Abstract
Autophagy, a fundamental and evolutionarily-conserved eukaryotic pathway, coordinates a complex balancing act for achieving both nutrient and energetic requirements for proper cellular function and homeostasis. We have discovered that soluble proteoglycans evoke autophagy in endothelial cells and mitophagy in breast carcinoma cells by directly interacting with receptor tyrosine kinases, including VEGF receptor 2 and Met. Under these circumstances, autophagic regulation is considered “non-canonical” and is epitomized by the bioactivity of the small leucine-rich proteoglycan, decorin. Soluble matrix-derived cues being transduced downstream of receptor engagement converge upon a newly-discovered nexus of autophagic machinery consisting of Peg3 for endothelial cell autophagy and mitostatin for tumor cell mitophagy. In this thematic mini-review, we will provide an overview of decorin-mediated autophagy and mitophagy and propose that regulating intracellular catabolism is the underlying molecular basis for the versatility of decorin as a potent oncosuppressive agent.
Keywords: Peg3, Mitostatin, Small leucine rich proteoglycans, Receptor tyrosine kinases, VEGFR2, Met
Introduction
Matrix biology has gone through an extensive revitalization over the last few years. A vast number of studies have demonstrated a nuanced complexity of interactions among extracellular matrix (ECM) components and cell surface receptors that are essential for multiple biological activities [1]. Moreover, matrix microenvironmental clues [2–4] together with the increasing complexity of the matrisome [5,6] and its proteolytic processing [7,8], have expanded our view of the extracellular matrix. Genetic and biochemical studies have characterized matrix components as signaling molecules, able to modulate cellular behavior during development and disease progression. Decorin is a ubiquitous small-leucine rich proteoglycan localized to the ECM and is associated with all major collagen-rich matrices [9,10]. Although decorin is mostly known for its function in collagen fibrillogenesis and for maintaining structural integrity of various organs [11–13,13,14,14–18,18], it is also involved in regulating physiological and pathological processes as a secreted proteoglycan [19–23]. These include roles in carcinogenesis [24–29], calcium homeostasis [30], fetal membrane signaling [31], fibrosis, wound healing and angiogenesis [32–38], myogenesis [39], myocardial homeostasis [40,41], osteoarthritis [42], renal diseases [43,44], growth factor bioactivity and signaling [45–48], and immune disorders [22,49]. Recent work has focused on the role of decorin in tumorigenesis and angiogenesis through its ability to bind with high affinity to various cell surface signaling receptors. We have characterized decorin as a pan-receptor tyrosine kinase (RTK) antagonist acting as a guardian from the matrix [50] in analogy to the tumor suppressor, p53. This definition is based on several key observations deriving from the complex binding repertoire of decorin [51]. The innovative discovery that decorin impedes tumor cell growth by binding with high affinity (KD~75 nM) and down-regulating the epidermal growth factor receptor (EGFR), with induction of p21WAF1 coincident with cell cycle arrest [52,53] broke new ground to a series of studies on the decorin interactome and its intracellular signaling effects. We found that soluble decorin binds directly and with high affinity (KD~2 nM) to Met, the receptor for hepatocyte growth factor (HGF) [54]. This interaction induces transient receptor activation followed by lysosomal degradation of Met and ultimately downregulation of the potent oncogenes, β-catenin and Myc. Biologically, activation of this signaling pathway leads to a reduction in primary tumor growth and metastatic spreading [55]. Both EGFR and Met activate the MAPK and PI3K/Akt pathways thereby controlling cell growth and survival. Moreover, transcriptional down-regulation of Myc and its targets involved in the pathogenesis of multiple cancers, contributes to the oncostatic effects of decorin. Decorin also antagonizes the angiogenic network by suppressing HIF-1α, VEGFA and evokes secretion of the anti-angiogenic factors Thrombospondin 1 and TIMP3 [56]. Recently, it was found that HIF-1α suppresses Thrombospondin 2 [57]. As decorin also decreases HIF-1α, it is possible that decorin may also regulate the levels of Thrombospondin 2. Moreover, the anti-oncogenic properties of decorin have been confirmed in various preclinical [58–60] and genetic studies utilizing Dcn−/− mice whereupon lack of decorin is permissive for tumorigenesis [26,27,61,62]. Notably, systemic or adenovirus-mediated delivery of decorin in animals bearing various types of solid malignant xenografts markedly suppresses tumor growth and metastasis [21,58–60,63–65]. Collectively, these studies point toward an intrinsic decorin “promiscuity” for various RTKs. This will allow decorin to target both transformed and genetically-stable cells such as endothelial cells, thereby providing an advantage as a soluble tumor repressor. Ultimately, these properties will make decorin and decorin-like molecules novel therapeutic agents against both cancer growth and tumor angiogenesis. In this review, we will illustrate the “intracellular remodeling”, i.e., the profound downstream effects mediated by this versatile proteoglycan as a result from binding multiple RTKs in the tumor microenvironment. We will dissect its critical activity in inducing and regulating two new sophisticated processes involving the degradation of cellular components: autophagy and mitophagy.
The role of decorin in autophagy
The ability of decorin to affect the trafficking of several receptor tyrosine kinases by triggering receptor internalization, degradation, and downstream signaling is just the tip of the iceberg concerning decorin biology. Recently, we have discovered that soluble decorin induces autophagy in endothelial cells [66] and mitophagy in breast carcinoma cells [67]. More than 40 years ago, Christian de Duve, discovered that, upon glucagon perfusion of rat liver, mitochondria and other intracellular structures undergo lysosomal degradation. Consequently he coined the term autophagy, Greek for “self-eating”. Few knew the profound importance of this homeostatic mechanism, a self-degradative process by which damaged organelles, misfolded, or aggregated protein, and other cytoplasmic components are degraded and recycled through the lysosomal pathway [68]. Basal levels of autophagy are now known to be necessary for normal cell functions, signaling, and proliferation. The role of autophagy in cancer is controversial, and it can be either beneficial for tumor progression or it can actually suppress metastatic spreading [69]. Studies have suggested that autophagy may prevent cancer initiation by protecting healthy cells from accumulating damaged organelles and metabolic products [70].
We serendipitously discovered that nanomolar concentrations of soluble decorin induced the formation of bubble-like structures in endothelial cells structurally reminiscent of autophagosomes. These structures, observed by differential interference contrast (DIC) microscopy, were strongly immunoreactive for Beclin 1 and LC3, two key components of the autophagic machinery [71]. Thus, decorin became the first soluble proteoglycan known to influence a catabolic intracellular process in normal endothelium. Recently, decorin has been shown to inhibit migration of glioma cells by activating autophagy and inhibiting TGFβ signaling [72]. Moreover, decorin overexpression evokes autophagy and protects intestinal cells in mice with inflammatory bowel disease [73]. These independent reports suggest that decorin might evoke autophagic flux not only in endothelial cells but also in epithelial and glioma cells.
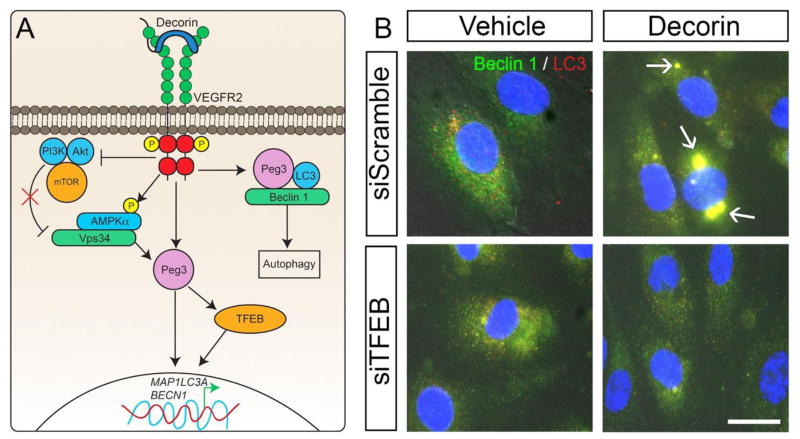
The autophagic process arises from the interaction of decorin with VEGFR2, and subsequent activation of AMPKα at Thr172 (Fig. 1A), an energy sensor kinase involved in cancer inhibition [74]. This, in turn, stimulates the synthesis of Peg3 (Paternally expressed gene 3), a gene previously found to be upregulated exclusively in the microenvironment of orthotopic tumor xenografts systemically treated with decorin [66]. Peg3 encodes a Krupple-like zinc finger-containing transcription factor, of the C2H2 variety. Initially identified as a tumor suppressor gene, Peg3 is now a key player involved in the regulation of endothelial cell autophagy upon decorin exposure [66,75]. The functional connection between Peg3 and decorin is based on the fact that both genes converge on inhibiting the Wnt/β-catenin pathway in a non-canonical fashion [76], Moreover, Peg3 expression is silenced in multiple solid tumors due to promoter methylation and/or loss of heterozygosity [77,78], strengthening its role as an onco-suppressor. We found that in microvascular and macrovascular endothelial cells, Peg3 physically bound Beclin 1 and localized to LC3-positive autophagosomes (Fig. 1A). This was confirmed by a series of confocal scanning laser microscopy experiments that illustrated the co-localization of Peg3 with Beclin 1 and LC3 in the bubble-like structures upon decorin stimulation [66]. This interaction was followed up and biochemically validated with co-immunoprecipitation studies. Furthermore, we discovered that Peg3 is required for decorin-evoked autophagy and for the transcriptional induction of the two major autophagic markers, Beclin 1 and LC3 [75,79]. The decorin/Peg3 axis may also regulate mitochondrial mass and genes involved in mitochondrial homeostasis in endothelial cells [80]. Peg3 has been shown to directly bind the promoters of Ndufs7, Ndufs8, and Sdhb [81,82], therefore decorin may compromise endothelial cell mitochondrial dynamics in a manner similar to tumor cell mitophagy (see below).
Fig. 1.
Schematic representation of decorin-evoked endothelial cell autophagy. (A) Mechanistic depiction of decorin-evoked endothelial cell autophagy downstream of VEGFR2. (B) Differential interference contrast (DIC) images of HUVEC treated for 6 hours of decorin (200 nM) upon silencing of TFEB, utilizing a specific siRNA, and probed for Beclin 1 (green) and LC3 (red). Arrows, in the top right panel, indicate the dually positive Beclin 1/LC3 autophagosomes as evoked by decorin in scramble transfected cells. Nuclei have been stained with DAPI (blue). All images were taken with the same exposure, gain, and intensity. Scale bar ~ 10 μm.
Mechanistically, Peg3 functions downstream of VEGFR2, as knocking down this receptor tyrosine kinase, completely abolishes the autophagic machinery in response to decorin [75]. Decorin binds to the VEGFR2 ectodomain and triggers the pro-autophagic AMPKα/Vps34 signaling pathway [83,84] (Fig. 1A). In addition, the decorin/VEGFR2 interaction reduces the anti-autophagic BCL-2/Beclin 1 protein complex [85]. Decorin is able of concurrently suppressing VEGFA levels through binding to VEGFR2 and blocking the mTOR pathway in a canonical fashion similar to rapamycin. Moreover, by promoting the activation of AMPK, decorin further induces autophagy. Ultimately, the decorin/VEGFR2/Peg3 signaling cascade leads to induction of the pro-autophagic genes BECN1 and MAP1LC3A (gene encoding LC3). These activities are specifically dependent on Peg3 and VEGFR2 since this signaling can be blocked by siRNA directed against Peg3 or VEGFR2, respectively [66,85]. Therefore, the ultimate effect of decorin is inhibition of angiogenesis through enhanced autophagy within the endothelial compartment. The formation of autophagosomes leads, eventually, to the degradation of the organelles, proteins, and other cellular components that they are carrying through lysosomal fusion and subsequent formation of autophagolysosomes [86]. One of the key components involved in the autophagic flux and lysosomal machinery is TFEB (Transcription Factor EB). TFEB specifically recognizes and binds the CLEAR-box sequences present in the regulatory region of many lysosomal genes, leading to the activation of their expression [87]. It thereby acts as a positive regulator of the autophagic process [88–90]. Intriguingly, TFEB is normally inactive, is directly phosphorylated by mTOR and sequestered via 14-3-3 scaffolding proteins. However, following an autophagic signal (such as decorin), TFEB is dephosphorylated by the protein phosphatase, calcineurin, and translocates to the nucleus where it initiates lysosomal gene transcription [87] (Fig. 1A). In the presence of decorin, mTOR is inactivated and TFEB expression and nuclear translocation is induced. We found that Peg3 is necessary for driving TFEB induction [91]. Indeed, Peg3 depletion is sufficient to inhibit decorin-mediated TFEB expression at both transcriptional and protein levels. Moreover, increasing concentrations of Peg3 are able to boost TFEB expression. Therefore, Peg3 functions as an upstream regulator of decorin-induced TFEB expression. Furthermore, blocking VEGFR2/AMPKα signaling with Compound C (Dorsomophin) abolishes decorin-mediated TFEB induction, thereby placing TFEB under the control of the decorin/VEGFR2/Peg3 autophagic axis [91]. This was the first study that linked decorin, a stroma derived secreted proteoglycan, to lysosomal homeostasis: a crucial step towards terminal autophagic progression. TFEB possesses a compelling role in the formation of autophagosomes upon decorin stimulation of endothelial cells. The induction of dually positive Beclin 1/LC3 autophagosomes by decorin, is indeed abolished when TFEB expression is silenced (Fig. 1B), further underscoring the key role of TFEB in the induction of crucial autophagic genes and in the structural formation of autophagosomes.
As the anti-tumorigenic “guardian of the matrix”, decorin executes its role in two ways: by triggering growth inhibition and angiostasis via direct and high-affinity binding to multiple RTKs, governed by a hierarchy of binding constants, and indirectly and non-canonically stimulating endothelial autophagy under nutrient-rich conditions. This is important as nutrient status is a critical regulator of autophagic activity, and aberrations in both autophagy and metabolism are implicated in many cardiac disorders. Notably, decorin mRNA and protein levels are induced by fasting in mouse cardiac tissue through inhibition of the canonical mTOR pathway [92,93]. This posits decorin as the first proteoglycan whose expression is mediated by an autophagic stimulus. We note that both Peg3 and p21WAF1, two genes that are induced by decorin, are also induced by fasting [94]. Thus, a secreted proteoglycan can remodel the intracellular machineries linked to self-eating, Peg3 and ATF4, and cell cycle control, p21WAF1 (see below). Furthermore, Dcn−/− mice have impaired cardiac autophagy when compared to wild-type animals [92]. Indeed, MAP1LC3A mRNA failed to increase in Dcn−/− mice undergoing a 25-h fast, whereas a 3.5-fold induction of MAP1LC3A was noticed in fasted wild-type animals belonging to the same genetic strain. Moreover, in GFP-LC3 mice, following fasting, a significant increase in decorin immunostaining as well as GFP-LC3 puncta was observed compared to fed animals. However, compound GFP-LC3;Dcn−/− animals did not show an increase in the number of GFP-LC3 puncta in starved animals compared to the fed cohort.
A number of important genes are upregulated in the heart of wild type animals under poor-nutrient conditions. Among them, Cdkn1a, which encodes for the cyclin dependent kinase inhibitor p21WAF1, is significantly increased in wild-type starved hearts but not in the Dcn−/− animals, indicating that decorin is necessary for in vivo p21WAF1induction upon fasting. Interestingly, the response to nutrient deprivation is not uniform across SLRP family members. Biglycan is the closest family member to decorin and share similar molecular architectures (~67% homology). However, upon fasting, Bgn mRNA is not induced, suggesting that this particular response to nutrient restriction is unique to decorin [92].
Decorin evokes tumor cell mitophagy
As a key constituent of the multi-faceted manner for abrogating tumorigenesis and attenuating angiogenesis (that is, an overall effect of differentially modulating pro- and anti-angiogenic cues concurrent with the sustained and excessive induction of endothelial cell autophagy), decorin directly influences catabolic programs and organelle turnover within the tumor proper itself. Therefore, extrapolating this concept to the tumor parenchyma, we found prolonged respiratory complex turnover (up to 24 h) and mitochondrial DNA (mtDNA) depletion, key properties of mitochondrial autophagy (mitophagy) [67]. Engagement of this fundamental pathway may functionally and biologically unify canonical oncosuppresive effects of decorin (e.g. suppression of potent oncogenes such as Myc, β-catenin and concomitant induction of p21WAF1) with the paradigmatic discovery of matrix-mediated autophagy for impeding tumorigenesis and angiogenesis.
In a mechanism that parallels pro-autophagic VEGFR2 signaling, decorin requires the presence and kinase activity of Met for proper mitophagy and respiratory chain turnover in triple negative and luminal breast carcinoma cells [67] (Fig. 2A). Conceptually, both forms of autophagic activation require cell surface receptors (VEGFR2 or Met) and the inherent kinase activity of the appropriate receptor. Again, this is considered non-canonical autophagic (mitophagic) induction insofar as this form of catabolism occurs irrespective of nutrient conditions. Indeed, the autophagic programs evoked by soluble decorin are operative in nutrient-rich conditions.
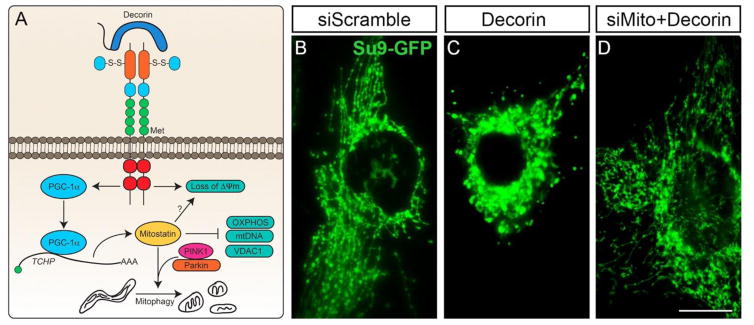
Fig. 2.
Schematic representation of decorin-evoked tumor cell mitophagy. (A) Mechanistic depiction of decorin-evoked tumor cell mitophagy downstream of Met. (B–D) Gallery of fluorescence images of FACS sorted MDA-MB-231 cells stably expressing the Su9-GFP transgene for mitochondrial network visualization. Cells were then super-transfected with RNAi against mitostatin to ascertain the effect on the mitochondrial network as shown for the siScramble transfectants (B), treatment with 200 nM decorin (C), or decorin following the depletion of mitostatin (D). All images were taken with the same exposure, gain, and intensity. Scale bar ~10 μm.
Mitostatin is required for decorin-evoked mitophagy
A central factor for decorin-evoked mitophagy is a poorly understood tumor suppressor gene referred to as mitostatin or trichoplein (TCHP), located on chromosome 12q24.1). Originally known as Ts12q, for tumor suppressor at 12q, the resulting ORF product of TCHP has since been eponymously renamed as mitostatin, for mitochondrial protein with oncostatic activity [95]. Mitostatin was identified as a decorin-inducible gene via differential hybridization of cDNA libraries (subtractive hybridization) with probes that were obtained from logarithmically growing (mock-transferred) or from cells transfected with decorin (growth-suppressed) [95]. Mitostatin is ubiquitously expressed in many normal tissues and is evolutionarily conserved among several species. Importantly, mitostatin is frequently lost and/or mutated at highly conserved residues throughout its coding region in bladder and breast carcinomas [95,96], suggesting it might function as a tumor suppressor gene.
Mitostatin staining reveals a punctate and primarily mitochondrial localization [96] and is significantly enriched at specialized membrane: membrane juxtapositions of endoplasmic reticulum and mitochondria, where mitostatin directly interacts with mitofusion-2 [97]. Over-expression of mitostatin results in ultrastructural changes in mitochondrial architecture such as loss of the mitochondrial matrix, abnormal cristae, and a swollen, stouter appearance when viewed under electron microscopy [95]. Intriguingly, phosphorylation of Hsp27 at Ser82 was significantly diminished by mitostatin while total Hsp27 levels inversely correlated with the levels of mitostatin expression [95].
In the initial stages of mitophagy and downstream of Met signaling, PGC-1α, a master regulator of mitochondrial biogenesis [98], is mobilized and binds TCHP mRNA directly for rapid stabilization coincident with accumulating amounts of mitostatin [67] (Fig. 2A). Functionally, the interaction between PGC-1α protein and TCHP mRNA occurs via the C-terminal RNA recognition motif [67]. Truncating this domain or silencing protein arginine methyltransferase 1, for proper protein arginine methylation, significantly abrogates TCHP mRNA stabilization [67]. Elucidating the determinants of this pathway revealed a unique and unlikely molecular cooperation between a mitophagic effector (mitostatin) and a known oncogene (PGC-1α). Indeed, PGC-1α mediates B-Raf-mediated oxidative metabolism [99] for a subset of aggressive melanomas characterized by heightened mitochondrial capacity for augmented resistance to various oxidative stressors [100].
The underlying properties of decorin-evoked mitophagy require the presence of and yet-to-be-discovered function of mitostatin [67] (Fig. 2A). Preliminary data have suggested that RNAi-mediated depletion of mitostatin precludes the turnover of various respiratory chain components, decreased TFAM and mtDNA content, VDAC1 clearance, and ultimately the collapse and fragmentation of the mitochondrial network [67] (Fig. 2B–D), all known markers of mitophagy [101]. The latter findings of mitochondrial network fragmentation are consistent with the results of simply over-expressing mitostatin, and may represent a critical initial step for mitophagic clearance following mitochondrial depolarization (Fig. 2A, see below). This further reinforces that mitostatin is a downstream effector of decorin-evoked mitochondrial fragmentation and turnover. Importantly, as measured by incorporation of Su9-GFP, a well-established mitochondrial matrix protein, laser confocal microscopy and conventional immunoblotting techniques reveal a decrease in the total amounts of Su9-GFP, suggesting clearance via the autophagosome-lysosome system.
Preceding the collapse and subsequent aggregation of the tubular mitochondrial network, decorin triggers mitochondrial depolarization [67], with a magnitude comparable to that of an established depolarization agent (the protonophore, FCCP). This loss of mitochondrial membrane potential across the outer and inner mitochondrial membrane (OMM/IMM) is an early harbinger of mitochondrial dysfunction and an effective signal for Parkin-mediated turnover [102,103]. We note that decorin has been shown to regulate intracellular Ca2+ levels in squamous cell carcinomas [30,104]. Thus, the appearance of depolarized mitochondria may represent the terminal product of increased cytosolic Ca+2 levels that occur downstream of decorin/EGFR. As mitostatin is a mitochondrial-associated membrane component and interacts with MFN-2, it may permit an efflux of stored Ca+2 from the ER directly into the mitochondria as the initial, triggering event for decorin-evoked mitophagy.
Depolarized (e.g. damaged) mitochondria trigger the recruitment of cytosolic Parkin, an RBR-domain containing E3-ubiquiting ligase, to the OMM where PINK1 is stabilized due to loss of membrane potential. Binding its OMM receptors (Bnip3/Nix, FUNDC1, and NDP52), Parkin utilizes phospho-ubiquitin to ubiquitinate several OMM components, such as MFN2. Recognition by several ubiquitin binding autophagy receptors (p62/SQSTM1, optineurin, or NBR1), the organelle is engulfed by LC3-positive autophagosomes. Parkin is strictly required for proper mitochondrial homeostasis, as recessive mutations in Parkin are responsible for the familial form of the devastating neurodegenerative disease, Parkinson’s [102,105,106].
It is plausible that decorin may promote the mobilization and recruitment of Parkin to the outer mitochondrial membrane for mitochondrial turnover in a mitostatin-dependent manner (Fig. 2A). Moreover, mitostatin, harboring five coiled-coil domains (as predicted by bioinformatics analyses) may mediate recruitment and/or binding of Parkin to cognate receptors on the mitochondrial surface. This appears plausible as decorin triggers loss of membrane potential, a central requirement for PINK1 stabilization, Parkin recruitment, and organelle turnover. Mitostatin may therefore indirectly stimulate inherent PINK1 activity (following stabilization and dimerization) for proper targeting, ubiquitin activation [107,108], and/or Parkin-mediated ubiquitination of mitochondrial proteins [109–111]. This axis is key for recycling respiratory chain complexes [112,113].
Collectively, the above findings culminate in a transduction pathway that conveys biological information via Met for mitophagic induction, in a mitostatin-dependent manner, within the tumor parenchyma of breast and prostate carcinomas [65,114]. This catabolic process, coupled with the discovery of endothelial cell autophagy, may form a molecular nexus for integrating the various outputs of decorin-mediated RTK regulation. This concept can be extended insofar as decorin suppression of Myc and HIF-1α may reverse the chemotherapeutic resistance of triple negative breast cancers via decreased mitochondrial OXPHOS [115]. Therefore, this newly-found activity may lie at the crossroads of controlling tumorigenic growth and unchecked tumor vascularization.
Conclusion
Soluble proteoglycans are rapidly evolving within the realm of matrix biology (and related fields) as pivotal components for understanding fundamental, homeostatic cellular processes and as novel targets for discerning complex pathological conditions [116] when these pathways malfunction. As such, the goal of fully realizing the diverse intricacies and nuances of “dynamic reciprocity” between the cell and matrix components has been extensively facilitated by an exhaustive proteomics approach, organized into an invaluable public resource encompassing various normal and disease states, that is constantly being updated [5,6].
However, before the inception of such a robust database, the seminal discoveries involving decorin revealed encrypted signaling information intrinsic to members of soluble proteoglycans that could only be decoded by interactions with a cell-surface RTK. Since this groundbreaking finding, similar mechanisms of action have been proposed as the underlying molecular explanation for a variety of biological processes [1] across diverse tissues, microenvironments, and matrix components. This concept is further reinforced by the ever-expanding decorin interactome [50,117] that encompasses a myriad of matrix-bound and cell-localized binding partners that decode and curbs pro-tumorigenic and pro-angiogenic signaling cues [118]. Despite these properties, however, it is not unique to proteoglycans to function as a nexus of regulating cellular behavior, as large, modular heparan-sulfate proteoglycans (such as perlecan) are intrinsically multivalent [119].
Concurrent with the “canonical” roles ascribed to decorin, evolutionarily conserved and fundamental intracellular catabolic processes [51,120] are being engaged for the summary degradation of bulk cytosolic contents or the targeted turnover of critical organelles. This perspective resulted in a paradigm shift where soluble matrix components, via cell surface signaling receptors, are capable of overriding the ancient constraints of catabolic metabolism and activating degradative pathways, even when nutrients are in abundance. The ultimate result of this activity is protracted and excessive autophagic and mitophagic outcomes that impair cellular function to curb tumorigenesis and angiogenesis. Collectively, the multifaceted bioactivity of decorin [121] manifests as a long-lasting oncosuppression [55,122] that is efficacious and clinically relevant in a variety of solid tumor models.
Highlights.
Regulating intracellular catabolism is an emerging theme among secreted matrix constituents
Soluble decorin evokes endothelial cell autophagy and tumor cell mitophagy independent of nutrient deprivation
Decorin induces autophagy via VEGFR2/AMPKα activation and simultaneous mTOR inhibition
Peg3 is required for upstream TFEB expression and proper autophagic induction
Decorin causes mitochondrial depolarization and mitophagy via mitostatin and parkin downstream of Met
Acknowledgments
This work was supported, in part, by National Institutes of Health Grants, CA39481, and CA47282 (R.V.I.). We wish to thank all past and present members of the laboratory and apologize for not referencing many valuable contributions due to space limitations.
Abbreviations used
- AMPKα
AMP-activated protein kinase catalytic α subunit
- Ca
calcium
- CLEAR
coordinated lysosomal expression and regulation
- DIC
Differential interference contrast
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- FCCP
Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
- GFP
green fluorescent protein
- HIF-1α
hypoxia inducible factor-1α
- HUVEC
human umbilical vein endothelial cells
- IMM
inner mitochondrial matrix
- LC3
microtubule-associated proteins 1A/1B light chain 3A
- MAPK
mitogen activated protein kinase
- MFN-2
mitofusion-2
- mTOR
mammalian/mechanistic target of rapamycin
- PEG3
Paternally expressed gene 3
- PGC-1α
peroxisome proliferator activated receptor γ co-activator 1α
- PINK1
PTEN-induced putative kinase 1
- OMM
outer mitochondrial matrix
- RTK
receptor tyrosine kinase
- Ser
serine
- SLRP
small leucine rich proteoglycan
- TFAM
transcription factor A, mitochondrial
- TFEB
Transcription Factor EB
- Thr
threonine
- TIMP3
tissue inhibitor of metalloproteinases 3
- VDAC1
voltage dependent anion channel 1
- VEGFA
vascular endothelial growth factor A
- VEGFR2
vascular endothelial growth factor receptor 2
Footnotes
Disclosure
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iozzo RV, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells A, Nuschke A, Yates CC. Skin tissue repair: Matrix microenvironmental influences. Matrix Biol. 2016;49:25–36. doi: 10.1016/j.matbio.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prockop DJ. Inflammation, fibrosis, and modulation of the process by mesenchymal stem/stromal cells. Matrix Biol. 2016;51:7–13. doi: 10.1016/j.matbio.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young MF. Skeletal biology: Where matrix meets mineral. Matrix Biol. 2016;52–54:1–6. doi: 10.1016/j.matbio.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naba A, Clauser KR, Mani DR, Carr SA, Hynes RO. Quantitative proteomic profiling of the extracellular matrix of pancreatic islets during the angiogenic switch and insulinoma progression. Sci Rep. 2017;7:40495. doi: 10.1038/srep40495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016;49:10–24. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckhard U, Huesgen PF, Schilling O, Bellac CL, Butler GS, Cox JH, et al. Active site specificity profiling of the matrix metalloproteinase family: Proteomic identification of 4300 cleavage sites by nine MMPs explored with structural and synthetic peptide cleavage analyses. Matrix Biol. 2016;49:37–60. doi: 10.1016/j.matbio.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Muir AM, Massoudi D, Nguyen N, Keene DR, Lee SJ, Birk DE, et al. BMP1-like proteinases are essential to the structure and wound healing of skin. Matrix Biol. 2016;56:114–131. doi: 10.1016/j.matbio.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed CC, Iozzo RV. The role of decorin in collagen fibrillogenesis and skin homeostasis. Glycoconj J. 2002;19:249–255. doi: 10.1023/A:1025383913444. [DOI] [PubMed] [Google Scholar]
- 10.Robinson KA, Sun M, Barnum CE, Weiss SN, Huegel J, Shetye SS, et al. Decorin and biglycan are necessary for maintaining collagen fibril structure, fiber realignment, and mechanical properties of mature tendons. Matrix Biol. 2017 doi: 10.1016/j.matbio.2017.08.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielson KG, Baribault H, Holmes DF, Graham H, Kadler KE, Iozzo RV. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol. 1997;136:729–743. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keene DR, San Antonio JD, Mayne R, McQuillan DJ, Sarris G, Santoro SA, et al. Decorin binds near the C terminus of type I collagen. J Biol Chem. 2000;275:21801–21804. doi: 10.1074/jbc.C000278200. [DOI] [PubMed] [Google Scholar]
- 13.Dunkman AA, Buckley MR, Mienaltowski MJ, Adams SM, Thomas SJ, Satchell L, et al. Decorin expression is important for age-related changes in tendon structure and mechanical properties. Matrix Biol. 2013;32:3–13. doi: 10.1016/j.matbio.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Young MF, Chakravarti S, Birk DE. Interclass small leucine-rich repeat proteoglycan interactions regulate collagen fibrillogenesis and corneal stromal assembly. Matrix Biol. 2014;35:103–111. doi: 10.1016/j.matbio.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rühland C, Schönherr E, Robenek H, Hansen U, Iozzo RV, Bruckner P, et al. The glycosaminoglycan chain of decorin plays an important role in collagen fibril formation at the early stages of fibrillogenesis. FEBS J. 2007;274:4246–4255. doi: 10.1111/j.1742-4658.2007.05951.x. [DOI] [PubMed] [Google Scholar]
- 16.Kalamajski S, Oldberd Å. The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 2010;29:248–253. doi: 10.1016/j.matbio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Reese SP, Underwood CJ, Weiss JA. Effects of decorin proteoglycan on fibrillogenesis, ultrastructure, and mechanics of type I collagen gels. Matrix Biol. 2013;32:414–423. doi: 10.1016/j.matbio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunkman AA, Buckley MR, Mienaltowski MJ, Adams SM, Thomas SJ, Kumar A, et al. The injury response of aged tendons in the absence of biglycan and decorin. Matrix Biol. 2014;35:232–238. doi: 10.1016/j.matbio.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldoni S, Owens RT, McQuillan DJ, Shriver Z, Sasisekharan R, Birk DE, et al. Biologically active decorin is a monomer in solution. J Biol Chem. 2004;279:6606–6612. doi: 10.1074/jbc.M310342200. [DOI] [PubMed] [Google Scholar]
- 20.Goldoni S, Iozzo RV. Tumor microenvironment: Modulation by decorin and related molecules harboring leucine-rich tandem motifs. Int J Cancer. 2008;123:2473–2479. doi: 10.1002/ijc.23930. [DOI] [PubMed] [Google Scholar]
- 21.Goldoni S, Seidler DG, Heath J, Fassan M, Baffa R, Thakur ML, et al. An anti-metastatic role for decorin in breast cancer. Am J Pathol. 2008;173:844–855. doi: 10.2353/ajpath.2008.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhão JG, et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and microRNA-21. Sci Signal. 2011;4:ra75. doi: 10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer L, Tredup C, Gubbiotti MA, Iozzo RV. Proteoglycan neofunctions: regulation of inflammation and autophagy in cancer biology. FEBS J. 2017;284:10–26. doi: 10.1111/febs.13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santra M, Skorski T, Calabretta B, Lattime EC, Iozzo RV. De novo decorin gene expression suppresses the malignant phenotype in human colon cancer cells. Proc Natl Acad Sci USA. 1995;92:7016–7020. doi: 10.1073/pnas.92.15.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moscatello DK, Santra M, Mann DM, McQuillan DJ, Wong AJ, Iozzo RV. Decorin suppresses tumor cell growth by activating the epidermal growth factor receptor. J Clin Invest. 1998;101:406–412. doi: 10.1172/JCI846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iozzo RV, Chakrani F, Perrotti D, McQuillan DJ, Skorski T, Calabretta B, et al. Cooperative action of germline mutations in decorin and p53 accelerates lymphoma tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3092–3097. doi: 10.1073/pnas.96.6.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath Z, Kovalszky I, Fullar A, Kiss K, Schaff Z, Iozzo RV, et al. Decorin deficiency promotes hepatic carcinogenesis. Matrix Biol. 2014;35:194–205. doi: 10.1016/j.matbio.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan MB. Extracellular matrix transcriptome dynamics in hepatocellular carcinoma. Matrix Biol. 2013;32:393–398. doi: 10.1016/j.matbio.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel S, Santra M, McQuillan DJ, Iozzo RV, Thomas AP. Decorin activates the epidermal growth factor receptor and elevates cytosolic Ca2+ in A431 cells. J Biol Chem. 1998;273:3121–3124. doi: 10.1074/jbc.273.6.3121. [DOI] [PubMed] [Google Scholar]
- 31.Wu Z, Horgan CE, Carr O, Owens RT, Iozzo RV, Lechner BE. Biglycan and decorin differentially regulate signaling in the fetal membranes. Matrix Biol. 2014;35:266–275. doi: 10.1016/j.matbio.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Järveläinen HT, Iruela-Arispe ML, Kinsella MG, Sandell LJ, Sage EH, Wight TN. Expression of decorin by sprouting bovine aortic endothelial cells exhibiting angiogenesis in vitro. Exp Cell Res. 1992;203:395–401. doi: 10.1016/0014-4827(92)90013-x. [DOI] [PubMed] [Google Scholar]
- 33.Grant DS, Yenisey C, Rose RW, Tootell M, Santra M, Iozzo RV. Decorin suppresses tumor cell-mediated angiogenesis. Oncogene. 2002;21:4765–4777. doi: 10.1038/sj.onc.1205595. [DOI] [PubMed] [Google Scholar]
- 34.Baghy K, Iozzo RV, Kovalszky I. Decorin-TGFβ axis in hepatic fibrosis and cirrhosis. J Histochem Cytochem. 2012;60:262–268. doi: 10.1369/0022155412438104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Järveläinen H, Puolakkainen P, Pakkanen S, Brown EL, Höök M, Iozzo RV, et al. A role for decorin in cutaneous wound healing and angiogenesis. Wound Rep Reg. 2006;14:443–452. doi: 10.1111/j.1743-6109.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 36.Järveläinen H, Sainio A, Wight TN. Pivotal role for decorin in angiogenesis. Matrix Biol. 2015;43:15–26. doi: 10.1016/j.matbio.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Järvinen TAH, Ruoslahti E. Target-seeking antifibrotic compound enhances wound healing and suppresses scar formation in mice. Proc Natl Acad Sci USA. 2010;107:21671–21676. doi: 10.1073/pnas.1016233107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Häkkinen L, Strassburger S, Kahari VM, Scott PG, Eichstetter I, Iozzo RV, et al. A role for decorin in the structural organization of periodontal ligament. Lab Invest. 2000;80:1869–1880. doi: 10.1038/labinvest.3780197. [DOI] [PubMed] [Google Scholar]
- 39.Brandan E, Gutierrez J. Role of skeletal muscle proteoglycans during myogenesis. Matrix Biol. 2013;32:289–297. doi: 10.1016/j.matbio.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Weis SM, Zimmerman SD, Shah M, Covell JW, Omens JH, Ross J, Jr, et al. A role for decorin in the remodeling of myocardial infarction. Matrix Biol. 2005;24:313–324. doi: 10.1016/j.matbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Barallobre-Barreiro J, Gupta SK, Zoccarato A, Kitazume-Taneike R, Fava M, Yin X, et al. Glycoproteomics Reveals Decorin Peptides With Anti-Myostatin Activity in Human Atrial Fibrillation. Circulation. 2016;134:817–832. doi: 10.1161/CIRCULATIONAHA.115.016423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gronau T, Kruger K, Prein C, Aszodi A, Gronau I, Iozzo RV, et al. Forced exercise-induced osteoarthritis is attenuated in mice lacking the small leucine-rich proteoglycan decorin. Ann Rheum Dis. 2017;76:442–449. doi: 10.1136/annrheumdis-2016-209319. [DOI] [PubMed] [Google Scholar]
- 43.Merline R, Lazaroski S, Babelova A, Tsalastra-Greul W, Pfeilschifter J, Schluter KD, et al. Decorin deficiency in diabetic mice: aggravation of nephropathy due to overexpression of profibrotic factors, enhanced apoptosis and mononuclear cell infiltration. J Physiol Pharmacol. 2009;60(suppl 4):5–13. [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer L, Macakova K, Raslik I, Micegova M, Grone HJ, Schonherr E, et al. Absence of decorin adversely influences tubulointerstitial fibrosis of the obstructed kidney by enhanced apoptosis and increased inflammatory reaction. Am J Pathol. 2002;160:1181–1191. doi: 10.1016/S0002-9440(10)64937-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferdous Z, Wei VM, Iozzo RV, Höök M, Grande-Allen KJ. Decorin-transforming growth factor-β interaction regulates matrix organization and mechanical characteristics of three-dimensional collagen matrices. J Biol Chem. 2007;282:35887–35898. doi: 10.1074/jbc.M705180200. [DOI] [PubMed] [Google Scholar]
- 46.Ferdous Z, Peterson SB, Tseng H, Anderson DK, Iozzo RV, Grande-Allen KJ. A role for decorin in controlling proliferation, adhesion, and migration of murine embryonic fibroblasts. J Biomed Mater Res Part A. 2010;93:419–428. doi: 10.1002/jbm.a.32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iozzo RV, Buraschi S, Genua M, Xu SQ, Solomides CC, Peiper SC, et al. Decorin antagonizes IGF receptor I (IGF-IR) function by interfering with IGF-IR activity and attenuating downstream signaling. J Biol Chem. 2011;286:34712–34721. doi: 10.1074/jbc.M111.262766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morcavallo A, Buraschi S, Xu SQ, Belfiore A, Schaefer L, Iozzo RV, et al. Decorin differentially modulates the activity of insulin receptor isoform A ligands. Matrix Biol. 2014;35:82–90. doi: 10.1016/j.matbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seidler DG, Mohamed NA, Bocian C, Stadtmann A, Hermann S, Schäfers K, et al. The role for decorin in delayed-type hypersensitivity. J Immunol. 2011;187:6108–6199. doi: 10.4049/jimmunol.1100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neill T, Schaefer L, Iozzo RV. Decorin, a guardian from the matrix. Am J Pathol. 2012;181:380–387. doi: 10.1016/j.ajpath.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neill T, Schaefer L, Iozzo RV. Decoding the matrix: Instructive roles of proteoglycan receptors. Biochemistry. 2015;54:4583–4598. doi: 10.1021/acs.biochem.5b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Luca A, Santra M, Baldi A, Giordano A, Iozzo RV. Decorin-induced growth suppression is associated with upregulation of p21, an inhibitor of cyclin-dependent kinases. J Biol Chem. 1996;271:18961–18965. doi: 10.1074/jbc.271.31.18961. [DOI] [PubMed] [Google Scholar]
- 53.Seidler DG, Goldoni S, Agnew C, Cardi C, Thakur ML, Owens RA, et al. Decorin protein core inhibits in vivo cancer growth and metabolism by hindering epidermal growth factor receptor function and triggering apoptosis via caspase-3 activation. J Biol Chem. 2006;281:26408–26418. doi: 10.1074/jbc.M602853200. [DOI] [PubMed] [Google Scholar]
- 54.Goldoni S, Humphries A, Nyström A, Sattar S, Owens RT, McQuillan DJ, et al. Decorin is a novel antagonistic ligand of the Met receptor. J Cell Biol. 2009;185:743–754. doi: 10.1083/jcb.200901129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buraschi S, Pal N, Tyler-Rubinstein N, Owens RT, Neill T, Iozzo RV. Decorin antagonizes Met receptor activity and downregulates β-catenin and Myc levels. J Biol Chem. 2010;285:42075–42085. doi: 10.1074/jbc.M110.172841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neill T, Painter H, Buraschi S, Owens RT, Lisanti MP, Schaefer L, et al. Decorin antagonizes the angiogenic network. Concurrent inhibition of Met, hypoxia inducible factor-1α and vascular endothelial growth factor A and induction of thrombospondin-1 and TIMP3. J Biol Chem. 2012;287:5492–5506. doi: 10.1074/jbc.M111.283499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacLauchlan SC, Calabro NE, Huang Y, Krishna M, Bancroft T, Sharma T, et al. HIF-1alpha represses the expression of the angiogenesis inhibitor thrombospondin-2. Matrix Biol. 2017 doi: 10.1016/j.matbio.2017.07.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reed CC, Gauldie J, Iozzo RV. Suppression of tumorigenicity by adenovirus-mediated gene transfer of decorin. Oncogene. 2002;21:3688–3695. doi: 10.1038/sj.onc.1205470. [DOI] [PubMed] [Google Scholar]
- 59.Tralhão JG, Schaefer L, Micegova M, Evaristo C, Schönherr E, Kayal S, et al. In vivo selective and distant killing of cancer cells using adenovirus-mediated decorin gene transfer. FASEB J. 2003;17:464–466. doi: 10.1096/fj.02-0534fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed CC, Waterhouse A, Kirby S, Kay P, Owens RA, McQuillan DJ, et al. Decorin prevents metastatic spreading of breast cancer. Oncogene. 2005;24:1104–1110. doi: 10.1038/sj.onc.1208329. [DOI] [PubMed] [Google Scholar]
- 61.Bi X, Tong C, Dokendorff A, Banroft L, Gallagher L, Guzman-Hartman G, et al. Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis. 2008;29:1435–1440. doi: 10.1093/carcin/bgn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bi X, Pohl NM, Yang GR, Gou Y, Guzman G, Kajdacsy-Balla A, et al. Decorin-mediated inhibition of colorectal cancer growth and migration is associated with E-cadherin in vitro and in mice. Carcinogenesis. 2012;33:326–330. doi: 10.1093/carcin/bgr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shintani K, Matsumine A, Kusuzaki K, Morikawa J, Matsubara T, Wakabayashi T, et al. Decorin suppresses lung metastases of murine osteosarcoma. Oncology Reports. 2008;19:1533–1539. [PubMed] [Google Scholar]
- 64.Araki K, Wakabayashi H, Shintani K, Morikawa J, Matsumine A, Kusuzaki K, et al. Decorin suppresses bone metastasis in a breast cancer cell line. Oncology. 2009;77:92–99. doi: 10.1159/000228253. [DOI] [PubMed] [Google Scholar]
- 65.Xu W, Neill T, Yang Y, Hu Z, Cleveland E, Wu Y, et al. The systemic delivery of an oncolytic adenovirus expressing decorin inhibits bone metastasis in a mouse model of human prostate cancer. Gene Therapy. 2015;22:31–40. doi: 10.1038/gt.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, et al. Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci U S A. 2013;110:E2582–E2591. doi: 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neill T, Torres A, Buraschi S, Owens RT, Hoek J, Baffa R, et al. Decorin induces mitophagy in breast carcinoma cells via peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) and mitostatin. J Biol Chem. 2014;289:4952–4968. doi: 10.1074/jbc.M113.512566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morselli E, Galluzzi L, Kepp O, Marino G, Michaud M, Vitale I, et al. Oncosuppressive functions of autophagy. Antioxid Redox Signal. 2011;14:2251–2269. doi: 10.1089/ars.2010.3478. [DOI] [PubMed] [Google Scholar]
- 70.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 71.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao T, Zhang CG, Gong MT, Zhang M, Wang L, Ding W. Decorin-mediated inhibition of the migration of U87MG glioma cells involves activation of autophagy and suppression of TGF-beta signaling. FEBS Open Bio. 2016;6:707–719. doi: 10.1002/2211-5463.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao H, Xi H, Wei B, Cai A, Wang T, Wang Y, et al. Expression of decorin in intestinal tissues of mice with inflammatory bowel disease and its correlation with autophagy. Exp Ther Med. 2016;12:3885–3892. doi: 10.3892/etm.2016.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zadra G, Photopulos C, Tyekucheva S, Heidari P, Weng QP, Fedele G, et al. A novel direct activator of AMPK inhibits prostate cancer growth by blocking lipogenesis. EMBO Mol Med. 2013;6:519–538. doi: 10.1002/emmm.201302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neill T, Torres AT, Buraschi S, Iozzo RV. Decorin has an appetite for endothelial cell autophagy. Autophagy. 2013;9:1626–1628. doi: 10.4161/auto.25881. [DOI] [PubMed] [Google Scholar]
- 76.Jiang X, Yu Y, Yang HW, Agar NYR, Frado L, Johnson MD. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J Biol Chem. 2010;285:8472–8480. doi: 10.1074/jbc.M109.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li LL, Tada M, et al. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nature Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 78.Kohda T, Asai A, Kuroiwa Y, Kobayashi S, Aisaka K, Nagashima G, et al. Tumour suppressor activity of human imprinted gene PEG3 in a glioma cell line. Genes Cells. 2001;6:237–247. doi: 10.1046/j.1365-2443.2001.00412.x. [DOI] [PubMed] [Google Scholar]
- 79.Torres A, Gubbiotti MA, Iozzo RV. Decorin-inducible Peg3 Evokes Beclin 1-mediated Autophagy and Thrombospondin 1-mediated Angiostasis. J Biol Chem. 2017;292:5055–5069. doi: 10.1074/jbc.M116.753632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Theka I, Sottile F, Aulicino F, Garcia AC, Cosma MP. Reduced expression of Paternally Expressed Gene-3 enhances somatic cell reprogramming through mitochondrial activity perturbation. Sci Rep. 2017;7:9705. doi: 10.1038/s41598-017-10016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thiaville MM, Huang JM, Kim H, Ekram MB, Roh TY, Kim J. DNA-binding motif and target genes of the imprinted transcription factor PEG3. Gene. 2013;512:314–320. doi: 10.1016/j.gene.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee S, Ye A, Kim J. DNA-Binding Motif of the Imprinted Transcription Factor PEG3. PLoS One. 2015;10:e0145531. doi: 10.1371/journal.pone.0145531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex- at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goyal A, Neill T, Owens RT, Schaefer L, Iozzo RV. Decorin activates AMPK, an energy sensor kinase, to induce autophagy in endothelial cells. Matrix Biol. 2014;34:46–54. doi: 10.1016/j.matbio.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ling Y, Yang Y, Lu N, You QD, Wang S, Gao Y, et al. Endostar, a novel recombinant human endostatin, exerts antiangiogenic effect via blocking VEGF-induced tyrosine phosphorylation of KDR/Flk-1 of endothelial cells. Biochem Biophys Res Commun. 2007;361:79–84. doi: 10.1016/j.bbrc.2007.06.155. [DOI] [PubMed] [Google Scholar]
- 87.Palmieri M, Impey S, Kang H, di RA, Pelz C, Sardiello M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 88.Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Settembre C, Ballabio A. TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy. 2011;7:1379–1381. doi: 10.4161/auto.7.11.17166. [DOI] [PubMed] [Google Scholar]
- 91.Neill T, Sharpe C, Owens RT, Iozzo RV. Decorin-Evoked Paternally Expressed Gene 3 (PEG3) is an Upstream Regulator of the Transcription Factor EB (TFEB) in Endothelial Cell Autophagy. J Biol Chem. 2017;292:16211–16220. doi: 10.1074/jbc.M116.769950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gubbiotti MA, Neill T, Frey H, Schaefer L, Iozzo RV. Decorin is an autophagy-inducible proteoglycan and is required for proper in vivo autophagy. Matrix Biol. 2015;48:14–25. doi: 10.1016/j.matbio.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gubbiotti MA, Iozzo RV. Proteoglycans regulate autophagy via outside-in signaling: An emerging new concept. Matrix Biol. 2015;48:6–13. doi: 10.1016/j.matbio.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ebert SM, Monteys AM, Fox DL, Bongers KS, Shields BE, Malmberg SE, et al. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol. 2010;24:790–799. doi: 10.1210/me.2009-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vecchione A, Fassan M, Anesti V, Morrione A, Goldoni S, Baldassarre G, et al. MITOSTATIN, a putative tumor suppressor on chromosome 12q24.1, is downregulated in human bladder and breast cancer. Oncogene. 2009;28:257–269. doi: 10.1038/onc.2008.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fassan M, D’Arca D, Letko J, Vecchione A, Gardiman MP, McCue P, et al. Mitostatin is down-regulated in human prostate cancer and suppresses the invasive phenotype of prostate cancer cells. PLoS ONE. 2011;6:e19771. doi: 10.1371/journal.pone.0019771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cerqua C, Anesti V, Pyakurel A, Liu D, Naon D, Wiche G, et al. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Reports. 2010;11:854–860. doi: 10.1038/embor.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ventura-Clapier R, Garnier A, Veksler W. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1α. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 99.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dagda R, Cherra SJI, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kubli DA, Gustafsson ÅB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res. 2012;111:1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Csordás G, Santra M, Reed CC, Eichstetter I, McQuillan DJ, Gross D, et al. Sustained down-regulation of the epidermal growth factor receptor by decorin. A mechanism for controlling tumor growth in vivo. J Biol Chem. 2000;275:32879–32887. doi: 10.1074/jbc.M005609200. [DOI] [PubMed] [Google Scholar]
- 105.Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- 106.Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 109.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, Kimura M, et al. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem. 2013;288:22019–22032. doi: 10.1074/jbc.M113.467530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 112.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y, Xu WW, Neill T, Hu Z, Wang CH, Xiao X, et al. Systemic Delivery of an Oncolytic Adenovirus Expressing Decorin for the Treatment of Breast Cancer Bone Metastases. Hum Gene Ther. 2015;26:813–825. doi: 10.1089/hum.2015.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee KM, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab. 2017;26:633–647. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rønnov-Jessen L, Bissell MJ. Breast cancer by proxy: can the microenvironment be both the cause and consequence? Trends Mol Med. 2009;15:5–13. doi: 10.1016/j.molmed.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gubbiotti MA, Vallet SD, Ricard-Blum S, Iozzo RV. Decorin interacting network: A comprehensive analysis of decorin-binding partners and their versatile functions. Matrix Biol. 2016;55:7–21. doi: 10.1016/j.matbio.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Neill T, Schaefer L, Iozzo RV. An oncosuppressive role for decorin. Mol Cell Oncol. 2015;2:e975645. doi: 10.4161/23723556.2014.975645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gubbiotti MA, Neill T, Iozzo RV. A current view of perlecan in physiology and pathology: A mosaic of functions. Matrix Biol. 2017;57–58:285–298. doi: 10.1016/j.matbio.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neill T, Schaefer L, Iozzo RV. Instructive roles of extracellular matrix on autophagy. Am J Pathol. 2014;184:2146–2153. doi: 10.1016/j.ajpath.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neill T, Schaefer L, Iozzo RV. Decorin as a multivalent therapeutic agent against cancer. Adv Drug Deliv Rev. 2016;97:174–185. doi: 10.1016/j.addr.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buraschi S, Neill T, Owens RT, Iniguez LA, Purkins G, Vadigepalli R, et al. Decorin protein core affects the global gene expression profile of the tumor microenvironment in a triple-negative orthotopic breast carcinoma xenograft model. PLoS ONE. 2012;7:e45559. doi: 10.1371/journal.pone.0045559. [DOI] [PMC free article] [PubMed] [Google Scholar]