Abstract
Cetacean health may be potentially affected by anthropogenic sound. We have initiated investigations on the effect of low-frequency underwater sound on immunological gene transcript profiles of captive bottlenose dolphins (Tursiops truncatus) using a probe-based quantitative gene expression assay. Six immunologic genes (IL-2Rα, -4, -10, -12, TNFα and IFNγ) were selected for analysis using two validated housekeeping genes (PGK1 and HPRT1) as reference genes. Twenty-four blood samples from six clinically healthy individuals and six blood samples from individuals after sound exposures were available. The gene transcript profile of sound-exposed dolphins was consistent with a stress-induced TH2 shift profile as compared to controls. This study may lead to better understanding of the effects of anthropogenic sound on immune responses of cetaceans.
Keywords: cytokine, dolphin, immune response, qRT-PCR, sound
Sound is vital of importance for marine animals as a means of communication and orientation [8], and to find prey [16] and socialize [22]. Marine animals may be susceptible to undesirable effects from noise in their environment. In addition to natural noises, anthropogenic noise generated by industrial construction and maintenance activities, such as pile driving in offshore wind farm construction, may have negative physiological effects or behavioral effects on marine fauna [10]. Furthermore, high-intensity underwater noise has been supposed to cause an increasing level of stress for the animals [26]. Exposure to high or cumulative noise levels in cetaceans can cause elevated stress levels, which have the ability to lead to a compromised immune system [11].
Induction of leukocyte cytokine transcription is commonplace in innate, adaptive, and various inflammatory responses. Leukocyte transcriptional biomarkers, such as cytokine genes, may be useful for cetacean health assessment because they have a broad range of functions and are associated with diseases in humans and animals [5]. Therefore, a set of cytokine transcripts in animals within a specific microenvironment may serve as indicators in many clinical and basic research studies [9]. The resulting data can provide important insights into the immune response of marine animals subjected to environmental insults such as noise, and the potential etiology of infectious diseases or stress-related illnesses. In addition to causing masking and behavioral changes, underwater noise has been proposed to cause increased levels of stress for cetaceans by measuring neural-immune changes (increased norepinephrine, epinephrine, dopamine, and aldosterone levels with increasing sound levels) [26]. The purpose of this study was to investigate anthropogenic sound as a stressor and potential impacts in dolphins by monitoring changes in the immunological gene transcripts. Although determinations of protein expression are more indicative of cytokine activities than transcript analyses, the required monoclonal antibodies are not available in all cases. The experiment was designed to measure differential transcription of immunological genes in blood samples of bottlenose dolphin (Tursiops truncatus) after exposure to low-frequency multiple impulsive sound using species-specific probe-based qRT-PCR assays. The study is intended to provide preliminary information to an understanding of the effects of sound on cetacean health and facilitating future studies on characterizing dolphin immune response to a range of anthropogenic noises. Besides, the data collected from this study in bottlenose dolphins may provide a foundation for the application of transcriptional biomarkers as diagnostic tools that reflect pathogenic processes and provide information on the status of the immune system in captive and free-ranging cetaceans.
The present target genes were selected to cover a broad range of immunological events in dolphins, and their protein products are all powerful regulators that play key roles in the interplay between innate and adaptive immune responses. The general approach in this study was to measure transcripts representing the following activities: (I) pro-inflammatory features including inducing proliferation and differentiation of T cells (IL-12) [12] and mediating cytotoxicity and cytokine secretion (TNFα) [7] (II) establishing the TH1 phenotype (IFNγ) [27] (III) suppression of TH1 cells (IL-4) [31] (IV) T cell activation (TAC) receptor (IL-2 receptor α subunit, IL-2Rα) [15] and (V) regulatory/anti-inflammatory features (IL-10) [19] that inhibits the expression of inflammatory cytokines such as IL-2, TNFα and IFNγ, and specifically promotes TH2 responses by suppressing IL-12 expression. Two validated reference genes (PGK1 and HPRT1) [6] were included for normalization.
Voluntary blood collection of dolphins by human caretakers in Farglory Ocean Park was performed in accordance with international guidelines [25]. The protocol has been reviewed and approved by Council of Agriculture of Taiwan (Approval number 1020727724 and 1031701206). Twenty-four blood samples from six clinically healthy (no obvious abnormal findings in laboratory tests, physical examination and behaviors) non-pregnant female individuals were collected on a monthly basis (six samples collected from Dolphin A; five from B, C and D, respectively; two from E; and one from F) to establish reference values. Six blood samples from two clinically healthy individuals (three each from B and C) were obtained after a noise exposure experiment. Dolphins B and C were arranged in a pool (15 × 10 m, 4.5-m deep) and received sound exposures at the same time. The noise exposure was an 800-Hz pure-tone sound (40 strikes/min, duration 150 msec) lasting 30 min, which resembles pile-driving sound. The received sound pressure level (SPL) at 1 m from the underwater transducer (LL9162T, Lubell, Whitehall, OH, U.S.A.) was 153 dB re 1 µPa and the estimated mean received SPL (measured at 31 locations in the pool) was 140 dB re 1 µPa, which are both lower than the thresholds for onset of permanent and temporary threshold shift in dolphins [21]. The exposure interval was 2 days and the exposure was repeated three times. Whole blood was collected from dolphins 5 min after noise exposure and 500-µl EDTA-anticoagulated samples were preserved in 1.3 ml of RNAlater® (Ambion, Applied Biosystems, Foster City, CA, U.S.A.) immediately, and were stored at −20°C until analysis.
RNA was isolated from frozen blood samples using RiboPure™-Blood Kits (Ambion) according to the manufacturer’s instructions. The RNase inhibitor (RNA ArmorTM Reagent, Protech, Taipei, Taiwan) was then added to samples to prevent degradation and RNA integrity was assessed using denaturing gel electrophoresis. RNA concentrations were then measured using Quant-iT™ RNA Assay Kits with a Qubit™ fluorometer (Invitrogen, Carlsbad, CA, U.S.A.). Subsequently, genomic DNA (gDNA) was removed using gDNA wipeout solution (Qiagen, Valencia, CA, U.S.A.), and was confirmed using quantitative PCR (qPCR) before reverse transcription reactions with 73–444 ng of RNA and reverse transcription kits (QuantiTect®, Qiagen). Excess RNA and cDNA samples were stored at −80°C. DNA sequences of the cytokines IL-2Rα, IL-4, IL-10, IL-12, IFNγ and TNFα from bottlenose dolphins were obtained from GenBank (Table 1) and specific primers were designed for qRT-PCR assays using web-based software (ProbeFinder, v.2.49, Roche, Pleasanton, CA, U.S.A.) with the Universal ProbeLibrary (Roche; Table 1). Primer specificity was confirmed using PCR with Fast-Run Hotstart PCR kit (Protech) and the resulting DNA fragments were subjected to gel electrophoresis.
Table 1. Name, accession number, primer sequence, probe number, amplicon size, efficiency and R2 of 6 immunologically relevant genes.
| Gene name | Accession number | Primer Sequence (5′-3′) | UPL Probe number |
Amplicon size (bp) |
Threshold | Efficiency (%) ± SD |
R2 |
|---|---|---|---|---|---|---|---|
| IL-2Rα | XM_004313501 | F-TGAACCTTTGAAGAGAATTTACCA | 112 | 72 | 0.015 | 96.43 ± 0.269 | 0.999 |
| R-CTGAATCCCTGAATGCACTG | |||||||
| IL-4 | NM_001280657.1 | F-GCATGGAGCTGCCTGTAGA | 140 | 69 | 0.01 | 95.85 ± 0.16 | 0.995 |
| R-TGCAGAAAGTTTCCTTCTCAGTT | |||||||
| IL-10 | AB775207.1 | F-AAGCCCTGTCGGAGATGAT | 25 | 86 | 0.012 | 99.49 ± 0.344 | 0.997 |
| R-CACGTGCTCTTTGATGTTGG | |||||||
| IL-12 | XM_004324402.1 | F-CAGAAGGAGCTCTTTTATGACGA | 98 | 71 | 0.015 | 97.34 ± 0.672 | 0.996 |
| R-CCATGTGGTACATCTTCAAGTCC | |||||||
| TNF-α | NM_001280615.1 | F-CCAACTGGCTACTCCATCATC | 106 | 76 | 0.01 | 95.44 ± 0.281 | 0.996 |
| R-CGGGCTTGTTACTTGAGGTT | |||||||
| IFN-γ | AB022044.2 | F-TTTTCAGCTATGCGTGATTTTG | 129 | 94 | 0.012 | 90.54 ± 2.451 | 0.997 |
| R-TGCATTAAAATATTCCTTTAGGTTTTG |
All PCR protocols were performed in accordance with the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [4]. Briefly, qPCR analyses of cDNA samples were performed using FastStart Essential DNA Probes Master (Roche) and an Eco Real Time PCR system (Illumina, San Diego, CA, U.S.A.) according to the manufacturer’s protocols. Thermal cycling conditions for all target genes were as follows: 95°C for 10 min, followed by 45 cycles of 95°C for 10 sec and 60°C for 30 sec. Reactions for samples, controls, and blanks were all performed in triplicate. Plate controls include identical reaction materials on every run. A stable quantification cycle (Cq) value from all plate controls allowed data from multiple plates to be consolidated into a single data set. Cycling thresholds were set manually (Table 1) and baseline values were assigned for all plates using Eco Software V4.0 (Illumina). PCR amplification efficiencies (E) were calculated from slopes of standard curves for each gene using E=(10(−1/slope) −1) ×100%, and are presented as means from at least three E values for each gene. Data from qRT-PCR were analyzed as normalized values (NV) as described by Pfaffl et al. [24] with minor modifications. Briefly, NVs were calculated using the equation Log2 (ETCqT/Geomean (ER1CqR1 & ER2CqR2)), where ET, ER1 and ER2 are efficiencies of target and reference genes, and CqT, CqR1 and CqR2 are Cq values of target and reference genes, so that NVs are inversely correlated with transcription levels. The Mann–Whitney U test was used for group comparison.
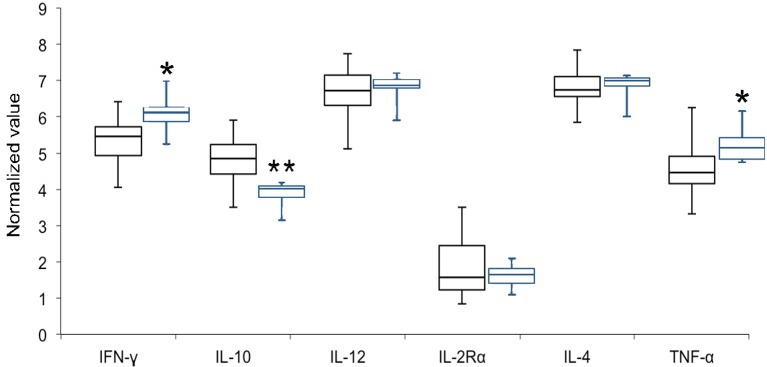
E values for the six cytokine genes ranged from 90.54 to 99.49% with R2 values of >0.99 in all cases (Table 1), and were all included in analyses of reference NVs (Fig. 1). Among 24 samples from healthy animals, median NVs of the six genes ranged from 1.58 for IL-2Rα to 6.74 for IL-4, and whereas IL-2Rα transcription levels were high (median NV<2), those of IL-10, IFNγ and TNFα were moderate and those of IL-4 and IL-12 were low (median NV>6). Normality of NV data for each gene was confirmed using Shapiro–Wilk tests (P>0.05), indicating the potential for outlier diagnostics. Two-tailed Mann-Whitney U tests indicated that gene transcript levels of IL-10 (P<0.01) were increased and IFNγ (P<0.05) and TNFα (P<0.05) were decreased in sound-exposure samples relative to both all healthy samples and “Dolphin B and C” healthy samples, whereas the other three genes did not differ significantly between sound-exposure samples and healthy samples.
Fig. 1.
Box plot of normalized value of 6 immunologically relevant genes for healthy samples (n=24, black) and noise-exposed samples (n=6, blue). Significant differences between the two sample groups are indicated by stars (*P<0.05; **P<0.01). Standard deviation/coefficient of variation of tested genes for healthy samples: IFNγ 0.68/0.13, IL-10 0.64/0.13, IL-12 0.76/0.12, IL-2Rα 0.76/0.41, IL-4 0.50/0.07 and TNFα 0.74/0.16.
The present study showed that increased IL-10 gene transcription corresponded with decreased transcript levels of IFNγ and TNFα in animals subjected to sound exposure, indicative of the stress-induced TH2 shift. Such stress-induced immune perturbations have the potential to have a profound effect by increasing their susceptibility to microbial insults [23], neoplastic changes, and immune-mediated disorders [18]. Because of experimental limitations, only 6 female bottlenose dolphins with 24 samples as reference were used in these studies. Although the appropriate non-parametric statistics was implemented, some statistically significant differences might be spurious because a number of measurements after noise exposure were evaluated in this study. More samples are needed to fully elucidate the impact of immunological perturbations in dolphins subjected to stress induced by multiple impulsive low-frequency sound. Additional parameters (such as neural-immune factors, complete blood cell counts, levels, duration and sources of sound required for TH2 shift onset, and species, age, gender, and recovery rates from TH2 shift) will need to be considered for future investigations of the noise impact on the dolphin immune response. A previous study suggested that harbor porpoises (Phocoena phocoena) become agitated and may flee from locations where they are exposed to playbacks of pile driving sound of above ~154 dB re 1 µPa [14]. Although in the present study the trainers noticed no obvious behavior change during and after the sound exposure, a behavioral response study in the pool is highly recommended accompanying with sound-exposure immunological experiment in the future.
The analysis of cetacean gene transcripts has been performed using quantitative reverse transcription PCR (qRT-PCR) with SYBR Green and a variety of different house-keeping genes (HKGs) in multiple tissues and species, including skin biopsies in killer whales (Orcinus orca) [3] and blood samples in harbor porpoises (Phocoena phocoena) [2, 20], bottlenose dolphins (T. truncatus) [17, 28, 29], killer whales [30], beluga whales (Delphinapterus leucas) and Pacific white-sided dolphins (Lagenorhynchus obliquidens) [28]. qRT-PCR has been suggested to be superior for reliable quantification in vivo [13]. While such studies have provided new and valuable information on cetacean immunologic health, probe-based qRT-PCR with validated reference genes would be a preferable method for accurate quantification of transcript abundance and detection of small changes in gene transcription [34]. The improved specificity of the probe-based qRT-PCR assays used in the current study was markedly helpful because many cytokines are produced in low abundance [32], and facilitated the identification of noise-induced immunologic perturbations in the present study.
It has been suggested that using appropriate controls (PGK1 and HPRT1) in the data normalization step is important for accurate mRNA measurements of immune-inducible genes with typical low transcript level in bottlenose dolphins [6]. The authors also emphasized that proper amplification efficiency (E) adjustment can improve qPCR data analysis with greater accuracy because the gene quantification requires corrected when efficiency is not close to 100%. In the present study, two validated reference genes and calculated PCR amplification efficiencies were implemented, which were helpful to detect small perturbations in target gene expression with high sensitivity. Furthermore, we also exploited the relative convenience of RNAlater, which requires smaller blood volumes (0.5 ml) and facilitates serial collection of samples for temporal blood transcriptome profiling studies with high quality of isolated RNA [6, 33]. The resulting accuracy was notable given the high variability of clinical samples and small differences in gene transcription.
Efforts to develop immunological research programs for assessing and maintaining cetacean health are encouraged not only because free-ranging cetaceans may serve as ideal sentinels of ecosystem health [1] but because the cetaceans under human care are important for the purposes of public display, rehabilitation, education programs and naval defense. This pioneering work on the immunologic gene transcript profiles of bottlenose dolphins subjected to sound exposure paves the way for future studies on measuring the stress of dolphins and efficacy of current noise mitigation attempts. The present data demonstrate the use of a probe-based qRT-PCR protocol using the validated reference genes PGK1 and HPRT1 for accurate and reliable analyses of IL-2Rα, -4, -10, -12, TNFα and IFNγ mRNA expression in bottlenose dolphins. The present qRT-PCR protocol was developed with specific chemistry and temperature profiles, and can be used for simple and highly sensitive determinations of gene transcript profiles. This essential tool for evaluating peripheral blood cytokine gene transcription in cetaceans will better facilitate identification of immune system perturbations induced by environmental insults, and provide diagnostic tools for characterizing immune responses to foreign antigens and vaccines.
Acknowledgments
We thank the staffs of Farglory Ocean Park for sample collection. This work was supported by Ministry of Science and Technology in Taiwan (MOST 104-3113-E-002-012). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Beineke A., Siebert U., Müller G., Baumgärtner W.2007. Increased blood interleukin-10 mRNA levels in diseased free-ranging harbor porpoises (Phocoena phocoena). Vet. Immunol. Immunopathol. 115: 100–106. doi: 10.1016/j.vetimm.2006.09.006 [DOI] [PubMed] [Google Scholar]
- 2.Beineke A., Siebert U., van Elk N., Baumgärtner W.2004. Development of a lymphocyte-transformation-assay for peripheral blood lymphocytes of the harbor porpoise and detection of cytokines using the reverse-transcription polymerase chain reaction. Vet. Immunol. Immunopathol. 98: 59–68. doi: 10.1016/j.vetimm.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 3.Buckman A. H., Veldhoen N., Ellis G., Ford J. K. B., Helbing C. C., Ross P. S.2011. PCB-associated changes in mRNA expression in killer whales (Orcinus orca) from the NE Pacific Ocean. Environ. Sci. Technol. 45: 10194–10202. doi: 10.1021/es201541j [DOI] [PubMed] [Google Scholar]
- 4.Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T.2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 5.Chaussabel D.2015. Assessment of immune status using blood transcriptomics and potential implications for global health. Semin. Immunol. 27: 58–66. doi: 10.1016/j.smim.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 6.Chen I. H., Chou L. S., Chou S. J., Wang J. H., Stott J., Blanchard M., Jen I. F., Yang W. C.2015. Selection of suitable reference genes for normalization of quantitative RT-PCR in peripheral blood samples of bottlenose dolphins (Tursiops truncatus). Sci. Rep. 5: 15425. doi: 10.1038/srep15425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark I. A.2007. How TNF was recognized as a key mechanism of disease. Cytokine Growth Factor Rev. 18: 335–343. doi: 10.1016/j.cytogfr.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 8.David J. A.2006. Likely sensitivity of bottlenose dolphins to pile-driving noise. Water Environ. J. 20: 48–54. doi: 10.1111/j.1747-6593.2005.00023.x [DOI] [Google Scholar]
- 9.de Jager W., Bourcier K., Rijkers G. T., Prakken B. J., Seyfert-Margolis V.2009. Prerequisites for cytokine measurements in clinical trials with multiplex immunoassays. BMC Immunol. 10: 52. doi: 10.1186/1471-2172-10-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erbe C.2013. International Regulation of Underwater Noise. Acoust. Aust. 41: 12–19. [Google Scholar]
- 11.Fair P. A., Becker P. R.2000. Review of stress in marine mammals. J. Aquat. Ecosyst. Stress Recovery 7: 335–354. doi: 10.1023/A:1009968113079 [DOI] [Google Scholar]
- 12.Hsieh C. S., Macatonia S. E., Tripp C. S., Wolf S. F., O’Garra A., Murphy K. M.1993. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260: 547–549. doi: 10.1126/science.8097338 [DOI] [PubMed] [Google Scholar]
- 13.Huggett J., Dheda K., Bustin S., Zumla A.2005. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6: 279–284. doi: 10.1038/sj.gene.6364190 [DOI] [PubMed] [Google Scholar]
- 14.Kastelein R. A., van Heerden D., Gransier R., Hoek L.2013. Behavioral responses of a harbor porpoise (Phocoena phocoena) to playbacks of broadband pile driving sounds. Mar. Environ. Res. 92: 206–214. doi: 10.1016/j.marenvres.2013.09.020 [DOI] [PubMed] [Google Scholar]
- 15.Liao W., Lin J. X., Leonard W. J.2011. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 23: 598–604. doi: 10.1016/j.coi.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madsen P. T., Wahlberg M., Tougaard J., Lucke K., Tyack P.2006. Wind turbine underwater noise and marine mammals: implications of current knowledge and data needs. Mar. Ecol. Prog. Ser. 309: 279–295. doi: 10.3354/meps309279 [DOI] [Google Scholar]
- 17.Mancia A., Warr G. W., Chapman R. W.2008. A transcriptomic analysis of the stress induced by capture-release health assessment studies in wild dolphins (Tursiops truncatus). Mol. Ecol. 17: 2581–2589. doi: 10.1111/j.1365-294X.2008.03784.x [DOI] [PubMed] [Google Scholar]
- 18.Moberg G. P.1987. Problems in defining stress and distress in animals. J. Am. Vet. Med. Assoc. 191: 1207–1211. [PubMed] [Google Scholar]
- 19.Mosser D. M., Zhang X.2008. Interleukin-10: new perspectives on an old cytokine. Immunol. Rev. 226: 205–218. doi: 10.1111/j.1600-065X.2008.00706.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller S., Lehnert K., Seibel H., Driver J., Ronnenberg K., Teilmann J., van Elk C., Kristensen J., Everaarts E., Siebert U.2013. Evaluation of immune and stress status in harbour porpoises (Phocoena phocoena): can hormones and mRNA expression levels serve as indicators to assess stress? BMC Vet. Res. 9: 145. doi: 10.1186/1746-6148-9-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NMFS 2016. N. M. F. S. Technical Guidance for Assessing the Effects of Anthropogenic Sound on Marine Mammal Hearing: Underwater Acoustic Thresholds for Onset of Permanent and Temporary Threshold Shifts., U.S. Dept. of Commer., NOAA. NOAA Technical Memorandum NMFS-OPR-55.
- 22.Nowacek D. P., Thorne L. H., Johnston D. W., Tyack P. L.2007. Responses of cetaceans to anthropogenic noise. Mammal Rev. 37: 81–115. doi: 10.1111/j.1365-2907.2007.00104.x [DOI] [Google Scholar]
- 23.Padgett D. A., Glaser R.2003. How stress influences the immune response. Trends Immunol. 24: 444–448. doi: 10.1016/S1471-4906(03)00173-X [DOI] [PubMed] [Google Scholar]
- 24.Pfaffl M. W., Horgan G. W., Dempfle L.2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. doi: 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez K.1999. Animal Training: Successful Animal Management Through Positive Reinforcement, Shedd Aquarium Society, Chicago. [Google Scholar]
- 26.Romano T. A., Keogh M. J., Kelly C., Feng P., Berk L., Schlundt C. E., Carder D. A., Finneran J. J.2004. Anthropogenic sound and marine mammal health: measures of the nervous and immune systems before and after intense sound exposure. Can. J. Fish. Aquat. Sci. 61: 1124–1134. doi: 10.1139/f04-055 [DOI] [Google Scholar]
- 27.Schroder K., Hertzog P. J., Ravasi T., Hume D. A.2004. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75: 163–189. doi: 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- 28.Sitt T., Bowen L., Blanchard M. T., Smith B. R., Gershwin L. J., Byrne B. A., Stott J. L.2008. Quantitation of leukocyte gene expression in cetaceans. Dev. Comp. Immunol. 32: 1253–1259. doi: 10.1016/j.dci.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 29.Sitt T., Bowen L., Blanchard M. T., Gershwin L. J., Byrne B. A., Dold C., McBain J., Stott J. L.2010. Cellular immune responses in cetaceans immunized with a porcine erysipelas vaccine. Vet. Immunol. Immunopathol. 137: 181–189. doi: 10.1016/j.vetimm.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 30.Sitt T., Bowen L., Lee C. S., Blanchard M. T., McBain J., Dold C., Stott J. L.2016. Longitudinal evaluation of leukocyte transcripts in killer whales (Orcinus Orca). Vet. Immunol. Immunopathol. 175: 7–15. doi: 10.1016/j.vetimm.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 31.Sokol C. L., Barton G. M., Farr A. G., Medzhitov R.2008. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat. Immunol. 9: 310–318. doi: 10.1038/ni1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai M. A., Chen I. H., Wang J. H., Chou S. J., Li T. H., Leu M. Y., Ho H. K., Yang W. C.2017. A probe-based qRT-PCR method to profile immunological gene expression in blood of captive beluga whales (Delphinapterus leucas). PeerJ 5: e3840. doi: 10.7717/peerj.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber D. G., Casjens S., Rozynek P., Lehnert M., Zilch-Schöneweis S., Bryk O., Taeger D., Gomolka M., Kreuzer M., Otten H., Pesch B., Johnen G., Brüning T.2010. Assessment of mRNA and microRNA stabilization in peripheral human blood for multicenter studies and biobanks. Biomark. Insights 5: 95–102. doi: 10.4137/BMI.S5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong W., Farr R., Joglekar M., Januszewski A., Hardikar A.2015. Probe-based real-time PCR approaches for quantitative measurement of microRNAs. J. Vis. Exp. 98: e52586. [DOI] [PMC free article] [PubMed] [Google Scholar]