.e. density) can be easily adjusted depending on the experimental needs. For example, it is possible to serially dilute the retrovirus to achieve infection of a single or few dividing progenitor cells for lineage analysis at the clonal level. Notably, the broad tropism of retroviruses can also be achieved by envelope pseudotyping. These remarkable features have made retroviruses attractive in studying cell lineages in vivo, particularly in the mammalian nervous system [27,28].
There are, however, several limitations of this method that should be kept in mind. First, retrovirus infection-mediated marker gene genome integration is considered to be mostly random [29], and thus may result in insertional mutagenesis of endogenous genes. The invention of replication-incompetent retroviral vectors limits additional infection and mutagenesis in the host cell genome. Second, a stable expression of the marker gene is required to detect the infected cells. Retroviral vectors can suffer gene silencing that compromises the detection of transfected cells [30]. Retroviral silencing occurs stochastically, in an individual locus-specific manner. It may lead to an underestimation of the overall lineage size and complexity, but should not systematically skew the experimental results. To circumvent this limitation, retroviral vectors can be optimized to reduce the probability of silencing. For example, insulator and locus control region elements can be employed to reduce vector insertion site position effects and epigenetic-mediated silencing [31]. In addition, high-throughput next-generation sequencing may be considered to detect the marker gene, instead of the marker protein, for lineage tracing.
The third main disadvantage of using retroviruses for lineage analysis is that they lack the intrinsic resolution of true clonality (i.e. progeny originated from the same dividing progenitor cells), as all cells are labeled similarly. In a typical clonal analysis experiment, a low titer retrovirus is used to sparsely infect individual dividing progenitor cells. In this way, the labeled progenitors and their progeny (i.e. individual clones) are scattered throughout a tissue or organism and may therefore be distinguished. In other words, spatial segregation of labeled cells is used to infer clonal relationship. However, a degree of uncertainty always remains, especially when cells within a clone can disperse widely. To gain additional resolution in lineage relationship, retroviral libraries harboring a marker gene as well as a short but variable DNA fragment as a barcode tag have been developed [32–38]. The rich repertoire of DNA barcode sequences in principle allows the clonality of labeled cells to be inferred at the single-cell resolution. On the other hand, the accuracy and capacity of using barcoded retroviral libraries for a definitive clonal analysis critically depend on the single representation of individual barcodes in the library and the success of barcode recovery [38,39]. Any overrepresentation of individual barcodes would compromise data interpretation (e.g. two clones being considered as one). Current methods for recovering barcodes through PCR amplification from individual cells dissected from tissue sections also appear to be challenging [34,35,38]. With the rapid advances in next-generation sequencing, it is now possible to access barcodes by high-throughput single cell sequencing.
Sparse retroviral labeling has been extensively used for clonal analysis of the mammalian nervous system, especially in the retina and the cerebral cortex [14,26,40–43]. Sibling cells arising from individual progenitor cells have been found to form prominent radially aligned clusters in the developing retina as well as the cortex, supporting the concept of ontogenetic units [15]. It has also been reported that some clones in the cortex disperse widely [36,44,45]. The exact nature of these dispersed clones remains to be investigated.
Mouse genetics for lineage tracing
Mouse genetic engineering is a powerful means to drive the expression of a marker gene in a cell and temporal specific manner for lineage tracing (Figure 3c). Cell specificity can be achieved by using specific promoters alone in transgenics or in the context of gene recombination mediated by two sequence-specific recombinases, Cre that recognizes a loxP sequence and Flp that recognizes FRT sequence [46–48]. Mice are engineered to express the recombinase under the control of a cell type- or region-specific promoter. These mouse lines are then crossed with a reporter line in which a marker gene, such as β-Gal or GFP, is preceded by a loxP or FRT-flanked transcriptional/translational stop cassette. The conditional marker gene is usually inserted into a ubiquitously expressed locus, such as Rosa26. Upon recombinase-driven recombination, the stop sequence is excised and the marker gene is expressed. Additional temporal control of recombination (e.g. genetic inducible fate mapping, GIFM) can be achieved by using an inducible Cre or Flp system, in which the Cre or Flp recombinase is fused with a mutant form of estrogen receptor (CreER or FlpER) that binds Tamoxifen, but not its endogenous ligands [49]. Upon Tamoxifen binding, Cre recombinase translocates from the cytoplasm to the nucleus and triggers recombination and reporter gene expression. The timing and dose of Tamoxifen administration can be adjusted to label progenitor cells at a defined time point in different densities for lineage analysis.
Neural tissues are exceedingly complex and often demand additional cellular resolution. Intersectional approaches have been developed to achieve greater cell type specificity. The split-Cre system expresses two inactive but complementary Cre fragments [50,51]. Alternatively, Cre and Flp recombinases that recognize distinct sequences can be combined [52–54]. These intersectional approaches not only provide additional cell type specificity but also reduce background labeling due to any leakage in reporter gene expression in the absence of recombination.
More sophisticated multi-color reporter gene designs have also been incorporated by mouse engineering to improve the capacity and resolution of lineage tracing. The Brainbow mice takes advantage of stochastic Cre-mediated recombination and incompatible loxP sites to drive the combinatorial expression of four fluorescent reporter genes, including CFP, GFP, YFP, and RFP, under the control of the Thy1 promoter [55]. Upon Cre activation, cells expressing a particular combination of fluorescent proteins share a common progenitor cell origin. As many as 90 different colors can be generated. Sophisticated microscopic analysis is, however, necessary to distinguish the precise expression of different fluorescent proteins at distinct levels. The Confetti mice are an example of a modified version of the Brainbow mice for analysis in any tissue [56]. Stochastic Cre-mediated recombination generates the expression of four distinct markers for lineage tracking; however, the recombination frequency of each marker is not equal.
Mosaic Analysis with Double Markers (MADM) is another remarkable mouse genetic technique for lineage tracing. Analogous in principle to Drosophila Mosaic Analysis with a Repressible Cell Marker (MARCM) [57], MADM relies on the Cre-loxP system to catalyze inter-chromosomal recombination to reconstitute two otherwise nonfunctional split marker genes (i.e. GFP and RFP) [58]. Depending on the chromosomal segregation pattern, the two reconstituted fluorescent marker genes are inherited by the same or two different daughter cells of the labeled progenitor cells, resulting in the generation of yellow or green/red cell lineages, respectively. A great advantage of this approach is that in addition to conventional lineage tracing, the permanent and separate green or red labeling of two daughter cells of one initial dividing progenitor cell offers important insights into progenitor cell division patterns, such as whether they are symmetric or asymmetric.
In conjunction with the available inducible CreER mouse lines, MADM can be implemented in a cell type and temporally specific manner [59–62]. Moreover, MADM can be used to delete endogenous genes located on the same chromosome as the MADM allele selectively in certain labeled cells at the clonal level, whereas the other labeled cells serve as a wild type control. Notably, a recent study utilizing MADM revealed that radial glial progenitors (RGPs) in the developing mouse neocortex exhibit highly deterministic behaviors in proliferation, neurogenesis, and gliogenesis [59]. An alternative method for combining Flp-inducible lineage analysis and Cre-mediated cell mutagenesis is mosaic mutant analysis with spatial and temporal control of recombination (MASTR), which has the advantage that a loxP-flanked gene can be mutated on any chromosome [63,64].
CRISPR-Cas9 genome-editing for lineage tracing
The rapid advance of the clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 genome-editing technology has offered new strategies for large scale lineage tracing even at the whole-organism level (Figure 3d). Several methods have been developed to harness diverse, permanent edits generated by genome-editing at designated genomic regions for lineage reconstruction, such as genome editing of synthetic target arrays for lineage tracing (GESTALT) [65], memory by engineered mutagenesis with optical in situ readout (MEMOIR) [66], and Scartrace [67]. GESTALT introduces synthetic arrays of 9–12 CRISPR/Cas9 target sites as the barcode, which can be progressively and stochastically edited over cell divisions. The accumulated combinatorial barcode edits are systematically examined by targeted single cell sequencing of either DNA or RNA. Lineage relationships can then be reconstructed by relating the patterns of recovered edits. GESTALT has been successfully applied to the zebrafish to evaluate the lineage relationship of ~200,000 cells. A similar approach has recently been applied in C. elegans [68].
While powerful, GESTALT does not preserve the information about the actual anatomical location of each queried cell. On the other hand, MEMOIR provides a versatile platform for CRISPR/Cas9-mediated barcode editing and in situ, single-cell level of readout through multiplexed single-molecule RNA fluorescence hybridization (smFISH). It is also compatible with same-cell measurements of endogenous gene expression through additional rounds of smFISH or other in situ sequencing technologies [69]. Therefore, it has the potential to provide both lineage and endpoint cell state (e.g. cell identity) information for the queried cells.
Additional optimizations on the design of barcode targets, the delivery of editing reagents including Cas9 and guide RNAs (gRNAs), and the efficiency and accuracy in recovering and analyzing edits will further empower genome-editing based techniques towards faithfully constructing complete maps of cell lineages in complex nervous tissues. Furthermore, integrating them with other single cell analysis methods such as RNA sequencing (RNA-Seq) or assay for transposase-accessible chromatin (ATAC)-Seq will add rich cellular information to lineage studies to build comprehensive atlases of neural cell types and brain development.
Naturally occurring somatic mutations for lineage tracing
Naturally occurring somatic mutations in individual precursor cells are transmitted into their progeny, thereby serving as indelible genetic signatures for lineage reconstruction (Figure 3e). For example, highly mutated microsatellite DNA sequences have been used to build cell lineage trees in normal and disease human tissues [70]. A challenge in using naturally occurring somatic mutations as lineage markers is to discover the mutations reliably in individual cells. Due to the low frequency, it is often difficult to identify somatic mutations by sequencing a mixture of cells at conventional depths. Recent advances in next-generation single cell sequencing have made it possible to discover rare mutations or variants at the single cell level that carry lineage information [71–73]. These variants, from the least to the most frequently occurring, include retrotransposons, copy-number variants (CNVs), single-nucleotide variants (SNVs), and microsatellites. The rates at which these variants occur in somatic tissues dictate the resolution of lineage relationship reconstruction.
In principle, this strategy allows lineage tracing in any organisms, including humans. Endogenous retroelements such as long interspersed nuclear element 1 (LINE-1 or L1) retain the mobilization ability to insert into a new genomic location during somatic cell division [74–76]. While the actual rate of L1 retrotransposition in the human brain is debated [77–80], L1 insertion has been used to assess lineage relationship of neurons in the human cerebral cortex [81]. Similarly, somatic CNVs and SNVs are potentially useful lineage-tracing markers [82–85]. The relative readiness in identifying CNVs on the basis of low-coverage sequencing makes the sequencing of many single cells for CNV variant discovery and lineage analysis possible. Microsatellites are the most frequently mutated somatic loci [86]. The analysis of all microsatellite locations in the genome may have the potential for being used to reconstruct the complete cell lineage tree of an entire organism.
One important condition of using somatic mutations for reliable lineage analysis is that the mutations are sufficiently abundant and functionally neutral. In addition, whole-genome single cell sequencing currently requires genome amplification prior to sequencing to generate sufficient material, which can introduce technical artifacts and complication. It is therefore necessary to consider the types and frequencies of potential errors. Finally, single-cell sequencing based lineage tracing does not provide actual information on the precise progenitor cell origin, as the exact location and timing at which the somatic mutations are introduced is unknown.
Concluding remarks
Lineage tracing provides fundamental information on cell production and specification that cannot be obtained by other studies. When designing a lineage tracing experiment, it is important to consider the strengths and weaknesses of all possible techniques and select one that is best suited to the biological question under study. Notably, different techniques are not mutually exclusive but can be integrated. Viral or transposon labeling can be combined with the Cre-loxP system and/or the multi-color reporter system; for example, retroviral labeling has been combined with the Cre-loxP system for lineage tracing of defined progenitor cells [11,87], and the transposon and Brainbow transgene methods have been integrated to develop a multiaddressable genome-integrative color (MAGIC) marker toolkit [88]. Future efforts harnessing multiple technologies in concert will permit systematic analyses of lineage relationships at an ever greater scale and with single cell resolution. In particular, rapid advances in genome-editing and single cell sequencing make it possible to decipher lineage relationships of the entire population of neural cells in a brain region under normal or pathological conditions.
Highlights.
Cell lineage underlies the developmental steps taken to acquire cell identity.
Genetic labeling allows lineage tracing with cell type and temporal specificity.
Genomic-editing and single-cell sequencing enable large scale lineage studies.
Somatic mutations offer signatures for lineage reconstruction in the human brain.
Acknowledgments
The authors thank Dr. Alexander L. Joyner for insightful comments. Our research is supported by NIH (R01DA024681, R01MH101382, R01NS085004), HFSP (RGP0053/2014), NYSTEM (N13G-232), HHMI, and the Natural Science Foundation of China (31228012). We apologize to all of our colleagues whose work we did not cite due to limited space but have been invaluable to our understanding of lineage tracing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared
References
- 1.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 2.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 3.Deppe U, Schierenberg E, Cole T, Krieg C, Schmitt D, Yoder B, von Ehrenstein G. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1978;75:376–380. doi: 10.1073/pnas.75.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu YC, He S, Chen S, Fu Y, Brown KN, Yao XH, Ma J, Gao KP, Sosinsky GE, Huang K, et al. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486:113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He S, Li Z, Ge S, Yu YC, Shi SH. Inside-Out Radial Migration Facilitates Lineage-Dependent Neocortical Microcircuit Assembly. Neuron. 2015;86:1159–1166. doi: 10.1016/j.neuron.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Lu H, Cheng PL, Ge S, Xu H, Shi SH, Dan Y. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 2012;486:118–121. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu HT, Han Z, Gao P, He S, Li Z, Shi W, Kodish O, Shao W, Brown KN, Huang K, et al. Distinct lineage-dependent structural and functional organization of the hippocampus. Cell. 2014;157:1552–1564. doi: 10.1016/j.cell.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi W, Xianyu A, Han Z, Tang X, Li Z, Zhong H, Mao T, Huang K, Shi SH. Ontogenetic establishment of order-specific nuclear organization in the mammalian thalamus. Nat Neurosci. 2017;20:516–528. doi: 10.1038/nn.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang XJ, Li Z, Han Z, Sultan KT, Huang K, Shi SH. Precise inhibitory microcircuit assembly of developmentally related neocortical interneurons in clusters. Nat Commun. 2017;8:16091. doi: 10.1038/ncomms16091. • This study for the first time demonstrated that the lineage relationship influences electrical synapse formation between interneurons and inhibitory microcircuit assembly in the mouse neocortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown KN, Chen S, Han Z, Lu CH, Tan X, Zhang XJ, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334:480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinosa JS, Luo L. Timing neurogenesis and differentiation: insights from quantitative clonal analyses of cerebellar granule cells. J Neurosci. 2008;28:2301–2312. doi: 10.1523/JNEUROSCI.5157-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legue E, Riedel E, Joyner AL. Clonal analysis reveals granule cell behaviors and compartmentalization that determine the folded morphology of the cerebellum. Development. 2015;142:1661–1671. doi: 10.1242/dev.120287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 15.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 16.Rawles ME. Origin of melanophores and their role in development of color patterns in vertebrates. Physiol Rev. 1948;28:383–408. doi: 10.1152/physrev.1948.28.4.383. [DOI] [PubMed] [Google Scholar]
- 17.Le Douarin N. A biological cell labeling technique and its use in expermental embryology. Dev Biol. 1973;30:217–222. doi: 10.1016/0012-1606(73)90061-4. [DOI] [PubMed] [Google Scholar]
- 18.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, Miyashita K, Saito TT, Yoneki T, Kakihara Y, Nabeshima K, Kishi YA, Shimoda C, Nojima H. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LoTurco J, Manent JB, Sidiqi F. New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb Cortex. 2009;19(Suppl 1):i120–125. doi: 10.1093/cercor/bhp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VandenDriessche T, Ivics Z, Izsvak Z, Chuah MK. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood. 2009;114:1461–1468. doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- 22.Cepko CL, Fields-Berry S, Ryder E, Austin C, Golden J. Lineage analysis using retroviral vectors. Curr Top Dev Biol. 1998;36:51–74. doi: 10.1016/s0070-2153(08)60495-0. [DOI] [PubMed] [Google Scholar]
- 23.Sanes JR. Analysing cell lineage with a recombinant retrovirus. Trends Neurosci. 1989;12:21–28. doi: 10.1016/0166-2236(89)90152-5. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 27.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci U S A. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cepko C. Retrovirus vectors and their applications in neurobiology. Neuron. 1988;1:345–353. doi: 10.1016/0896-6273(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 29.Panganiban AT. Retroviral DNA integration. Cell. 1985;42:5–6. doi: 10.1016/s0092-8674(85)80092-1. [DOI] [PubMed] [Google Scholar]
- 30.Klug CA, Cheshier S, Weissman IL. Inactivation of a GFP retrovirus occurs at multiple levels in long-term repopulating stem cells and their differentiated progeny. Blood. 2000;96:894–901. [PubMed] [Google Scholar]
- 31.Antoniou MN, Skipper KA, Anakok O. Optimizing retroviral gene expression for effective therapies. Hum Gene Ther. 2013;24:363–374. doi: 10.1089/hum.2013.062. [DOI] [PubMed] [Google Scholar]
- 32.Golden JA, Fields-Berry SC, Cepko CL. Construction and characterization of a highly complex retroviral library for lineage analysis. Proc Natl Acad Sci U S A. 1995;92:5704–5708. doi: 10.1073/pnas.92.12.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, Alvarez-Buylla A. Embryonic Origin of Postnatal Neural Stem Cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. •• This study used a retroviral library in conjunction with the inducible Cre-reporter system to determine the lineage relationship between embryonic neural progenitor cells and adult neural stem (B1) cells in the mouse forebrain. The authors showed that B1 cells share a common progenitor with cells in the cortex, striatum, and septum, and acquire regional specificity in olfactory bulb neuron subtype output at the ealry embryonic stage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer C, Jaglin XH, Cobbs LV, Bandler RC, Streicher C, Cepko CL, Hippenmeyer S, Fishell G. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron. 2015;87:989–998. doi: 10.1016/j.neuron.2015.07.011. •• This study and [35] used a retroviral library to examine lineage-related spatial organization of inhibitory interneurons in the mouse forebrain. Both studies suggested that clonally-related interneurons derived from individual progenitor cells in the medial ganglionic eminence and the preoptic area broadly disperse in the entire forebrain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron. 2015;87:999–1007. doi: 10.1016/j.neuron.2015.07.030. ••. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh C, Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science. 1992;255:434–440. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- 37.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sultan KT, Han Z, Zhang XJ, Xianyu A, Li Z, Huang K, Shi SH. Clonally Related GABAergic Interneurons Do Not Randomly Disperse but Frequently Form Local Clusters in the Forebrain. Neuron. 2016;92:31–44. doi: 10.1016/j.neuron.2016.09.033. •• This study and [10] performed in-depth analysis of the datasets in [34, 35] and demonstrated that clonally related interneurons originated from the same dividing progenitor cells in the medial ganglionic eminence and the preoptic area reliably form local clusters in the forebrain as well as in the cortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirkwood TB, Price J, Grove EA. The dispersion of neuronal clones across the cerebral cortex. Science. 1992;258:317–320. doi: 10.1126/science.1411530. [DOI] [PubMed] [Google Scholar]
- 40.Walsh C, Cepko CL. Clonally related cortical cells show several migration patterns. Science. 1988;241:1342–1345. doi: 10.1126/science.3137660. [DOI] [PubMed] [Google Scholar]
- 41.Williams BP, Read J, Price J. The generation of neurons and oligodendrocytes from a common precursor cell. Neuron. 1991;7:685–693. doi: 10.1016/0896-6273(91)90381-9. [DOI] [PubMed] [Google Scholar]
- 42.Luskin MB, Pearlman AL, Sanes JR. Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron. 1988;1:635–647. doi: 10.1016/0896-6273(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 43.Price J, Thurlow L. Cell lineage in the rat cerebral cortex: a study using retroviral-mediated gene transfer. Development. 1988;104:473–482. doi: 10.1242/dev.104.3.473. [DOI] [PubMed] [Google Scholar]
- 44.Walsh C, Cepko CL. Clonal dispersion in proliferative layers of developing cerebral cortex. Nature. 1993;362:632–635. doi: 10.1038/362632a0. [DOI] [PubMed] [Google Scholar]
- 45.Reid CB, Liang I, Walsh C. Systematic widespread clonal organization in cerebral cortex. Neuron. 1995;15:299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 46.Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 48.Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 49.Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- 50.Casanova E, Lemberger T, Fehsenfeld S, Mantamadiotis T, Schutz G. Alpha complementation in the Cre recombinase enzyme. Genesis. 2003;37:25–29. doi: 10.1002/gene.10227. [DOI] [PubMed] [Google Scholar]
- 51.Wang P, Chen T, Sakurai K, Han BX, He Z, Feng G, Wang F. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Sci Rep. 2012;2:497. doi: 10.1038/srep00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron. 2016;92:555. doi: 10.1016/j.neuron.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- 55.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 56.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 57.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 58.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. •• This study used MADM to examine the division behavior and progeny of individual radial progenitor cells in the developing mouse cortex and showed that radial glial progenitor cells display a highly deterministic behavior in proliferation, neurogenesis, and gliogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beattie R, Postiglione MP, Burnett LE, Laukoter S, Streicher C, Pauler FM, Xiao G, Klezovitch O, Vasioukhin V, Ghashghaei TH, et al. Mosaic Analysis with Double Markers Reveals Distinct Sequential Functions of Lgl1 in Neural Stem Cells. Neuron. 2017;94:517–533. e513. doi: 10.1016/j.neuron.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 61.Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wojcinski A, Lawton AK, Bayin NS, Lao Z, Stephen DN, Joyner AL. Cerebellar granule cell replenishment postinjury by adaptive reprogramming of Nestin+ progenitors. Nat Neurosci. 2017 doi: 10.1038/nn.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lao Z, Raju GP, Bai CB, Joyner AL. MASTR: a technique for mosaic mutant analysis with spatial and temporal control of recombination using conditional floxed alleles in mice. Cell Rep. 2012;2:386–396. doi: 10.1016/j.celrep.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353:aaf7907. doi: 10.1126/science.aaf7907. •• This study reported the GESTALT method that combines CRISPR/Cas9 genomic editing and targeted single-cell sequencing for large-scale lineage analysis in cell cultures and zebrafish embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frieda KL, Linton JM, Hormoz S, Choi J, Chow KK, Singer ZS, Budde MW, Elowitz MB, Cai L. Synthetic recording and in situ readout of lineage information in single cells. Nature. 2017;541:107–111. doi: 10.1038/nature20777. •• This study reported the MEMOIR method that combines CRISPR/Cas9 genomic editing and single-molecule fluorescence hibridization (smFISH) for in situ lineage analysis in embryonic stem cell cultures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Junker JP, Spanjaard B, Peterson-Maduro J, Alemany A, Hu B, Florescu M, van Oudenaarden A. Massivly parallel whole-organism lineage tracing using CRISPR/Cas9 induced genetic scars. bioRxiv. 2016 [Google Scholar]
- 68.Schmidt ST, Zimmerman SM, Wang J, Kim SK, Quake SR. Quantitative Analysis of Synthetic Cell Lineage Tracing Using Nuclease Barcoding. ACS Synth Biol. 2017;6:936–942. doi: 10.1021/acssynbio.6b00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 70.Frumkin D, Wasserstrom A, Kaplan S, Feige U, Shapiro E. Genomic variability within an organism exposes its cell lineage tree. PLoS Comput Biol. 2005;1:e50. doi: 10.1371/journal.pcbi.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 72.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 76.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016;5 doi: 10.7554/eLife.12966. •• This study discussed the discrepencies in the rate of somatic L1 retrotranspostion in human hippocampal and cortical neurons reported in different studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. •• This study performed single-cell retrotransposon capture sequencing (RC-seq) on individual human hippocampal neurons and glia, as well as cortical neurons, and reported pervasive L1 mosaicism at genomic loci in hippocmapal neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. •• This study used high-coverage whole-genome sequencing of individual human brain neurons and identified multiple lienages and sublineages of cells marked by different L1 retrotranspostion evens and subsequent mutation of poly-A microsatellites within L1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A, Walsh CA. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zafar H, Wang Y, Nakhleh L, Navin N, Chen K. Monovar: single-nucleotide variant detection in single cells. Nat Methods. 2016;13:505–507. doi: 10.1038/nmeth.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D'Gama AM, Cai X, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. •• This study surveyed somatic single-nucleotide variants (SNVs) in human cerebral cortex by single-cell sequencing and reconstructed neuronal lineage relationships based on somatic mutations including SNVs, long interspersed nuclear elements (LINE) insertions, and a TG-dinucleotide expansion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 87.Ciceri G, Dehorter N, Sols I, Huang ZJ, Maravall M, Marin O. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci. 2013;16:1199–1210. doi: 10.1038/nn.3485. [DOI] [PubMed] [Google Scholar]
- 88.Loulier K, Barry R, Mahou P, Le Franc Y, Supatto W, Matho KS, Ieng S, Fouquet S, Dupin E, Benosman R, et al. Multiplex cell and lineage tracking with combinatorial labels. Neuron. 2014;81:505–520. doi: 10.1016/j.neuron.2013.12.016. • This study reported the multiaddressable genome integrative color (MAGIC) markers that can be used for lineage analysis in various animal models. [DOI] [PubMed] [Google Scholar]