Summary
Clear-cell renal cell carcinoma (ccRCC) exhibits a broad range of metastatic phenotypes that have not been systematically studied to date. Here, we analyzed 575 primary and 335 metastatic biopsies across 100 patients with metastatic ccRCC, including two cases sampledat post-mortem. Metastatic competence was afforded by chromosome complexity, and we identify 9p loss as a highly selected event driving metastasis and ccRCC-related mortality (p = 0.0014). Distinct patterns of metastatic dissemination were observed, including rapid progression to multiple tissue sites seeded by primary tumors of monoclonal structure. By contrast, we observed attenuated progression in cases characterized by high primary tumor heterogeneity, with metastatic competence acquired gradually and initial progression to solitary metastasis. Finally, we observed early divergence of primitive ancestral clones and protracted latency of up to two decades as a feature of pancreatic metastases.
Keywords: renal cell cancer, metastasis, evolution of metastasis, oligometastasis, solitary metastasis, cytoreductive nephrectomy, metastasectomy, chromosome instability, loss of 9p
Graphical Abstract
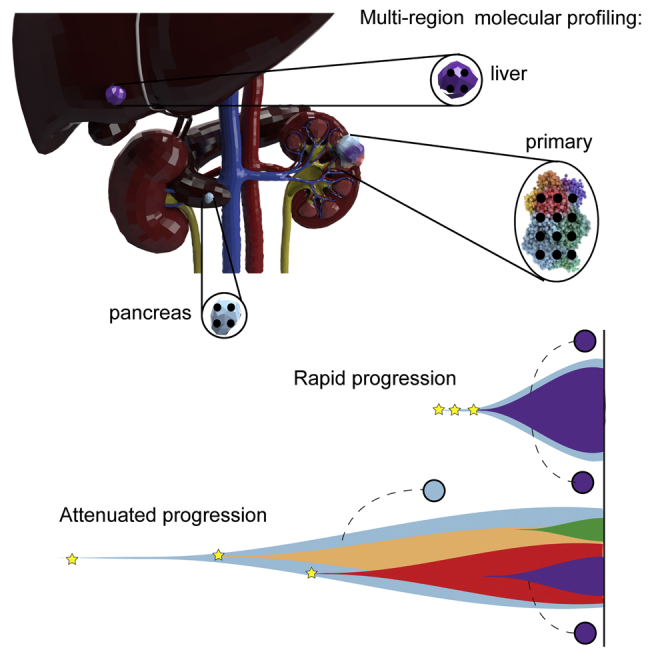
Highlights
-
•
Evolutionary study of matched primary metastasis biopsies from 100 ccRCC cases
-
•
Metastasis competence is afforded by chromosome complexity, but not driver mutation load
-
•
The hallmark genomic drivers of ccRCC metastasis are loss of 9p and 14q
-
•
Punctuated and branched evolution result in distinct patterns of metastases
A multi-center prospective study and two validation cohorts of matched primary metastasis biopsies from 100 patients with clear-cell renal cell carcinoma provides a comprehensive picture of the genetic underpinnings and the evolutionary patterns of metastasis.
Introduction
Large-scale sequencing initiatives, such as the cancer genome atlas (TCGA) and the international cancer genome consortium (ICGC), have profiled thousands of primary tumors across many cancer types. Similar large-scale studies of metastases have been limited (Robinson et al., 2017, Zehir et al., 2017) and have not included matched primary tumors. Understanding the evolution of metastatic disease requires simultaneous analysis of the primary tumor in order to distinguish between clones with and without metastatic potential. To date, a number of primary-metastasis pairs have been analyzed retrospectively (Shah et al., 2009, Campbell et al., 2010, Yachida et al., 2010, Ding et al., 2010, Haffner et al., 2013, Bashashati et al., 2013, Lee et al., 2014, Gerlinger et al., 2014, Brastianos et al., 2015, Gundem et al., 2015, Schwarz et al., 2015, McPherson et al., 2016, Zhao et al., 2016, Casuscelli et al., 2017, Yates et al., 2017). The well-established molecular landscape of primary clear-cell renal cell carcinoma (ccRCC), defined by the loss of 3p and VHL mutations/methylation as early events, provides an excellent model for the study of cancer evolution. Metastatic ccRCC, with its variable clinical presentation and natural history, is a compelling model for understanding the clonal evolution of metastasis. ccRCC spreads by both lymphatic and hematogenous routes, colonizing a range of sites, including lung, bone, liver, and brain; pancreas, adrenal, parotid, and thyroid glands; and muscle, skin, and soft tissue (Bianchi et al., 2012). Liver metastases confer a worse prognosis (McKay et al., 2014), whereas low-volume lung metastases are associated with a more indolent disease course. Some ccRCCs also grow intravascularly, forming a tumor thrombus (TT) in the renal vein/inferior vena cava. Approximately one-third of patients have metastases detected at pre-operative screening or surgery, termed “synchronous metastases.” Up to 50% develop metastases after the removal of the primary tumor (at least 3 months and as late as 30 years after primary surgery), termed “metachronous metastases.” The spatial distribution of metastatic disease in ccRCC varies from solitary (a single metastasis in a single location), oligo (limited in number and location; usually defined as ≤5 or ≤3 metastases) (Weichselbaum and Hellman, 2011), and widespread (multiple metastases in multiple sites).
The clinical relevance of solitary and oligometastases is that they can be managed by local strategies (surgery, stereotactic radiotherapy, and other ablative therapies) rather than systemic therapy. For metachronous metastases, the outcome of this approach generally depends on the time interval since the initial surgery (Dabestani et al., 2014). For patients presenting with synchronous solitary or oligometastases, a multi-modal strategy that involves cytoreductive nephrectomy (removal of the primary tumor), metastasectomy (complete resection of the metastasis) and systemic therapy can achieve an improved outcome (Bex et al., 2016). However, 20% of these patients progress as early as 1 month following surgery (Bex et al., 2017), sometimes failing to receive systemic therapy due to the rapid disease pace (Kutikov et al., 2010). Thus, there is a pressing need to understand which patients harbor more widespread occult metastases and would not benefit from surgery. In our analyses of 100 primary ccRCCs in the prospective longitudinal cohort study, TRAcking renal cell Cancer Evolution through Therapy (TRACERx Renal, clinical trials no. NCT03226886), we used conserved patterns of evolution to classify tumors into 7 distinct evolutionary subtypes (Turajlic et al., 2018a, Mitchell et al., 2018). Primary tumors with low intratumor heterogeneity (ITH) and a low fraction of the tumor genome affected by somatic copy-number alterations (SCNAs) had an overall low metastatic potential. Primary tumors with high ITH were associated with an attenuated pattern of progression, whereas primary tumors with low ITH but elevated SCNAs were associated with rapid progression at multiple sites (Turajlic et al., 2018a). A pre-defined endpoint in the TRACERx Renal study was to explore the contribution of subclonal dynamics to ccRCC metastasis. To distinguish metastasis-competent from incompetent clones and examine the routes and timing of metastases across multiple anatomic sites, we analyzed 463 primary and 169 matched metastatic regions from a subset of 38/100 patients in the TRACERx Renal Cohort (Turajlic et al., 2018a); 69 primary regions and 51 metastatic regions in an extension cohort of 26 patients; 34 matched primary metastasis pairs in a further validation cohort; and finally, 9 primary and 81 metastatic regions obtained at post-mortem in 2 patients. In total, we analyzed 5 primary and 335 metastatic biopsies from 100 patients.
Results
Overview of the Cohorts under Study
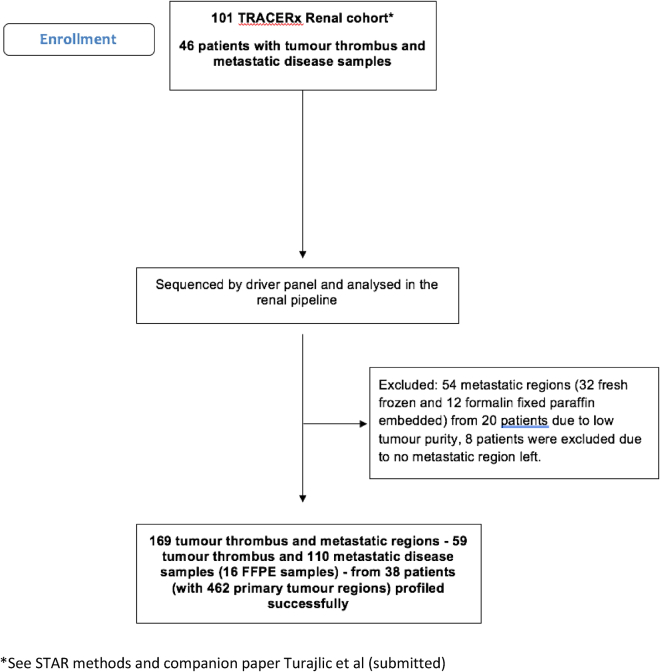
ccRCC tumors exhibit a variety of progressive phenotypes, including invasion of the peri-renal and renal sinus fat (T3a), direct invasion through the renal capsule (Gerota’s fascia) and the adrenal gland (T4), intravascular tumor growth (T3a-T3c), and lymph node (N1/N2) and visceral metastases (M1), including indirect spread to the adrenal gland. In 38 patients whose primary tumors were profiled in the TRACERx Renal cohort (Table S1A; Figure S1), we profiled multiple regions from matched TT, lymph node, and visceral metastases using a bespoke gene panel (STAR Methods).
Figure S1.
The flow diagram illustrates the phases of selection of metastatic samples included in the TRACERx Renal Primary-Metastasis cohort
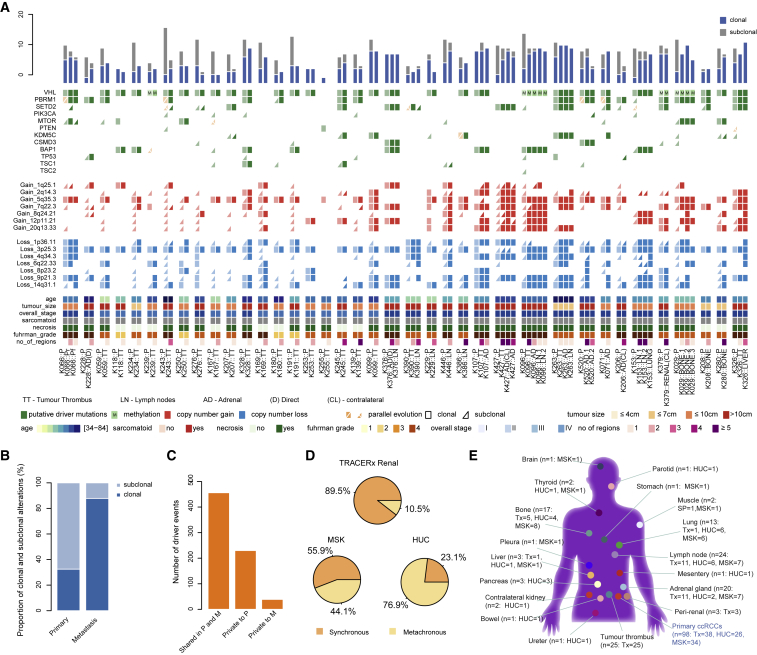
The overall number of driver events (mutations and SCNAs as presented in Figure 1A) was lower in metastases (mean = 9), compared to primary tumors (mean = 12, p = 0.05, adjusted for the varying number of profiled regions; STAR Methods) (Figure 1A). Consistent with evolutionary bottlenecking, metastases were significantly more homogeneous (proportion of clonal variants = 0.87) compared to primary tumors (proportion of clonal variants = 0.32, p = 3.6 × 10−13, adjusted for the varying number of profiled regions) (Figure 1B). Across all primary-metastasis pairs, 456 driver events were shared between primary tumors and metastases, 230 were private to primary tumors, and 39 driver events were private to metastases (Figure 1C). Driver phylogenies were reconstructed to infer clonal relationships between primary tumors and metastases (STAR Methods).
Figure 1.
Overview
(A) An overview of somatic alterations detected in matched primary and metastatic tumors across 38 TRACERx Renal patients. The top panel shows the proportion of clonal and subclonal alterations. In the middle panel alterations in primary tumors are indicated in a lighter shade and those detected in metastases in a darker shade. Clonal alterations are shown as rectangles and subclonal alterations as triangles. Parallel evolution is indicated in orange with a split indicating multiple events. Abbreviations for tumor sites: P, primary; TT, tumor thrombus; AD, adrenal gland, indirect metastasis; AD(D), direct invasion of adrenal gland; AD(CL), contralateral adrenal gland; Renal(CL), contralateral kidney; Pr, perirenal invasion; and Pf, peri-nephric fat and Gerota’s fascia invasion.
(B) The number of clonal and subclonal somatic alterations in primary and metastatic tumors.
(C) The number of somatic alterations (1) detected in both primary tumor (P) and the matched metastatic tumor (M), (2) detected in primary tumor but not the matched metastatic tumor, and (3) detected in the metastatic tumor but not the matched primary tumor.
(D) The proportions of synchronous and metachronous metastatic tumors profiled in the TRACERx Renal, HUC, and MSK cohorts.
(E) The range of the metastatic sites sampled across the TRACERx, HUC, and MSK cohorts. The total number of metastases sampled (n) and the number from each cohort are shown in brackets (Tx represents TRACERx Renal; HUC and MSK are extension cohorts).
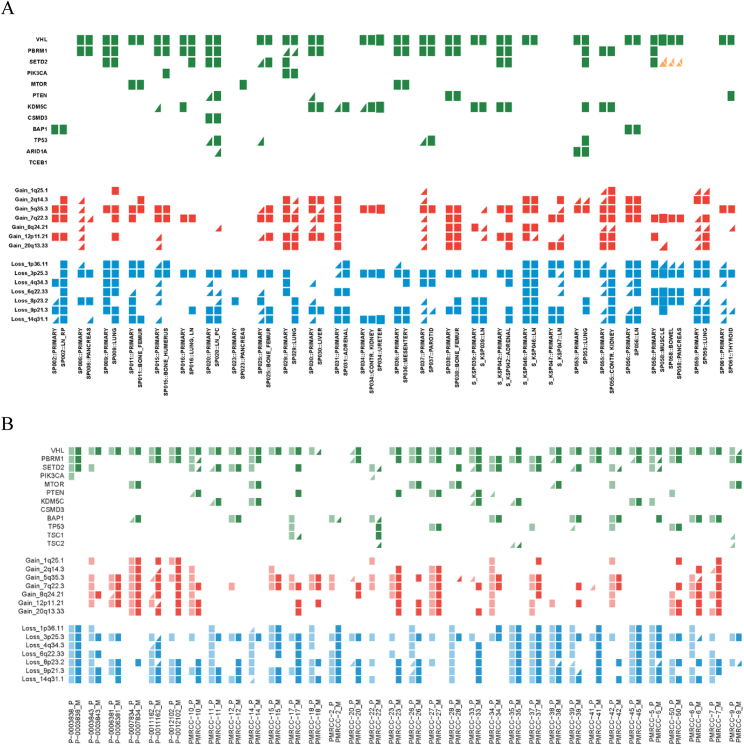
The TRACERx Renal cohort was enriched for synchronous metastases (Figure 1D), and to widen our investigation we analyzed two additional cohorts. Using the Driver Panel (STAR Methods), we multi-region profiled the “HUC” (Hospital Unversitario Cruces) cohort of archived formalin-fixed paraffin-embedded (FFPE) primary ccRCCs and matched synchronous (6 cases), and metachronous metastases (0 cases) (Table S1; STAR Methods). We successfully profiled 69 primary tumor regions and 51 metastatic regions in 26 patients (two patients contributed multiple metastases) (Figure S2). For the second cohort, “MSK” (Memorial Sloane Kettering), we reanalyzed the sequencing data from a study of primary-metastasis pairs (Becerra et al., 2017) (STAR Methods), to obtain both mutational and SCNA events in a total of 34 cases, including 19 synchronous, and 15 metachronous metastases (Figure S2; Table S1). As expected, we noted a difference in the overall frequency of driver events in the HUC and MSK cohorts compared to the TRACERx Renal cohort, owing to the increased sensitivity for detecting subclonal alterations in the latter (Table S2). There was a wide temporo-spatial representation of metastases across the three cohorts encompassing 18 distinct metastatic sites (Figure 1E), presenting 0–17 years after the removal of the primary tumor (Tables S1A–S1C). Finally, we profiled a wide range of metastatic tissues sampled at post-mortem in the context of the Cancer Research UK Posthumous Evaluation of Advanced Cancer Environment (PEACE) study (NCT03004755) in two cases of metastatic ccRCC (Table S1A).
Figure S2.
Driver Events in HUC and MSK Cohorts, Related to Figure 1
(A and B) This figure shows driver mutations and driver SCNAs detected in matched primary and metastatic tumours in HUC (A) and MSK (B) cohorts. Clonal alterations are shown as rectangles and subclonal alterations as triangles. Parallel evolution is indicated in orange with a split indicating multiple events.
Characterization of the Metastasizing Clone(s)
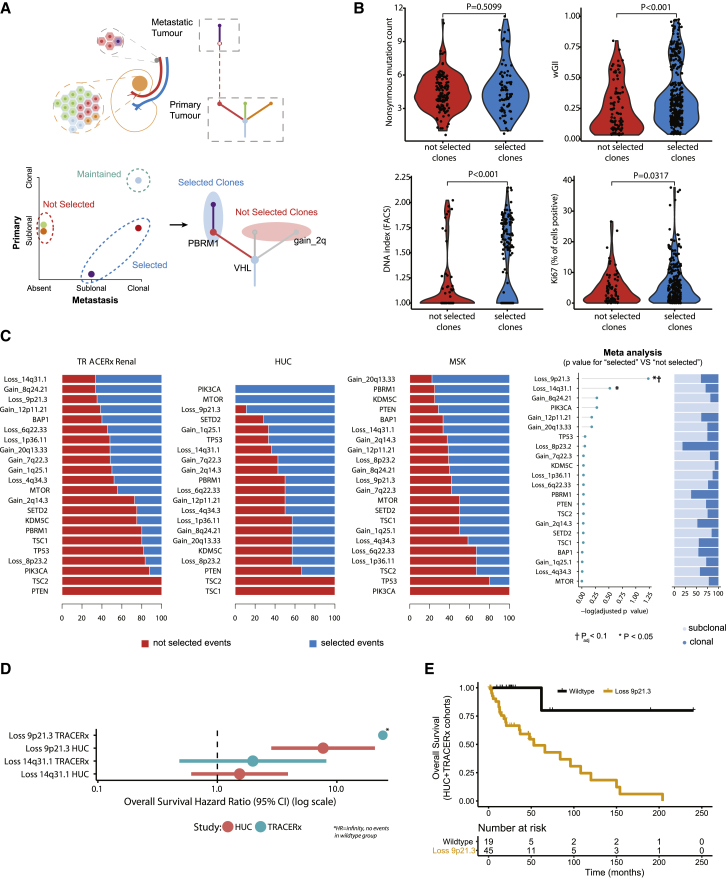
Taking advantage of the dense spatial sampling and phylogenetic reconstruction conducted in the TRACERx Renal cohort (Turajlic et al., 2018a), we analyzed the progression of individual clones from primary to metastatic sites. Across the 38 patients we observed 253 distinct tumor clones which we categorized into three groups based on the evidence of selection in the metastasis/metastases: (1) clones that are not selected (“not selected,” n = 130 clones, defined as subclonal in the primary and absent in metastasis), (2) clones that are maintained (“maintained,” n = 38 clones, defined as the most recent common ancestor (MRCA) clones, clonal in both primary and metastasis), and (3) clones that are selected (“selected,” n = 85 clones, defined as subclonal in the primary and clonal in metastasis; or absent in the primary and present in metastasis) (Figure 2A; STAR Methods). Clones that were private to the metastasis may have evaded detection as a minor subclone in the primary tumor or arisen de novo in the metastasis. The ability to differentiate the clones that appear to be selected versus not on a matched patient/tumor-specific background across the whole cohort allowed us to characterize the features associated with metastasis. We observed no difference in the number of non-synonymous mutations between the two groups (based on Driver Panel profiling, median value = 4 for both, p = 0.5099); however, wGII was significantly elevated in selected clones (median “selected” = 0.29 vs. “not selected” = 0.17, p<0.001 (Figure 2B). This was further supported by ploidy (determined by regional fluorescence activated cell sorting, FACS; STAR Methods) also being significantly elevated in selected clones (mean DNA index “selected” = 1.29, “not selected” = 1.16, p < 0.001 (Figure 2B). Multi-region immunohistochemistry staining for Ki67 (STAR Methods) demonstrated higher proliferation index in the clones that were selected, compared to those that were not (median Ki67 +40% higher in selected versus non-selected clones, p = 0.0317 (Figure 2B). Finally, we observed increased allelic imbalance at the human leukocyte antigen (HLA) locus in selected versus non-selected clones (HLA allelic imbalance observed in n = 12 “selected” vs. n = 2 “not selected” clones; Table S3), consistent with the recent findings in non-small-cell lung cancer (McGranahan et al., 2017).
Figure 2.
Characterization of a Metastasizing Clone(s)
(A) Illustration of the method used to categorize tumor clones.
(B) Four violin plots summarizing (starting at the top left and working clockwise): (1) non-synonymous mutation count, (2) wGII, (3) ploidy, and (4) Ki67. Values are compared between tumor clones “not selected” and “selected” in metastasis, with all region/clone values plotted per tumor (excluding MRCA “maintained” clones; see the STAR Methods). A linear mixed effects (LMEs) model was used to determine significance, to account for the non-independence of multiple observations from individual tumors.
(C) For each driver event the proportion of times it was observed in “not selected” and “selected” clones for TRACERx, HUC, and MSK cohorts. The far-right panel shows the log10 p value for each event for enrichment in “selected” versus “not selected” clones. Testing was performed using a binomial test with meta-analysis conducted using Fisher’s method of combining p values from independent tests. p values are corrected for multiple testing using Benjamini-Hochberg procedure.
(D) Overall survival hazard ratios for events with p < 0.05 in the analysis in (C). Data are shown for TRACERx and HUC cohorts separately, with the circle representing the hazard ratio value and the lines corresponding to the 95% confidence interval estimate.
(E) Overall survival results for TRACERx and HUC cohorts (combined), split into two groups based on SCNA status at chr 9p21.3 (either copy-number loss at chr 9p21.3 or normal wild-type copy number) in the metastasizing clone.
Next, we considered the individual driver events, mutational or SCNAs, that are selected during progression to metastasis, by comparing, for each event, the proportion of times it was found in “selected” versus “not selected” clones (Figure 2C). We conducted this analysis across TRACERx Renal (n = 38), HUC (n = 26), and MSK (n = 34) cohorts. Significance was calculated by comparing event selection proportions to null background rates as observed across all passenger events in each cohort (STAR Methods). “Selected” event frequencies were compared to "not selected", and one event was found to be significantly enriched in "selected" clones: loss of chromosome 9p21.3 (p = 0.0026, padj < 0.1 after adjustment for multiple testing (Figure 2C). We also note loss of chromosome 14q31.1 reached significance in the meta-analysis before correction for multiple testing (p = 0.0275, padj = 0.303), suggesting this and other driver events may also contribute to metastasis. We acknowledge the risk of illusion of clonality (i.e., subclonal events appearing clonal within a single region of a primary tumor) limited our power to detect metastatic selection in the MSK, and to a lesser extent HUC, cohorts. For example, 53% of events in the TRACERx Renal cohort were subclonal, compared to only 31% in HUC and 11% in MSK cohorts.
Metastatic ccRCC has a variable spectrum of survival outcomes, with overall survival (OS) times ranging from short (<6 months), to prolonged (>5 years). Accordingly, we conducted OS analysis for the two events that were enriched in metastasizing clones (p < 0.05 from Figure 2C), to understand if they were also driving early ccRCC-related mortality, based on their presence/absence within the metastasizing clone(s) of each case. OS data were not available for the MSK cohort. Hazard ratios (HR) were observed as follows (Figure 2D): 9p loss (HUC cohort HR = 7.7, [2.8–20.8] 95% confidence interval, TRACERx cohort HR = Infinity [no events in WT group], p = 0.0014 log-rank test across both cohorts) and 14q loss (HUC cohort HR = 1.5, [0.6–3.9] 95%, TRACERx cohort HR = 2.0, [0.5–8.2], non-significant). We note the strong association between reduced survival and 9p loss in the metastasizing clone remained significant after correction for known clinical variables (p = 0.046, adjusted for stage, grade, and cohort) (Figure 2E). 9p deletion has been reported to confer a poor prognosis (El-Mokadem et al., 2014, La Rochelle et al., 2010); however, the hazard ratios in our analysis (HR = 7.7; HR = infinity) are higher than reported in those studies (HR = 4.3 in El-Mokadem et al. [2014]; HR = 1.7 in La Rochelle et al. [2010]), which may reflect the greater sensitivity of profiling events within the metastasizing clones.
Evolution of Tumor Thrombus
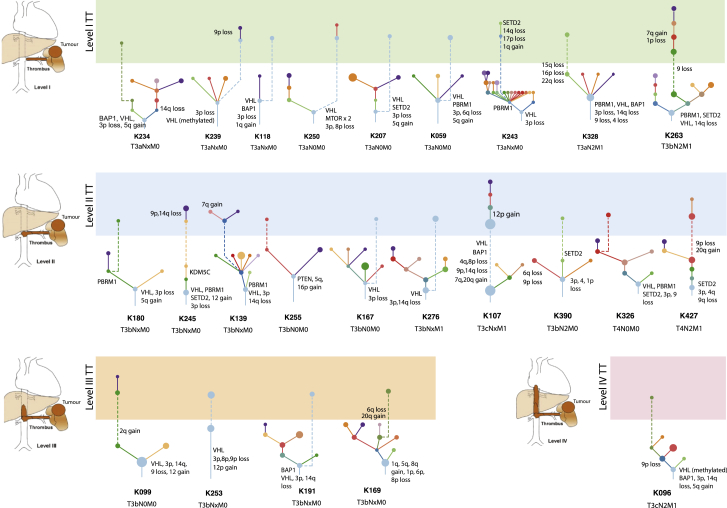
Intravascular tumor growth and formation of TT is observed in ∼15% of ccRCCs in the renal vein (level I), extending to the infrahepatic inferior vena cava (IVC) (level II), retrohepatic or suprahepatic IVC (level III), or reaching the right atrium (level IV) (Psutka and Leibovich, 2015) (Figure 3). Untreated TT is associated with a poor outcome (Reese et al., 2013), but aggressive surgical management involving a thrombectomy can result in long-term survival in some patients (Psutka and Leibovich, 2015). In the TRACERx Renal cohort 33/100 ccRCC cases presented with venous tumor extension (Turajlic et al., 2018a), only one of which was classified as a “VHL monodriver” tumor, which harbored 9p loss (K253) (Figure 3). Median survival in patients with TT was 17.8 months (Table S1) with 3 patients dying within 6 months of surgery due to disease progression (K328, K263, and K390); classified as “multiple clonal” driver (2 cases) and “VHL wild-type” (1 case) subtypes (Table S1).
Figure 3.
Tumor Thrombus
This figure shows tumor thrombus (TT) driver trees with primary clones in the lower panels and level I, level II, level III, and level IV TT clones in light green, blue, orange, and red, respectively, in the upper panels. Tumor TNM stage and driver events leading to TT are annotated. Length of branches connecting clones is not informative.
See also Figure S3.
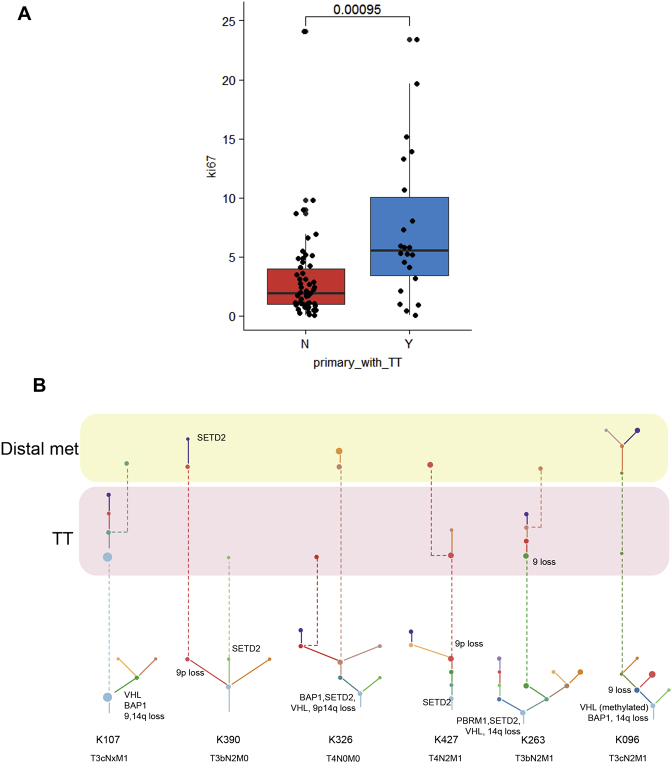
In 24/33 cases, we successfully profiled the TT along its length (Table S1) and reconstructed driver phylogenies to infer the clonal relationship between the primary tumor and the intravascular tumor extension (Figure 3). The TT was seeded directly by the most recent common ancestor (MRCA, the clone which harbors the full complement of alterations common to all the clones in the tumor; denoted by the first node in the phylogenetic tree) in 10 cases (K239, K118, K250, K207, K059, K167, K276, K107, K253, and K191) (Figure 3), suggesting intravascular growth was an early event. In other cases, the TT emerged from the more advanced subclones in the primary tumor, which harbored additional drivers, including 9p loss. Whereas most primary tumors had evidence of ongoing evolution, tumor thormbi harbored limited additional alterations (94.9% of TT events were shared with primary). Consistent with the propensity of TT to progress rapidly (Woodruff et al., 2013), we observed an elevated proliferation index within primary tumors presenting with TT compared to those without (p = 0.00095) (Figure S3A). Thus, the lack of fixation of new driver events in TTs may be due to their rapid extension and/or limited selective pressure in the intravascular space.
Figure S3.
Analysis of Tumor Thrombus, Related to Figure 3
(A and B) (A) shows Ki67 proliferation index data (mean % of cells staining positive for Ki67 across all primary tumour regions) for cases presenting with and without TT. (B) shows cases with TT and distal metastases.
An interesting biological and clinical question relates to the ability of TT to act as a source of other metastases, and in this context, we profiled six patients with venous tumor extension and concurrent lymph node and/or visceral metastases (Figure S3B). In some cases distinct clones in the primary tumor seeded the TT and the metastasis (K326 and K390; Figure S3B). Consistent with the poor prognosis conferred by lymph node involvement in ccRCC, the lymph node seeding clone in K390 harbored 9p loss, whereas the TT clone did not. The same primary clone seeded both TT and metastasis in K096 and K427 (Figure S3B), whereas in K107 and K263 (Figure S3B) the metastasizing clone appeared to first seed the thrombus and then lymph node and adrenal sites, respectively. These observations suggest that TT may act as a reservoir of metastases in some patients, consistent with the poor outcomes of untreated thrombus (Reese et al., 2013). The alternative explanation is that all the sites, including TT were seeded by a clone which evaded detection in the primary tumor.
Evolution of Progressive Disease
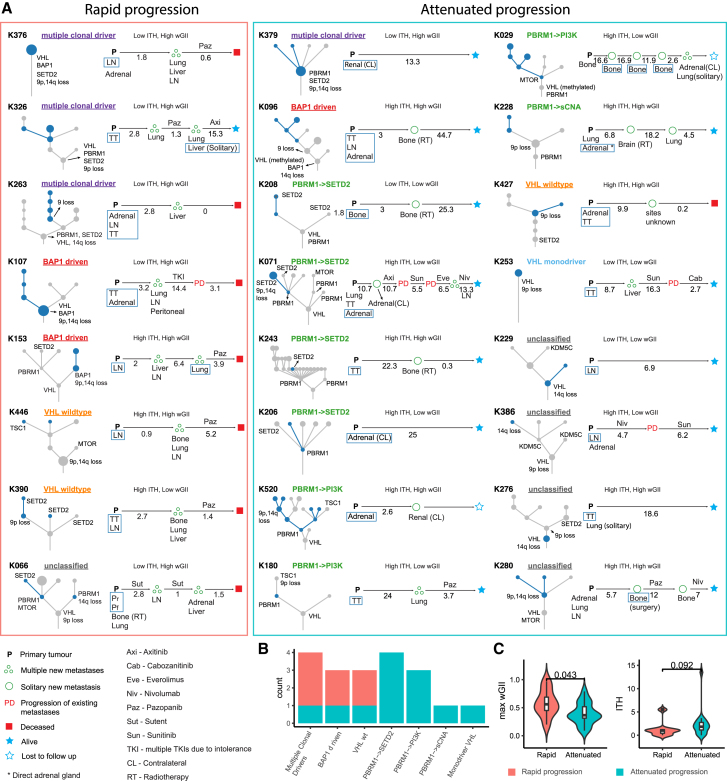
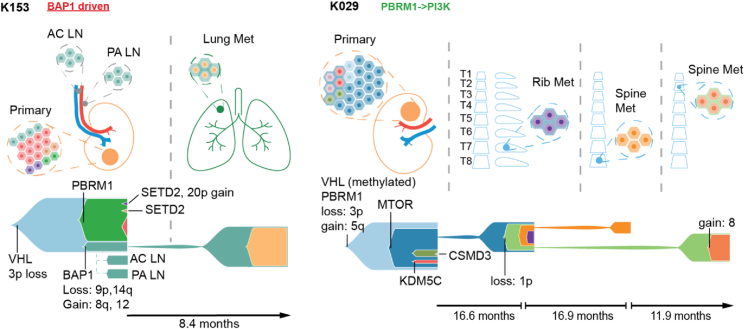
Within the 38 TRACERx Renal primary metastasis cohort, 25 patients developed progressive disease. The clinical outcomes in this group were variable, with overall survival time ranging from 1.5–54.4 months (Table 1A). Given that cytoreductive nephrectomy and metastasectomy are performed to achieve longer disease-free survival, we considered the evolutionary features of cases that progressed rapidly (i.e., multiple sites of disease progression within 6 months of surgery) versus those with attenuated progression (i.e., single-site progression < 6 months; or multi-site progression > 6 months), capturing both the speed and the extent of metastatic spread (Figure 4A; Table S1). One patient (K328) died from operative complications and was excluded from the analysis. Eight cases were classified as having “rapid progression”: K376, K326, K263, K107, K153, K446, K390 and K066 (Figure 4A). This group was enriched for “multiple clonal driver,” “VHL wild-type,” and “BAP1 driven” evolutionary subtypes (Figure 4B) and associated with lower ITH and elevated wGII relative to the cases with attenuated progression (Figure 4C). The association with ITH did not reach statistical significance, however we note that "low ITH, high wGII" tumours were linked to rapid progression in the larger cohort (p=0.01, Turajlic et al., 2018a). All primary tumors in this group harbored loss of 9p (Figure 4A). They were more likely to progress to liver metastases (6/8) compared to cases in the “attenuated progression” group (1/16) (p = 0.0013), and had a short overall survival (Figure 4A). Particularly notable was case K153 in which lymph node and lung metastases were seeded from the same “BAP1 driven” subclone, which had high wGII and harbored 9p loss, while the competing “PBRM1→SETD2” subclone failed to metastasize (Figure S4).
Figure 4.
Evolution of Progressive Disease
(A) Driver trees and the clinical course for cases with lymph node and distal metastases. Cases were grouped into those with “rapid progression” and “attenuated progression.” The primary tumor evolutionary subtype, primary tumor ITH/wGII classification, and select driver events on the tree (VHL, BAP1, PBRM1, MTOR, SETD2, TSC1, TSC2, chr 9p loss, and chr 14q loss) are annotated for each case. Metastasis seeding subclones and any subclones private to metastasis are highlighted in blue. Clinical course is shown from the time of nephrectomy to death or last follow-up. Pattern of disease progression is classified as multiple new metastases (multiple circles), solitary new metastasis (single circle), and progression of existing metastases (“PD”). Progression and follow-up times are shown in months. Systemic treatments are indicated. Synchronous and metachronous metastatic sites are listed under corresponding time points. Profiled metastases are highlighted in blue boxes. Abbreviation for tumor sites: P, primary; TT, tumor thrombus; AD, adrenal gland; AD(D), direct invasion of adrenal gland; AD(CL), contralateral adrenal gland; renal(CL), contralateral kidney; Pr, perirenal invasion; and Pf, peri-nephric fat and Gerota’s fascia invasion.
(B) The number of cases with “rapid progression” or “attenuated progression” in each evolutionary subtype.
(C) The maximum wGII and ITH in cases with “rapid progression” and “attenuated progression.”
Figure S4.
Fishplot Summary of Selected Cases, Related to Figure 4
This figure shows 2 example cases with distal metastases. Diagrams of the primary tumour and the involved tissue sites are illustrated. Fishplots are used to show disease evolution. Driver events are annotated on each fishplot.
16 cases were classified within the “attenuated progression” group: K379, K096, K208, K071, K243, K206, K520, K180, K029, K228, K427, K253, K229, K386, K276, K280 (Figure 4A). Disease progression interval was longer and often limited to a single metastatic site. Consequently, in some patients metastatic disease was controlled with further surgery (K029) or radiotherapy (K096, K228, K208 and K243), consistent with the lack of other occult metastases. This group was enriched for “PBRM1→SETD2” and “PBRM1→PI3K” evolutionary subtypes, with the primary tumors characterized by higher ITH index and lower wGII, as compared to the “rapid progression” group (Figure 4C). For example, case K029 (“PBRM1→PI3K”) presented with spatially separate bone metastases 3 years apart; the metastasizing clone harbored a PBRM1 mutation, but not 9p loss (Figure S4).
Evolution of Latent Metastases
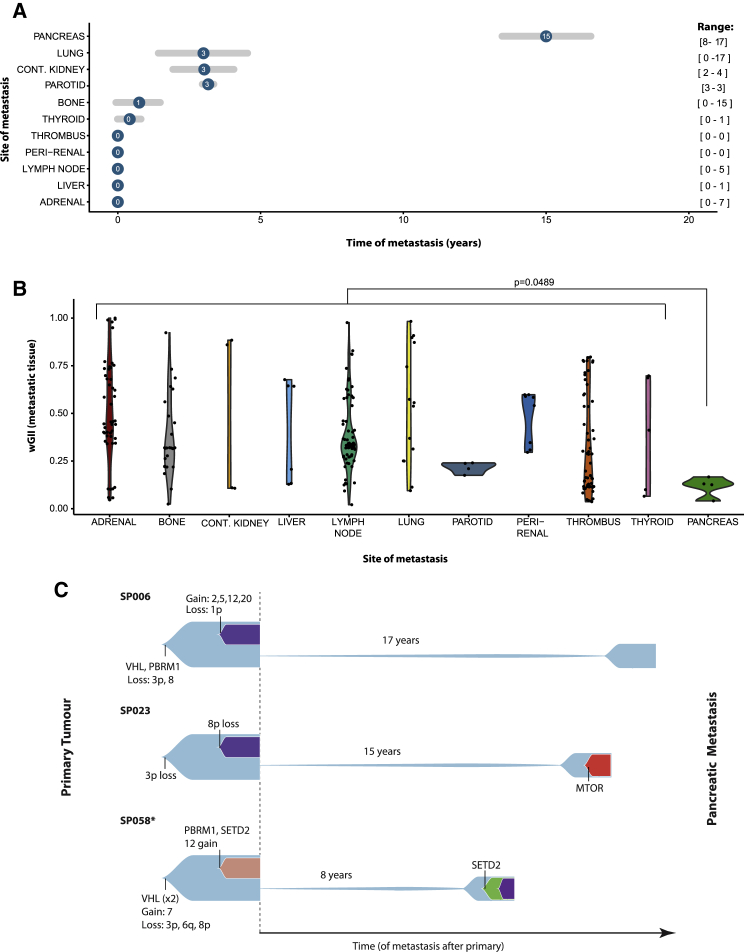
We compared the time from primary tumor to metastasis, by tissue site, across the combined TRACERx/HUC/MSK cohorts. In keeping with the known modes of late recurrence in ccRCC, we observed the pancreatic metastases to have the longest time to presentation (median 15 years, compared to 3 years for all other tissue sites) (Figure 5A). In an exploratory analysis, pancreatic metastases were found to have significantly lower wGII, as compared to all other metastatic tissue sites (p = 0.0489) (Figure 5B). A shared clonal ancestry was confirmed between primary and metastatic sites in all three cases, and we observed a low number of additional driver alterations in pancreatic metastases, despite the extended latency time (Figure 5C). In the case of SP006, the pancreatic metastasis was diagnosed 17 years after the primary tumor was resected, and the latent clone was mapped directly back to the founding MRCA clone, suggesting early divergence from the primitive ancestral clone. Similarly, in SP023, a case with pancreatic metastasis at 15 years, the latent clone derived from the primary MRCA and only acquired one additional driver mutation in MTOR (Figures 5C and S5). Finally, SP058 presented with pancreatic metastasis at 8 years, with a single additional driver event (SETD2 mutation) in metastasis, while we detected alternative subclones with a greater number of driver events in the primary tumor (Figure 5C). The seeding by the ancestral clone and the lack of 9p loss suggests that the pancreas may be a more permissive metastatic niche for ccRCC. The reasons for the characteristic latency of pancreatic metastases remain unknown, but are likely to involve interactions with the tumor microenvironment, the immune system and altered epigenetic states (Giancotti, 2013).
Figure 5.
Latent Metastases
(A) The distribution of times from nephrectomy to metastasis resection split by site of metastasis. The circle represents the median value, and the gray lines depict the median average deviation (MAD) value (i.e., plus/minus one MAD). The range (min to max) values are far right in brackets.
(B) wGII values per region split by site of metastasis. All regions are shown per metastasis, and a linear mixed effects (LMEs) model was used to determine significance (for pancreas vs. all other), to account for the non-independence of multiple observations from individual tumors.
(C) Fishplots for the three cases (SP006, SP023, and SP058) with latent pancreatic metastases.
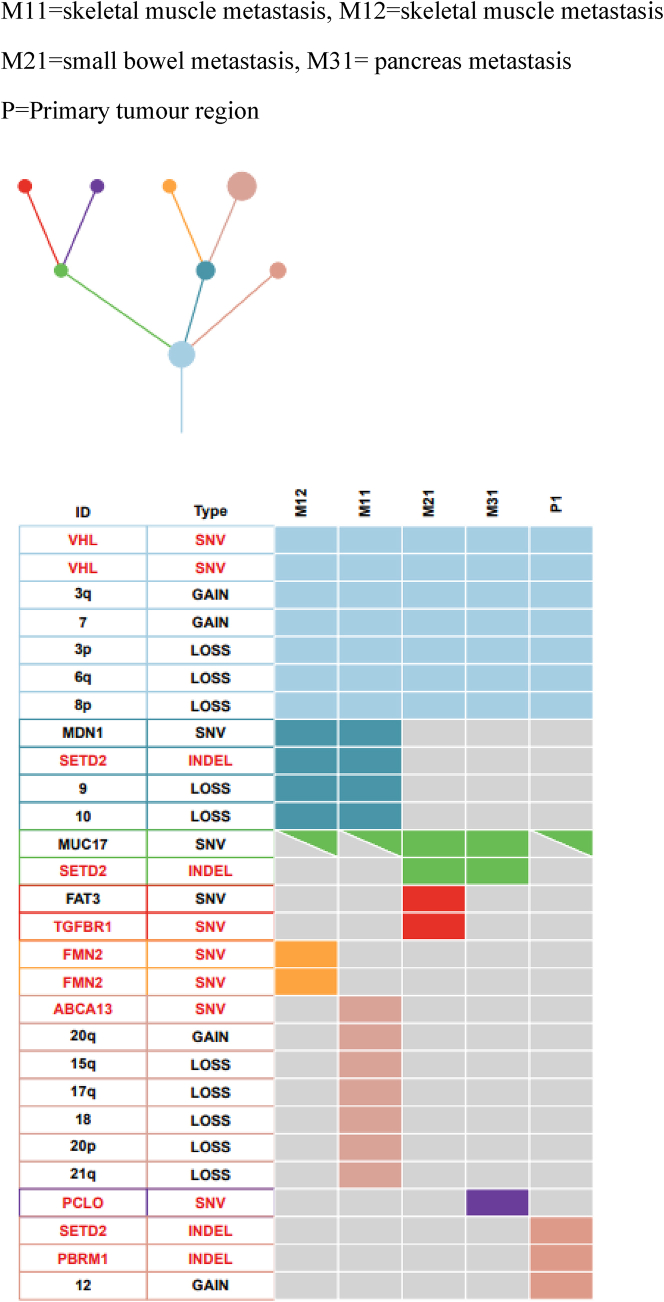
*Case SP058 had additional metastases to skeletal muscle (time = 0) and the small bowel (time = 7). See also Figure S5.
Figure S5.
Shown is the driver mutation and SCNA phylogenetic tree and heatmap illustrating the clonal relationship between the primary and metastasis for case SP58
Spatial Resolution of Metastases through Post-Mortem Sampling
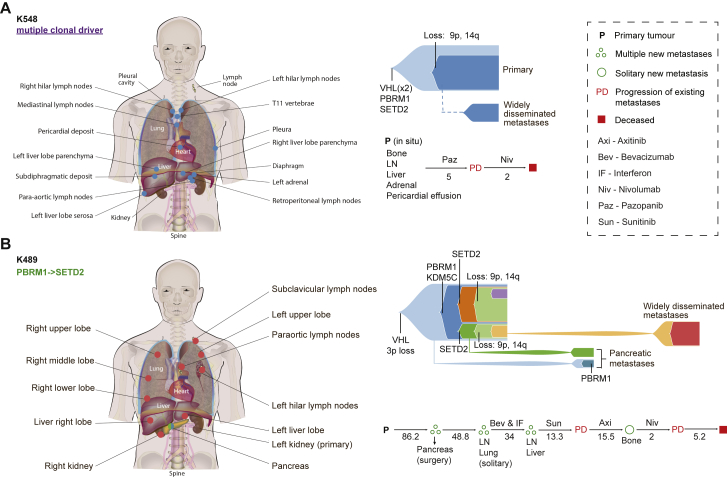
To explore the clonal dynamics of multiple metastases we sampled two cases at post-mortem(Figure 6; Table S1A) through the PEACE study (NCT03004755). Case K548 presented with a primary ccRCC which had already disseminated to multiple sites including adrenal, loco-regional and mediastinal lymph nodes, liver, and pleura (Table S1). All disease sites, including the primary tumor, were sampled at post-mortem (Figure 6A). Clonal mutations were detected in VHL, PBRM1, and SETD2 genes, and accordingly this case was categorized as a “multiple clonal driver” subtype. The primary tumor had low ITH and high wGII, and all 13 metastatic sites sampled were seeded by the dominant clone which was characterized by 9p and 14q loss (Figure 6A). We note this patient progressed rapidly through two lines of systemic therapy and died 6 months after the diagnosis of ccRCC (Table S1). The evolutionary features of the primary tumor are in keeping with those we observe in the TRACERx Renal cases with “rapid progression” (Figure 4A).
Figure 6.
Spatial Resolution of Metastases through Post-Mortem Sampling
(A and B) Shows cases K548 (A) and K489 (B), which were sampled at post-mortem with the extent of sampling. The clinical time course (in months) and therapuetic interventions are shown. Metastatic progression is illustrated using fish plots with the select driver events annotated (VHL, BAP1, PBRM1, MTOR, SETD2, TSC1, TSC2, 9p loss, and 14q loss). Metastasizing clone color matches that of the corresponding metastatic site.
See also Table S1.
In case K489 the patient presented with a primary ccRCC and underwent a nephrectomy with curative intent (Figure 6B). 7 years following surgery two pancreatic metastases were detected on imaging and the patient underwent a complete metastasectomy (Figure 6B; Table 1). 4 years later, they presented with lymph node and lung metastases (Figure 6B). They received multiple lines of systemic therapy, subsequently developing metastases at additional sites including liver and bone, and succumbing to their disease 17 years after the original diagnosis (Figure 6B; Table 1). We obtained fresh samples at post-mortem from multiple lymph node sites, liver, lung, and contralateral kidney metastases, and we accessed the primary tumor and the resected pancreatic metastases from archived FFPE material. The primary tumor harbored a clonal VHL mutation and 3p loss, and a subclonal PBRM1 and multiple SETD2 mutations, indicating parallel evolution. These features were consistent with the “PBRM1→SETD2” evolutionary subtype (Turajlic et al., 2018a). In accordance with our observations in the TRACERx renal cohort (Figure 4), the pattern of disease spread was consistent with “attenuated progression.” The two pancreatic metastases were seeded by separate clones (indicating potentially distinct waves of metastatic spread) neither of which harbored 9p loss. By contrast, subsequent metastases to the lymph nodes, liver, lung, and kidney were seeded by a subclone harboring additional SCNA events, including loss of 9p.
Discussion
We present an integrated analysis of 575 primary and 335 metastatic biopsies across 100 patients with metastatic ccRCC, including the first report of ccRCC profiled in a post-mortem setting. To our knowledge, this is the largest study of its kind to date, offering broad insights into the diverse spectrum of modes of progression from primary to metastatic disease. A key objective of the TRACERx Renal study is to reduce sampling bias and provide clonal resolution of the primary tumor, such that the metastasis seeding clones can be distinguished from metastasis incompetent clones more reliably. Clonal resolution facilitates an improved understanding of the genomic events, and broader clonal characteristics, that drive metastasis and mortality risk. In addition, the wide range of metastatic tissue sites sampled in this study allows detailed analysis of the varying metastatic phenotypes in ccRCC.
First, in characterizing metastases, we show profound evidence of evolutionary bottlenecking, with metastatic sites being both more homogeneous (proportion of clonal variants = 0.87) and harboring fewer driver somatic alterations (mean = 9), compared to their matched primary tumors (0.32 and 12). Furthermore, only a minority of driver events (5.4%) were found to be private (or de novo) in metastases, indicating that the majority of driver diversity accumulated at the primary tumor site, which then serves as the substrate for selection of metastasis-competent populations. Tumor clones that were “selected” and progressed from primary to metastatic sites of disease were characterized by elevated levels of somatic copy-number alterations, increased proliferation, and evidence of immune evasion (in the form of HLA LOH), but not by increased driver SNV/INDEL counts. 9p loss was found to be a potent driver of both metastasis and ccRCC mortality risk, even after adjustment for established clinically prognostic indicators. Loss of 14q also showed a trend towards significance, and taken together these two events represent hallmark genomic alterations in ccRCC metastasis (overall 36 of 38 TRACERx Renal cases had loss of at least one of these chromosome arms). Furthermore, 71% (n = 27) of the metastatic cases in the TRACERx renal cohort had loss of both 9p and 14q, compared to only 35% (n = 22) of cases without metastatic disease at presentation (n = 62) (Turajlic et al., 2018a), suggesting these events interact to drive metastatic risk. While investigation of functional mechanisms is beyond the scope of this study, we note that p16 (encoded by CDKN2A on 9p) has been shown to modulate VEGF expression via its interaction with HIF-1alpha, encoded by HIF1A on 14q (Zhang et al., 2010). Critically, in the context of their potential utility as biomarkers both 9p and 14q loss were predominantly subclonal in our multi-regional analysis of primary ccRCC (Turajlic et al., 2018a) and may be missed by single biopsy approaches.
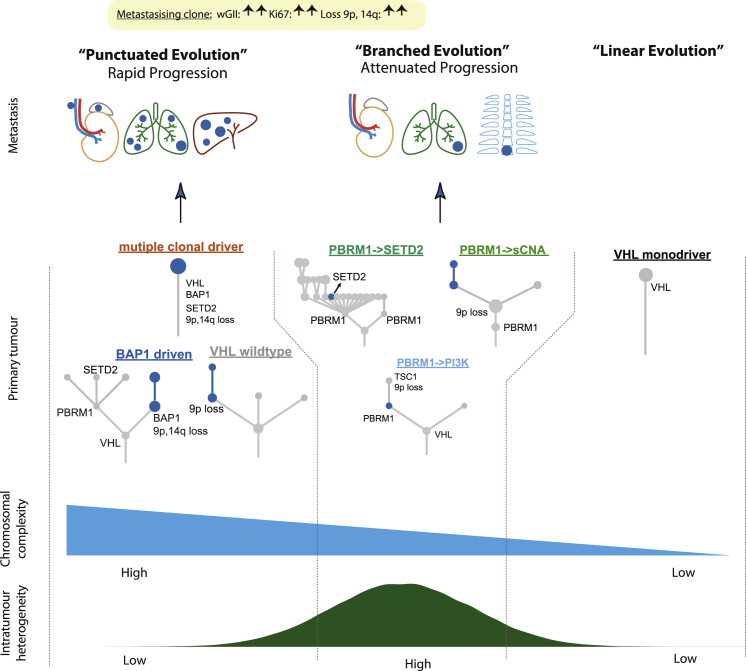
Secondly, our analyses highlight distinct modes of metastatic dissemination (Figure 7). In primary tumors characterized by low ITH and high wGII, metastatic competence is acquired within the most recent common ancestor, which drives rapid dissemination, leading to surgical failure, poor response to systemic therapy and early death from disease. These observations are consistent with the presence of occult micrometastases at the time of surgery. The “multiclonal driver” case K548, examined at post-mortem, is an exemplar of the disseminated metastatic seeding from such tumors. In this context, we note that these cases are a minority in the TRACERx Renal cohort which was weighted towards operable patients (Turajlic et al., 2018a); hence, low ITH/high wGII pattern may be prevalent in patients who are deemed inoperable. Acquisition of the metastatic potential at the early stage of tumor evolution has been reported in pancreatic (Notta et al., 2016) and breast cancers (Gao et al., 2016) and uveal melanoma (Field et al., 2018), consistent with the tendency of some tumors to metastasize rapidly. Improving outcomes in such cases presents a significant challenge.
Figure 7.
Summary of Key Conclusions from the Study
We observed a contrasting phenotype in primary tumors characterized by lower wGII, high ITH, and “attenuated progression” (Figure 4). Metastatic competence was acquired gradually and was limited to certain subpopulations in the primary tumor at the time of surgical resection. The clinical course was characterized by an initial solitary or oligometastatic pattern, with metastatic capacity increasing over time resulting in more efficient and widespread metastases, as exemplified by case K489. The marked latency of metastases in case K489 may have been mediated by effective immune surveillance. Our observations in this group underpin the role of cytoreductive nephrectomy in removing the “evolutionary sink of diversity” (Gerlinger et al., 2012) and thus minimizing the risk of future metastatic seeding from evolving primary tumors harboring clones of variable metastatic potential. We note that the evolutionary trajectories in this group are underpinned by PBRM1 alterations, in keeping with the observation by Brugarolas and colleagues that loss of PBRM1 expression is associated with an increased risk of metastasis but not with decreased survival (Joseph et al., 2016). Further OS analysis in larger metastatic cohorts will be required to comprehensively contrast the drivers of metastasis from the drivers of early mortality.
In an exploratory analysis of intravascular tumor growth, we observe that TT can be seeded from the ancestral clone. In this context, the TT conceivably formed shortly after the clonal sweep in the primary tumor, ascending rapidly through the IVC, resulting in clinical diagnosis (majority of patients presented with symptoms). This notion is consistent with the observation that TT is not prognostic in the absence of nodal or metastatic disease (Wszolek et al., 2008) and that thrombectomy can be curative. In contrast, isolated lymph node involvement portends an extremely poor prognosis in ccRCC, significantly worse than in other solid tumor types (Gershman et al., 2017). We observe that lymph node metastases are characterized by similar levels of wGII compared to distant metastases (no significant difference, p = 0.21) and frequently harbor 9p loss (21/22 cases), indicating that lymphatic and hematogenous spread require comparable metastatic competence. Our observations contrast the divergent lymph node/distant metastasis patterns reported in other tumor types (Brastianos et al., 2015, Yates et al., 2017), and are consistent with the frequent presentation of lymph node metastases with visceral metastases, and the lack of therapeutic benefit from lymph node clearance in ccRCC (Bhindi et al., 2018). Finally, in our limited analyses of late recurrences in the pancreas (3 HUC and 1 PEACE case), the metastasis-establishing clone diverged from the primary tumor early and harbored few additional events, consistent with protracted latency. The metastasizing clones lacked 9p loss, suggesting that less aggressive clones establish pancreatic metastasis in isolation in keeping with the excellent clinical outcome in these patients.
In summary, we demonstrate that the fitness attribute common to metastases and TT-seeding (sub)clones is chromosomal complexity. Chromosome-level alterations that simultaneously affect the expression of 100s of genes can support the complex metastatic cascade by altering many functional phenotypes (Santaguida and Amon, 2015) and potentially by impacting immune evasion (Davoli et al., 2017). The onset of chromosomal complexity in ccRCC provides a permissive genomic background for selection of 9p loss, previously linked to poor outcomes (El-Mokadem et al., 2014, Klatte et al., 2009, La Rochelle et al., 2010). While preliminary in nature, our collective observations point to the deterministic nature of the ccRCC evolutionary subtypes and their association with the mode and tempo of metastatic progression. The “rapid progression” group has echoes of the punctuated equilibrium model of rapid speciation events (Eldredge and Gould, 1997) and subsequent clonal stasis. In contrast, the evolution of the “attenuated progression” group is analogous to Darwin’s phyletic gradualism. Continuing longitudinal and post-mortem sampling opportunities in the TRACERx Renal and PEACE studies aim to validate these findings in larger cohorts.
In conclusion, evolutionary classification of metastatic potential could serve as an important biomarker for stratification of patients for surgical intervention (e.g., cytoreductive nephrectomy/metastasectomy) in the presence of metastatic disease, for management of patients following surgery with curative intent, including decisions on surveillance schedule and adjuvant therapy, and in the context of active surveillance of small renal masses.
STAR★Methods
Key Resources Table
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Charles Swanton (Charles.swanton@crick.ac.uk).
Experimental Model and Subject Details
Patients were recruited into TRACERx Renal, an ethically approved prospective cohort study (National Health Service Research Ethics Committee approval 11/LO/1996). The study sponsor is the Royal Marsden NHS Foundation Trust. The study is coordinated by the Renal Unit at the Royal Marsden Hospital NHS Foundation Trust. The study is open to recruitment at the following sites: Royal Marsden Hospital NHS Foundation Trust, Guy’s and St Thomas’ Hospital NHS Foundation Trust, Royal Free Hospital NHS Foundation Trust and Western General Hospital (NHS Lothian). Patients were recruited into the study according to the following eligibility criteria:
Inclusion criteria
-
•
Age 18- years or older
-
•
Patients with histologically confirmed renal cell carcinoma, or suspected renal cell carcinoma, proceeding to nephrectomy/metastectomy
-
•
Medical and/or surgical management in accordance with national and/or local guidelines
-
•
Written informed consent (permitting fresh tissue sampling and blood collection; access to archived diagnostic material and anonymised clinical data)
Exclusion criteria
-
•
Any concomitant medical or psychiatric problems which, in the opinion of the investigator, would prevent completion of treatment or follow-up
-
•
Lack of adequate tissue
Further eligibility criteria were applied to the cohort presented in this paper (it therefore follows that these patients do not have consecutive study ID numbers from 001 to 100):
-
•
Confirmed histological diagnosis of clear cell renal cell carcinoma.
-
•
No family history of renal cell carcinoma.
-
•
No known germline renal cell carcinoma predisposition syndrome (including VHL).
-
•
At least three primary tumour regions available for analysis.
The cohort was representative of patients eligible for curative or cytoreductive nephrectomy. Full clinical characteristics are provided in Table S1. Demographic data include: Sex, Age and Ethnicity. Clinical data include: Presenting symptoms, Smoking status, BMI, History of Previous RCC, Family History of RCC, Bilateral or Multi-focal RCC, Neoadjuvant therapy (6 patients received systemic therapy prior to nephrectomy). Histology data include: overall TNM Stage (based on Version 7 classification), Location of nephrectomy, Number of harvested and involved lymph nodes, presence of Microvascular Invasion, presence of Renal Vein Invasion, presence of IVC tumour thrombus, Size of primary tumour, Leibovich score, Fuhrman Grade, Time to nephrectomy (days). Clinical status of patients included: Relapse -free survival (months), Total follow up (months), Survival Outcome.
Extension cohort of primary and metastatic (P-M) pairs was accessed under the approval of Basque Country Research Ethics Committee, Hospital Universitario Cruces (Ref CEIC-Euskadi PI2015101).
Post-mortem sampling was performed in the context of the PEACE study (National Health Service Research Ethics Committee approval 13/LO/0972/AM05); https://clinicaltrials.gov/ct2/show/NCT03004755.
Method Details
Sample collection (TRACERx cohort and post-mortem sampling)
All surgically resected specimens were reviewed macroscopically by a pathologist to guide multi-region sampling for this study and to avoid compromising diagnostic requirements. Tumour measurements were recorded and the specimen were photographed before and after sampling. Primary tumours were dissected along the longest axes and spatially separated regions sampled from the “tumour slice” using a 6 mm punch biopsy needle. The punch was changed between samples to avoid contamination. The total number of samples obtained reflects the tumour size with a minimum of 3 biopsies that are non-overlapping and equally spaced. However, areas which are obviously fibrotic or haemorrhagic are avoided during sampling and every attempt is made to reflect macroscopically heterogeneous tumour areas. Primary tumour regions are labelled as R1, R2, R3…and locations are recorded. Normal kidney tissue was sampled from areas distant to the primary tumour and labelled N1. Each biopsy was split into two for snap freezing and formalin fixing respectively, such that the fresh frozen sample has its mirror image in the formalin-fixed sample which is subsequently paraffin embedded. Fresh samples were placed in a 1.8 ml cryotube and immediately snap frozen in liquid nitrogen for >30 seconds and transferred to -80 C for storage. Peripheral blood was collected at the time of surgery and processed to separate buffy coat.
Nucleic acid isolation from tissue and blood (TRACERx and PEACE cohorts)
DNA and RNA were co-purified using the AllPrep DNA/RNA mini kit. (Qiagen). Briefly, a 2mm3 piece of tissue was added to 900ul of lysis buffer and homogenised for five seconds using the TissueRaptor (Qiagen) with a fresh homogenisation probe being used for each preparation. Each lysate was applied to a QiaShredder (Qiagen) and then sequentially purified using the DNA and RNA columns according to the manufacturers protocol. Germline control DNA was isolated from whole blood using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturers protocol. DNA quality and yield was measured and accessed using the TapeStation (Agilent) and Qubit Fluorometric quantification (ThermoFisher Scientific).
Purification of DNA from Formalin Fixed Paraffin Embedded (FFPE) tissue
For a minority of TRACERx Renal cases (n = 8), tumour material was obtained from FFPE material (Table S4). An H&E section from all patient FFPE blocks is reviewed by a pathologist and tumour rich regions are identified for DNA purification. Either a 20uM sections is cut and the area of interest scraped from the slide using a blade alternatively a 2mm core is directly punched from the block. DNA is purified using the GeneRead DNA FFPE kit (Qiagen) with yields and quality being determined by Qubit quantification and TapeStation analysis.
Micro-dissection and nucleic acid isolation (HUC extension cohort)
H&E slides from each case were annotated by pathologists for regions of interest (ROI). Multiple ROIs within the primary tumour were selected on the bases of good tissue preservation avoiding areas of necrosis and haemorrhage, and to reflect microscopically distinct areas with regards to grade (high vs low) and morphology (clear vs. granular/eosinophilic), and sarcomatoid differentiation, where present, as well as areas of normal tissue. The annotated H&E was then used as a reference to guide the dissection of ROIs from serial sections. All tissue sections were cut to 10 μm thickness and deparaffinized with three, five minute incubations in xylene prior to dissection using the alpha AVENIO Millisect System (Roche Diagnostics, Indianapolis, IN) (Adey et al., 2013). The milling tip blade size for the dissection was selected based on the estimated area of the ROI, where small ROIs less than 200mm2 used small blade sizes (200 or 400μm) and ROIs larger than 200mm2 used larger blade sizes (800 μm). The milling buffer for all dissections was 1x TE buffer with 2% SDS, pH 7.5. Genomic DNA was isolated from each of the dissected FFPE tissue samples using a High Pure FFPE DNA Isolation kit (Roche).
Methylation specific PCR
Methylation of the VHL promoter was detected after bisulphite treatment of 500ng of patient DNA using the EZ DNA Methylation-Direct kit (Zymo Research). Bisulphite treated DNA was amplified in the PCR using methylation specific oligonucleotides (oligonucleotide sequences are detailed in Table S5), followed by Big Dye terminator Sanger sequencing. Methylation was confirmed by comparing and contrasting patient tumour and normal renal tissue for methylation protected CpG sequences.
Regional staining by Immunohistochemistry and Digital Image Analysis of Ki67
Tissue sections of 4μm were mounted on slides and immunohistochemical staining for Ki67 was performed using a fully automated immunohistochemistry (IHC) system and ready-to-use optimized reagents according to the manufacturer's recommendations (Ventana Discovery Ultra, Ventana, Arizona, USA). Primary antibody used was rabbit anti-Ki67 (AB16667, Abcam, Cambridge, UK) and secondary antibody was Discovery Omnimap anti-rabbit HRP RUO (760-4311, Roche, Rotkreuz, Switzerland). DAB kit was Discovery Chromomap DAB RUO (760-4311, Roche). After IHC procedure, slides were first evaluated for Ki67 staining quality using mouse intestine tissue as positive control. Regions containing tumour tissue were identified and marked by a pathologist and subsequently scanned in brightfield at 20x magnification using Zeiss Axio Scan.Z1 and ZEN lite imaging software (Carl Zeiss Microscopy GmbH, Jena, Germany). Digital images were then subjected to automated image analysis using StrataQuest version 5 (TissueGnostics, Vienna, Austria) for Ki67 quantification. Three different gates were set to quantify low, medium and high intensity DAB staining which corresponded to Ki67 expression levels. Results were depicted as total percentage of Ki67-positive nuclei.
Flow Cytometry Determination of DNA Content (FACS)
Fresh frozen tumour tissue samples, approximately 4mm3 in size, were mechanically disrupted and incubated in 2ml of 0.5% pepsin solution (Sigma, UK) at 37 ºC for 40 minutes to create a suspension of nuclei. The nuclei were washed with phosphate-buffered saline (PBS) and then fixed with 70% ethanol for a minimum of 90 minutes. The nuclei were washed again with PBS and stained with 200μl of propidium iodide (50μg/ml) overnight. Flow cytometric analysis of DNA content was performed using the LSR Fortessa Cell Analyzer (Becton Dickinson, San Jose, USA), BD Facs Diva™ software and FlowJo software (FlowJo LLC, Oregon, USA. A minimum of 10,000 events were recorded (typically up to 20,000 and up to 100,000 in complex samples). Analysis was performed using methods derived from the European Society for Analytical Cellular Pathology DNA Consensus in Flow Cytometry guidelines. Gating of forward and side scatter was applied to exclude debris and cell clumping. Samples with <7,500 events after gating were excluded from further analysis. The coefficient of variation (CV) was measured on each G1 peak. Samples with a CV>10% were excluded from further analysis. Each tumour sample was assumed to contain normal cells to act as internal standard. Where possible the position of the diploid peak was calculated with reference to the peak of diploid cells in a case matched normal tissue sample. The DNA index (DI) of any aneuploid peak present was calculated by dividing the G1 peak of the aneuploid population by the G1 peak of the normal diploid cells. Diploid samples were defined as having DI of 1.00. Any additional peak was defined as aneuploid. A tetraploid peak was defined as having a DI of 1.90-2.10 and containing >15% of total events unless a second peak corresponding to G2 was clear on the histogram. Similarly, aneuploid peaks near to G1 (DI 0.90-1.10) were only considered if there was a clear second peak containing >15% of total events.
Detection of allelic imbalance at the HLA locus
Allelic imbalance was detected using two polymorphic Sequence-Tagged Site (STR) markers located on the short arm of chr 6, close to the HLA locus - (D6S248 and ATA12D05), six STR markers located downstream of the HLA locus on the short arm of chr 6p - (D6S1960, GATA143B11, D6S1714, D6S1573, D6S438 and D6S257), and six STR markers located upstream of the HLA locus on the short arm of chr 6p – (D6S410, D6S2257, D6S1034, D6S202, D6S1617, D6S1668). 20ng of patient germline and tumor region DNA was amplified using the PCR. The PCR comprised of denaturing at 950C for 5mins, then 35 cycles of denaturing at 950C for 1min, followed by an annealing temperature of 550C for 1min, 720C for 1min and then a PCR extension at 720C for 10min. PCR products were separated on the ABI 3730xl DNA analyzer. Fragment length and area under the curve of each allele was determined using the Applied Biosystems software GeneMapper v5. When two separate alleles were identified for a particular marker, the fragments could be analyzed for allelic imbalance using the formula (Atumor/Atumor)/(Anormal/Anormal). The output of this formula was defined as the normalized allelic ratio.
Targeted Driver Panel (DP) design and validation
Driver gene panels (Panel_v3, Panel_v5 and Panel_v6) were used in this study. Panel_v3 was designed in 2014, including 110 putative driver genes. Panel_v5 and Panel_v6 were designed in 2015, including 119 and 130 putative driver genes respectively. Driver genes were selected from genes that were frequently mutated in TCGA (accessed in April 2015) or highlighted in relevant studies (Arai et al., 2014, Sato et al., 2013, Scelo et al., 2014). Only alterations in driver genes represented in all three panels were considered in the overall driver mutation analyses. All panels targeted potential driver SCNA regions. To prevent inter-patient samples swaps, we included the 24 SNPs that were previously identified by Pengelly et al in Panel_v5 and Panel_v6. Details of the 3 panels can be found in Table S6.
Driver Panel Library Construction and Targeted Sequencing
Following isolated gDNA QC, depending on the available yield, samples were normalised to either 1-3 μg or 200 ng for the Agilent SureSelectXT Target Enrichment Library Protocol; standard or low input sample preparation respectively. Samples were normalised using a 1X Low TE Buffer. Samples were sheared to 150-200bp using a Covaris E220 (Covaris, Woburn, MA, USA), following the run parameters outlined in the Agilent SureSelectXT standard 3 μg and low input 200 ng DNA protocols. Library construction of samples was then performed following the SureSelectXT protocols, using 6 pre-capture PCR cycles for the standard input samples and 10 pre-capture PCR cycles for the 200 ng low input samples. Hybridisation and capture were performed for each individual sample using the Agilent custom Renal Driver Panel target-specific capture library (versions 3, 5 & 6). The same version of the capture library being used for all samples from the same patient case. Captured SureSelect-enriched DNA libraries were PCR amplified using 14 post-capture PCR cycles in PCR reactions that included the appropriate indexing primer for each sample. Amplified, captured, indexed libraries passing final QC on the TapeStation 4200 were normalised to 2nM and pooled, ensuring that unique indexes were allocated to all final libraries (up to 96 single indexes available) in the pool. QC of the final library pools was performed using the Agilent Bioanalyzer High Sensitivity DNA Assay. Library pool QC results were used to denature and dilute samples in preparation for sequencing on the Illumina HiSeq 2500 and NextSeq 500 sequencing platforms. The final libraries were sequenced 101bp paired-end multiplexed on the Illumina HiSeq 2500 and 151bp paired-end multiplexed on the NextSeq 500, at the Advanced Sequencing Facility at the Francis Crick Institute. Equivalent sequencing metrics, including per sample coverage, was observed between platforms. Single nucleotide variants (SNVs), dinucleotide variants (DNVs), small insertions and deletions (INDELs) and somatic copy number alterations (SCNAs) were derived from 463 primary tumour regions and 169 matched metastatic regions from 56 primary-metastasis pairs in 38 patients (with some patients providing multiple metastases (Figures 1A and S1). Median sequencing coverage was 613x (range 166-1479x) across primary tumour regions and 567x (range 273-2661x) across metastatic regions.
Targeted DP library construction and sequencing (HUC cohort)
DP targeted hybrid-capture panel-
Solution-based hybridization capture probes (Roche Sequencing Solutions) were selected from a genome-wide database of pre-scored probes, which varied in size from 50 to 100 nucleotides. Capture design matched driver panel_v6. Probes were filtered for repetitiveness in the human genome by building a 15-mer histogram from the entire human genome, and then calculating the average 15-mer frequency of the probe by sliding a 15 bp window across the length of each probe. Probes with a score greater than 100 were filtered as repetitive. The remaining probes were scored for uniqueness in the human genome, using SSAHA (http://www.sanger.ac.uk/science/tools/ssaha). A match in the genome was defined as any 30-mer match in the genome, allowing up to 5 mismatches or indels along the length of the match. Additional scoring parameters included penalties for simple sequence repeats and penalties for deviation from a target Tm of 80 C. Target regions of interest were increased to a minimum size of 100 bp, and then tiled with an average overlap of 35 bp, allowing the probes to overhang the ends of the target regions. These tiled probes were selected from the aforementioned pre-scored database of probes by choosing the best scoring probe starting in a 15 bp window, moving 20 bp in the 3’ direction, and repeating. Probes were allowed to have up to 20 possible matches in the genome, though for this panel 99.5% of the probes had 5 or fewer matches. Selected probe sequences were manufactured into biotinylated sequence capture probe pools by Roche Sequencing Solutions – Madison.
Library construction
Libraries were constructed using the SeqCap EZ HyperCap Workflow User’s Guide, v1.0 (Roche Sequencing Solutions). The extracted DNA was enzymatically fragmented using the KAPA HyperPlus library prep kit according to manufacturer’s instructions (Roche Sequencing Solutions). Fragmentation time for DNA isolated from FFPE was linked to the mass of input DNA, and varied from 12 to 22 minutes depending on input amount (10 to 100 ng). To increase the efficiency of library prep, adapter volume was reduced to 3 l and the adapter ligation reaction was extended to 3 hours at 20oC for cases with 100ng of input DNA, and at 16 hours at 16oC for libraries with less than 100ng of input DNA.
Sequencing
Captured samples were pooled following post-capture amplification, and sequenced using an Illumina HiSeq 2500 instrument. Dual HiSeq SBS v4 (Illumina) runs at 101 base-paired-end reads generated the data for analysis.
SNV and INDEL calling from multi-region DP sequencing
Paired-end reads (2x100bp) in FastQ format sequenced by Hiseq or NextSeq were aligned to the reference human genome (build hg19), using the Burrows-Wheeler Aligner (BWA) v0.7.15. with seed recurrences (-c flag) set to 10000 (Li and Durbin, 2009). Intermediate processing of Sam/Bam files was performed using Samtools v1.3.1 and deduplication was performed using Picard 1.81 (http://broadinstitute.github.io/picard/) (Li and Durbin, 2009). Single Nucleotide Variant (SNV) calling was performed using Mutect v1.1.7 and small scale insetion/deletions (INDELs) were called running VarScan v2.4.1 in somatic mode with a minimum variant frequency (--min-var-freq) of 0.005, a tumour purity estimate (--tumor-purity) of 0.75 and then validated using Scalpel v0.5.3 (scalpel-discovery in - -somatic mode) (intersection between two callers taken)(Cibulskis et al., 2013, Fang et al., 2016, Koboldt et al., 2009). SNVs called by Mutect were further filtered using the following criteria: i) ≤5 alternative reads supporting the variant and variant allele frequency (VAF) ≤ 1% in the corresponding germline sample, ii) variants that falling into mitochondrial chr, haplotype chr, HLA genes or any intergenic region were not considered, iii) presence of both forward and reverse strand reads supporting the variant, iv) >5 reads supporting the variant in at least one tumour region of a patient, v) variants were required to have cancer cell fraction (CCF)>0.5 in at least one tumour region (see Subclonal deconstruction of mutations section for details of CCF calculation) , vi) variants were required to have CCF>0.1 to be called as present in a tumour region, vii) sequencing depth in each region need to be >=50 and ≤3000. Finally, suspected artefact variants, based on inconsistent allelic frequencies between regions, were reviewed manually on the Integrated Genomics Viewer (IGV), and variants with poorly aligned reads were removed. Dinucleotide substitutions (DNV) were identified when two adjacent SNVs were called and their VAFs were consistently balanced (based on proportion test, P>=0.05). In such cases the start and stop positions were corrected to represent a DNV and frequency related values were recalculated to represent the mean of the SNVs. To reduce sequencing artefacts from FFPE samples, we further filtered out variants that were significantly enriched for presence in FFPE compared with fresh frozen samples (Fisher’s exact test, P<0.001). A high artefact variant rate was observed in case K489, and to control for this a second germline sample underwent library preperation using the FFPE protocol, and somatic variants were called on a matched sample type basis (i.e. fresh frozen tumor samples compared to original germline, and FFPE tumor samples compared to the second FFPE protocol prepared germline). Variants were annotated using Annovar (Wang et al., 2010). Variants were annotated using Annovar (Wang et al., 2010). Deleterious mutations were defined if two out of three algorithms - SIFT, PolyPhen2 and MutationTaster - predicted the mutation as deleterious. Individual tumour biopsy regions were judged to have failed quality control and excluded from analysis based on the following criteria: i) sequencing coverage depth below 100X, ii) low tumour purity such that copy number calling failed. Mutations detected in high-confidence driver genes (VHL, PBRM1, SETD2, PIK3CA, MTOR, PTEN, KDM5C, CSMD3, BAP1, TP53, TSC1, TSC2) were defined as driver mutations. As TSC1 and TSC2 were not targeted in Panel v5, to check the mutation status in these two genes, patients were sequenced using Panel v5 were re-sequenced with Panel v6 and no new mutations were detected.
SCNA calling from multi-region DP sequencing
To estimate somatic copy number alterations, CNVkit v0.7.3 was performed with default parameter on paired tumour-normal sequencing data (Talevich et al., 2016). Outliers of the derived log2-ratio (logR) calls from CNVkit were detected and modified using Median Absolute Deviation Winsorization before case-specific joint segmentation to identify genomic segments of constant logR (Nilsen et al., 2012). Tumour sample purity, ploidy and absolute copy number per segment were estimated using ABSOLUTE v1.0.6 (Carter et al., 2012). In line with recommended best practice all ABSOLUTE solutions were reviewed by 3 researchers, with solutions selected based on majority vote. Copy number alterations were then called as losses or gains relative to overall sample wide estimated ploidy. Arm gain or loss was called when >50% of the chromosomal have copy number gain or loss. Driver copy number was identified by overlapping the called somatic copy number segments with putative driver copy number regions identified by Beroukhim et al. (2009). For a subset of TRACERx Renal patients, we compared SCNA calls between targeted panel and WGS datasets, and SCNA concordance was 87% (Turajlic et al., 2018a). The average proportion of the genome with aberrant copy number, weighted on each of the 22 autosomal chromosomes, was estimated as the weighted genome instability index (wGII).
MSK validation cohort
Matched tumour and normal aligned sequencing files (BAM format) for the MSK cohort were obtained directly from the authors (Becerra et al., 2017) and were then converted into FASTQ format files using bam2fastq in bedtools package (Quinlan and Hall, 2010). SNVs, INDELs and SCNAs were called using the same methods as TRACERx Renal data (see the sections SNV and INDEL calling from multi-region DP sequencing and SCNA calling from multi-region DP sequencing). Of the 49 cases with ccRCC histology, 15 cases (Pair 8, Pair 9, Pair 13, Pair 17, Pair 22, Pair 35, Pair 38, Pair 42, Pair 43, Pair 44, Pair 48, Pair 52, Pair 56, Pair 58, Pair 59) were excluded from the study as the ABSOLUTE v1.0.6 algorithm failed to find a stable SCNA solution. Clonality of SNVs and SCNAs were estimated using ABSOLUTE v1.0.6. Cancer cell fraction for INDELs were calculated using method described in the section Subclonal deconstruction of mutations. INDELs with CCF>0.5 were called clonal. ITH index for each patient was calculated as the measure of intratumour heterogeneity (ITH index = # subclonal drivers / # clonal drivers).
Quantification and Statistical Analysis
R 3.3.2 was used for all statistical analyses. We tested for difference in driver event count/clonal proportion between primary and metastatic samples using linear regression, including biopsy number per sample as an independent term in the regression model. The comparison of wGII, DNA index and Ki67 scores between “not selected” and “selected” clones was assessed using region values per case. Regions were classified as being within “not selected” or “selected” clones based on the clustering solution for each tumour. Regions found to be only within the founding MRCA clone, or polyclonal with both “not selected” and “selected” clones, were excluded. The comparison of non-synonymous mutation counts between “not selected” and “selected” clones was based directly on clonal clustering solution obtained for each case, again with founding MRCA clones excluded. For all “not selected” versus “selected” comparisons a linear mixed effect (LME) model was used to determine significance, to account for the non-independence of multiple observations from individual tumours. The comparison of maximum wGII (defined as the maximum regional wGII value per primary tumour) between “Rapid” and “Attenuated” metastatic progression groups was assessed using Mann-Whitney test. Comparison of ITH values (again one score per tumour) between “Rapid” and “Attenuated” metastatic progression groups was determined using Mann-Whitney test. The comparison of wGII between pancreatic and all other metastatic tissue sites was assessed conducted using region values per case, with significance determined using a LME model.
Subclonal deconstruction of mutations
To estimate the clonality of a mutation in a region, we used the following formula:
where is the variant allele frequency at the mutation base; is estimated tumour purity; and are the tumour locus specific copy number and the normal locus specific copy number which was assumed to be 2 for autosomal chromosomes; and is the fraction of tumour cells carrying the mutation. Consider is the number of chromosomal copies that carry the mutation, the possible is ranging from 1 to (integer number). We then assigned with one of the possible value: 0.01, 0.02, ..., 1, together with every possible to find the best fit cancer cell fraction of the mutation. Since we focused on driver genes in this study and the accuracy of the estimated CCF is limited by the size of the panel, we call mutations with CCF>0.5 as clonal mutations, mutations with CCF≤0.5 and CCF>0.1 are subclonal. To determine the clonality of a mutation in a tumour, we ask for the mutation to be clonal in all regions in a tumour. Exceptions were made for long INDELs that affect >6 bp of the genome, due to VAF under estimation. If a long INDEL is present in all regions of a tumour, we called it as clonal. To determine the clonality of a SCNA in a tumour, we ask for the SCNA to be presence in all tumour regions, otherwise it is called subclonal.
Driver tree reconstruction
A matrix with presence and absence of nonsynonymous and synonymous point mutations, DNVs, INDELs and arm level SCNAs was created for each tumour, and all the events were clustered based on the following rule: a valid cluster has to have at least two arm level SCNAs or one non-synonymous mutation. The driver events clusters were then ordered into a clonal hierarchy using TRONCO and presented as driver trees (De Sano et al., 2016).
In terms of limitations, we recognise that our Driver Panel phylogenies are based on fewer clonal markers, as compared to whole exome or genome derived phylogenetic trees. As a consequence, some tumour clones are based on only a limited number of genomic markers, and similarly the inferred modes of metastatic seeding (e.g. monoclonal vs polyclonal) are also based on a limited set of markers. However, two contingency measures are in place to mitigate against phylogenetic misconstruction: i) ultra-deep 500x sequencing coverage, which ensures stably derived cancer cell fraction estimates, ii) a bespoke gene panel which is enriched for driver events, increasing the likelihood that mutational markers are driving genuine clonal expansion.
Enrichment of events in metastases
All tumour clones were categorised into three groups based on evidence of selection in the metastasis/metastases: i) clone that are not selected (“no selection”, defined as subclonal in the primary and absent from metastasis), ii) clones that are maintained (“maintained”, defined as the most recent common ancestor (MRCA) clones, clonal in both primary and metastasis ), iii) clones that are selected (“selection”, defined as subclonal in the primary and clonal in metastasis; or absent in the primary and present in metastasis). In addition, we observed a small number of clones with alternative selection patterns: a) being subclonal in both primary and in metastases (i.e. polyclonal metastases), which we categorised as “maintained”, and b) being clonal in primary and sublconal in metastases categorised as “maintained” c) clonal in primary but absent in metasteses (i.e. illusion of clonality or events lost by secondary somatic changes), which we categorised as “no selection”. For each driver event (mutational or SCNA), the proportion of times it was found to be “not selected”, “maintained” and “selected” clones was calculated for each of the TRACERx, HUC and MSK cohorts. For comparison purposes, a background null distribution of proportions was determined for both mutations and SCNAs, based on all passenger events in each cohort. The proportion of “selected” events was compared to the “not selected” proportion, using a Binomial test, with probability of selection taken from the null model, and number of trials based on the number of patients with the given event in each cohort. Meta-analysis across the three cohorts was conducted using Fisher's method of combining p values from independent tests, and p-values were corrected for multiple testing using Benjamini–Hochberg procedure.
Survival analysis
Survival analysis was conducted using the Kaplan-Meier method, with p-value determined by a log-rank test. Hazard ratio and multivariate analysis adjusting for clinical parameters was determined through a Cox proportional hazards model.
Data and Software Availability
Sequencing data that supports this study will been deposited at the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI); accession number EGAS00001002793. Further supplementary data supporting this publication can be found at https://bitbucket.org/tracerxrenal/cell-paper-data-2018
Additional Resources
Clinical trial registry numbers:
TRACERx Renal: https://clinicaltrials.gov/ct2/show/NCT03226886;
PEACE: https://clinicaltrials.gov/ct2/show/NCT03004755
TRACERx Renal study website, detailing investigators, sponsors and collaborators: http://tracerx.co.uk/studies/renal/
Consortia
The members of the TRACERx Renal Consortium are Tim O’Brien, David Nicol, Ben Challacombe, Archana Fernando, Steve Hazell, Ashish Chandra, James Larkin, Martin Gore, Lisa Pickering, Sarah Rudman, Simon Chowdhury, Karen Harrison-Phipps, Mary Varia, Catherine Horsfield, Alexander Polson, Gordon Stamp, Marie O’Donnell, William Drake, Peter Hill, David Hrouda, Eric Mayer, Jonathon Olsburgh, Gordon Kooiman, Kevin O’Connor, Grant Stewart, Michael Aithcison, Maxine Tran, Nicos Fotiadis, Hema Verma, and Jose I. Lopez.
Acknowledgments
We thank Aida Murra, Naheed Shaikh, Justine Korteweg, Jeremy Tai, Kim Edmonds, Karla Lingard, Sarah Sarker, Nikki Hunter, Eleanor Carlyle, Emma Austin, Dilruba Kabir, Drashana Shah, Lyra Del Rosario, Lesley Cooper, Linda Shephard, Susie Slater, Catherine Rogers, Jo Laycock, and Diana Johnson for study support. We thank Axel Bex for the useful discussions regarding surgical outcomes in metastatic ccRCC. We thank Joe Brock for the excellent anatomical illustrations utilized in Figure 6. We thank the patients and their families.
S.T. and H.X. are funded by Cancer Research UK (CRUK) (C50947/A18176). S.T., T.C., J.L., and M.G. are funded by the NIH Research (NIHR) Biomedical Research Centre (BRC) at the Royal Marsden Hospital and Institute of Cancer Research (A109). J.I.L. is funded by the Ministerio de Economía y Competitividad (MINECO, SAF2016-79847-R). M.S., A.S., J.L., R.F., L.A., and L.S. are funded by the Royal Marsden Cancer Charity. K.L. is funded by UK Medical Research Council (MR/P014712/1). N.M. receives funding from CRUK, Rosetrees, and the NIHR BRC at University College London Hospitals. C.S is Royal Society Napier Research Professor. C.S. is funded by Cancer Research UK (TRACERx and CRUK Cancer Immunotherapy Catalyst Network), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees and Stoneygate Trusts, NovoNordisk Foundation (ID 16584), the Breast Cancer Research Foundation (BCRF), the European Research Council (THESEUS), Marie Curie Network PloidyNet, the NIHR BRC at University College London Hospitals, and the CRUK University College London Experimental Cancer Medicine Centre. The work presented in this manuscript was funded by Cancer Research UK (grant reference number C50947/A18176), Ventana Medical Systems (grant reference numbers 10467 and 10530), the Kidney Cancer Fund of The Royal Marsden Cancer Charity, NIHR BRC at the Royal Marsden Hospital and Institute of Cancer Research (grant reference number A109), and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001202), the UK Medical Research Council (FC001202), and the Wellcome Trust (FC001202). In particular, we acknowledge the support of the Advanced Sequencing Facility and the High-Performance Computing at the Francis Crick Institute. This project was enabled through access to the MRC eMedLab Medical Bioinformatics infrastructure, supported by the Medical Research Council (grant number MR/L016311/1).
Author Contributions
Conceptualization, S.T., A.R., J.L., D.N., N.A., T.O., J.I.L., and C.S.; Methodology, S.T., H.X., K.L., A.R., T.C., S. Horswell, M.S., S. Hazell, A.C., J.I.L., and C.S.; Software, H.X., S. Horswell, K.L., and T.B.K.W.; Validation, S.T., A.R., C.E.-J., F.J., J.I.L., T.C., G.F.M., T.A.R., S.M.H., C.D.M., C.J., H.R., S.S., D.L.B., and N.R.A.; Formal Analysis, S.T., H.X., K.L., S. Horswell, L.A., A.R., and C.S.; Investigation, S.T., A.R. T.C., S.W., J.I.L., H.X., K.L., S. Horswell., M.S., A. Soultati, S. Hazell, A.C., G.S., E.N., G.F.M., T.A.R., S.M.H., C.D.M., C.J., H.R., S.S., D.L.B., N.R.A., and C.S.; Resources, S.T., J.L., S.C., S.R., T.O., D.N., B.C., A.F., A. Stewart, L.P., M.G., R.G., I.P., M.F., M.J.-H., R.D., M.F.B., E.R., J.J.H., D.L.B., N.R.A., and C.S.; Data Curation, S.T., L.A., J.Ly., A. Soultati, M.S., S.L., and L.S.; Writing—Original Draft, S.T., K.L., H.X., S. H., A.R., and C.S.; Writing—Review and Editing, S.T., K.L., H.X., A.R., N.A., and C.S.; Visualization, H.X., K.L., S. Horswell, and S.T.; Supervision, S.T., N.A., and C.S.; Project Administration, S.T. and N.A.; Funding Acquisition, S.T., J.L., M.G., N.A., M.J.-H., and C.S.
Declaration of Interests
S.T., H.X., K.L., and C.S. have a patent pending on renal cell carcinoma biomarkers. C.S. reports grant support from Cancer Research UK, UCLH Biomedical Research Council, Rosetrees Trust, and AstraZeneca and personal fees from Boehringer Ingelheim, Novartis, Eli Lilly, Roche, GlaxoSmithKline, Pfizer, and Celgene. C.S. also reports stock options in GRAIL, APOGEN Biotechnologies, and EPIC Bioscience and has stock options from and is co-founder of Achilles Therapeutics.
Published: April 12, 2018
Footnotes
Supplemental Information includes five figures and six tables and can be found with this article online at https://doi.org/10.1016/j.cell.2018.03.057.
Supplemental Information
References
- Adey N., Emery D., Bosh D., Callahan S., Schreiner J., Chen Y., Greig A., Geiersbach K., Parry R. A mill based instrument and software system for dissecting slide-mounted tissue that provides digital guidance and documentation. BMC Clin. Pathol. 2013;13:29. doi: 10.1186/1472-6890-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai E., Sakamoto H., Ichikawa H., Totsuka H., Chiku S., Gotoh M., Mori T., Nakatani T., Ohnami S., Nakagawa T. Multilayer-omics analysis of renal cell carcinoma, including the whole exome, methylome and transcriptome. Int. J. Cancer. 2014;135:1330–1342. doi: 10.1002/ijc.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashashati A., Ha G., Tone A., Ding J., Prentice L.M., Roth A., Rosner J., Shumansky K., Kalloger S., Senz J. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J. Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra M.F., Reznik E., Redzematovic A., Tennenbaum D.M., Kashan M., Ghanaat M., Casuscelli J., Manley B., Jonsson P., DiNatale R.G. Comparative genomic profiling of matched primary and metastatic tumors in renal cell carcinoma. Eur. Urol. Focus. 2017 doi: 10.1016/j.euf.2017.09.016. S2405-4569(17)30237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R., Brunet J.P., Di Napoli A., Mertz K.D., Seeley A., Pires M.M., Linhart D., Worrell R.A., Moch H., Rubin M.A. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bex A., Ljungberg B., van Poppel H., Powles T., European Association of Urology The role of cytoreductive nephrectomy: European Association of urology recommendations in 2016. Eur. Urol. 2016;70:901–905. doi: 10.1016/j.eururo.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Bex A., Mulders P., Jewett M.A.S., Wagstaff J., van Velthoven R., Laguna Pes P.M., Wood L., van Melick H.H.E., Soetekouw P., Lattouf J.B. Immediate versus deferred cytoreductive nephrectomy (CN) in patients with synchronous metastatic renal cell carcinoma (mRCC) receiving sunitinib (EORTC 30073 SURTIME) Ann Oncol. 2017;28(Suppl 5):v605–v649. [Google Scholar]
- Bhindi B., Wallis C.J.D., Boorjian S.A., Thompson R.H., Farrell A., Kim S.P., Karam J.A., Capitanio U., Golijanin D., Leibovich B.C., Gershman B. The role of lymph node dissection in the management of renal cell carcinoma: a systematic review and meta-analysis. BJU Int. 2018 doi: 10.1111/bju.14127. Published online March 1, 2018. [DOI] [PubMed] [Google Scholar]
- Bianchi M., Sun M., Jeldres C., Shariat S.F., Trinh Q.D., Briganti A., Tian Z., Schmitges J., Graefen M., Perrotte P. Distribution of metastatic sites in renal cell carcinoma: a population-based analysis. Ann. Oncol. 2012;23:973–980. doi: 10.1093/annonc/mdr362. [DOI] [PubMed] [Google Scholar]
- Brastianos P.K., Carter S.L., Santagata S., Cahill D.P., Taylor-Weiner A., Jones R.T., Van Allen E.M., Lawrence M.S., Horowitz P.M., Cibulskis K. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P.J., Yachida S., Mudie L.J., Stephens P.J., Pleasance E.D., Stebbings L.A., Morsberger L.A., Latimer C., McLaren S., Lin M.L. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casuscelli J., Weinhold N., Gundem G., Wang L., Zabor E.C., Drill E., Wang P.I., Nanjangud G.J., Redzematovic A., Nargund A.M. Genomic landscape and evolution of metastatic chromophobe renal cell carcinoma. JCI Insight. 2017;2:2. doi: 10.1172/jci.insight.92688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabestani S., Marconi L., Hofmann F., Stewart F., Lam T.B., Canfield S.E., Staehler M., Powles T., Ljungberg B., Bex A. Local treatments for metastases of renal cell carcinoma: a systematic review. Lancet Oncol. 2014;15:e549–e561. doi: 10.1016/S1470-2045(14)70235-9. [DOI] [PubMed] [Google Scholar]
- Davoli T., Uno H., Wooten E.C., Elledge S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:355. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sano L., Caravagna G., Ramazzotti D., Graudenzi A., Mauri G., Mishra B., Antoniotti M. TRONCO: an R package for the inference of cancer progression models from heterogeneous genomic data. Bioinformatics. 2016;32:1911–1913. doi: 10.1093/bioinformatics/btw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Ellis M.J., Li S., Larson D.E., Chen K., Wallis J.W., Harris C.C., McLellan M.D., Fulton R.S., Fulton L.L. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mokadem I., Fitzpatrick J., Bondad J., Rauchhaus P., Cunningham J., Pratt N., Fleming S., Nabi G. Chromosome 9p deletion in clear cell renal cell carcinoma predicts recurrence and survival following surgery. Br. J. Cancer. 2014;111:1381–1390. doi: 10.1038/bjc.2014.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldredge N., Gould S.J. On punctuated equilibria. Science. 1997;276:338–341. doi: 10.1126/science.276.5311.337c. [DOI] [PubMed] [Google Scholar]
- Fang H., Bergmann E.A., Arora K., Vacic V., Zody M.C., Iossifov I., O’Rawe J.A., Wu Y., Jimenez Barron L.T., Rosenbaum J. Indel variant analysis of short-read sequencing data with Scalpel. Nat. Protoc. 2016;11:2529–2548. doi: 10.1038/nprot.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M.G., Durante M.A., Anbunathan H., Cai L.Z., Decatur C.L., Bowcock A.M., Kurtenbach S., Harbour J.W. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun. 2018;9:116. doi: 10.1038/s41467-017-02428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Davis A., McDonald T.O., Sei E., Shi X., Wang Y., Tsai P.C., Casasent A., Waters J., Zhang H. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 2016;48:1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C.R. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Rowan A.J., Horswell S., Math M., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman B., Moreira D.M., Thompson R.H., Boorjian S.A., Lohse C.M., Costello B.A., Cheville J.C., Leibovich B.C. Renal cell carcinoma with isolated lymph node involvement: long-term natural history and predictors of oncologic outcomes following surgical resection. Eur. Urol. 2017;72:300–306. doi: 10.1016/j.eururo.2016.12.027. [DOI] [PubMed] [Google Scholar]
- Giancotti F.G. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G., Van Loo P., Kremeyer B., Alexandrov L.B., Tubio J.M.C., Papaemmanuil E., Brewer D.S., Kallio H.M.L., Högnäs G., Annala M., ICGC Prostate Group The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner M.C., Mosbruger T., Esopi D.M., Fedor H., Heaphy C.M., Walker D.A., Adejola N., Gürel M., Hicks J., Meeker A.K. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R.W., Kapur P., Serie D.J., Parasramka M., Ho T.H., Cheville J.C., Frenkel E., Parker A.S., Brugarolas J. Clear cell renal cell carcinoma subtypes identified by BAP1 and PBRM1 expression. J. Urol. 2016;195:180–187. doi: 10.1016/j.juro.2015.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte T., Rao P.N., de Martino M., LaRochelle J., Shuch B., Zomorodian N., Said J., Kabbinavar F.F., Belldegrun A.S., Pantuck A.J. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J. Clin. Oncol. 2009;27:746–753. doi: 10.1200/JCO.2007.15.8345. [DOI] [PubMed] [Google Scholar]
- Koboldt D.C., Chen K., Wylie T., Larson D.E., McLellan M.D., Mardis E.R., Weinstock G.M., Wilson R.K., Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutikov A., Uzzo R.G., Caraway A., Reese C.T., Egleston B.L., Chen D.Y., Viterbo R., Greenberg R.E., Wong Y.N., Raman J.D., Boorjian S.A. Use of systemic therapy and factors affecting survival for patients undergoing cytoreductive nephrectomy. BJU Int. 2010;106:218–223. doi: 10.1111/j.1464-410X.2009.09079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rochelle J., Klatte T., Dastane A., Rao N., Seligson D., Said J., Shuch B., Zomorodian N., Kabbinavar F., Belldegrun A., Pantuck A.J. Chromosome 9p deletions identify an aggressive phenotype of clear cell renal cell carcinoma. Cancer. 2010;116:4696–4702. doi: 10.1002/cncr.25279. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Haq F., Kim D., Jun C., Jo H.J., Ahn S.M., Lee W.S. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS ONE. 2014;9:e90459. doi: 10.1371/journal.pone.0090459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N., Rosenthal R., Hiley C.T., Rowan A.J., Watkins T.B.K., Wilson G.A., Birkbak N.J., Veeriah S., Van Loo P., Herrero J. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R.R., Kroeger N., Xie W., Lee J.L., Knox J.J., Bjarnason G.A., MacKenzie M.J., Wood L., Srinivas S., Vaishampayan U.N. Impact of bone and liver metastases on patients with renal cell carcinoma treated with targeted therapy. Eur. Urol. 2014;65:577–584. doi: 10.1016/j.eururo.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A., Roth A., Laks E., Masud T., Bashashati A., Zhang A.W., Ha G., Biele J., Yap D., Wan A. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet. 2016;48:758–767. doi: 10.1038/ng.3573. [DOI] [PubMed] [Google Scholar]
- Mitchell T.J., Turajlic S., Rowan A., Nicol D., Farmery J.H.R., O’Brien T., Martincorena I., Tarpey P., Angelopoulos N., Yates L. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer. Cell. 2018;173:611–623. doi: 10.1016/j.cell.2018.02.020. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Van Loo P., Wedge D.C., Alexandrov L.B., Greenman C.D., Lau K.W., Raine K., Jones D., Marshall J., Ramakrishna M. The life history of 21 breast cancers. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen G., Liestøl K., Van Loo P., Moen Vollan H.K., Eide M.B., Rueda O.M., Chin S.F., Russell R., Baumbusch L.O., Caldas C. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., Chan-Seng-Yue M., Lemire M., Li Y., Wilson G.W., Connor A.A., Denroche R.E., Liang S.B., Brown A.M., Kim J.C. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psutka S.P., Leibovich B.C. Management of inferior vena cava tumor thrombus in locally advanced renal cell carcinoma. Ther. Adv. Urol. 2015;7:216–229. doi: 10.1177/1756287215576443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese A.C., Whitson J.M., Meng M.V. Natural history of untreated renal cell carcinoma with venous tumor thrombus. Urol. Oncol. 2013;31:1305–1309. doi: 10.1016/j.urolonc.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Robinson D.R., Wu Y.M., Lonigro R.J., Vats P., Cobain E., Everett J., Cao X., Rabban E., Kumar-Sinha C., Raymond V. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- Scelo G., Riazalhosseini Y., Greger L., Letourneau L., Gonzàlez-Porta M., Wozniak M.B., Bourgey M., Harnden P., Egevad L., Jackson S.M. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat. Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- Schwarz R.F., Ng C.K., Cooke S.L., Newman S., Temple J., Piskorz A.M., Gale D., Sayal K., Murtaza M., Baldwin P.J. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12:e1001789. doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S.P., Morin R.D., Khattra J., Prentice L., Pugh T., Burleigh A., Delaney A., Gelmon K., Guliany R., Senz J. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Talevich E., Shain A.H., Botton T., Bastian B.C. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S., Xu H., Litchfield K., Rowan A., Horswell S., Chambers T., O’Brien T., Lopez J.I., Watkins T.B.K., Nicol D. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell. 2018;173:595–610. doi: 10.1016/j.cell.2018.03.043. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P., Nordgard S.H., Lingjaerde C., Russnes H.G., Rye I.H., Sun W., Weigman V.J., Marynen P., Zetterberg A., Naume B. Allele-specific copy number analysis of tumors. PNAS. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum R.R., Hellman S. Oligometastases revisited. Nat. Rev. Clin. Oncol. 2011;8:378–382. doi: 10.1038/nrclinonc.2011.44. [DOI] [PubMed] [Google Scholar]
- Woodruff D.Y., Van Veldhuizen P., Muehlebach G., Johnson P., Williamson T., Holzbeierlein J.M. The perioperative management of an inferior vena caval tumor thrombus in patients with renal cell carcinoma. Urol. Oncol. 2013;31:517–521. doi: 10.1016/j.urolonc.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Wszolek M.F., Wotkowicz C., Libertino J.A. Surgical management of large renal tumors. Nat. Clin. Pract. Urol. 2008;5:35–46. doi: 10.1038/ncpuro0963. [DOI] [PubMed] [Google Scholar]
- Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates L.R., Knappskog S., Wedge D., Farmery J.H.R., Gonzalez S., Martincorena I., Alexandrov L.B., Van Loo P., Haugland H.K., Lilleng P.K. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32:169–184.e7. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lu A., Li L., Yue J., Lu Y. p16 Modulates VEGF expression via its interaction with HIF-1alpha in breast cancer cells. Cancer Invest. 2010;28:588–597. doi: 10.3109/07357900903286941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.M., Zhao B., Bai Y., Iamarino A., Gaffney S.G., Schlessinger J., Lifton R.P., Rimm D.L., Townsend J.P. Early and multiple origins of metastatic lineages within primary tumors. Proc. Natl. Acad. Sci. USA. 2016;113:2140–2145. doi: 10.1073/pnas.1525677113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.