Abstract
Interferon regulatory factors (IRFs) are a group of closely related proteins collectively referred to as the IRF family. Members of this family were originally recognized for their roles in inflammatory responses; however, recent research has suggested that they are also involved in tumor biology. This review focusses on current knowledge of the roles of IRF-1 and IRF-2 in human cancer, with particular attention paid to the impact of IRF-1 inactivation. The different mechanisms underlying IRF-1 inactivation and their implications for human cancers and the potential importance of IRF-1 in immunotherapy are also summarized.
Keywords: Cancer, Interferon Regulatory Factor, Oncogene, Tumor Suppressor
Introduction
In human cancers, the accumulation of genetic aberrations is known to affect the normal functions of several genes that control cell proliferation and survival. Amongst these genes is IRF-1, a member of the interferon regulatory factor (IRF) family. IRF-1 was first identified in 1988 as a transcription factor able to induce expression of the gene interferon β (IFN-B) [1]. The following year, IRF-2 was identified and found to suppress the function of IRF-1 [2]. Currently, the IRF family comprises ten members: IRF-1, IRF-2, IRF-3, IRF-4/Pip/ICSAT/LSIRF, IRF-5, IRF-6, IRF-7, IRF-8/interferon consensus sequence binding protein (ICSBP), IRF-9/ISGF3γ/p48, and IRF-10 [3–5]. IRF-1 to IRF-9 are present in mammals, but IRF-10 appears to be restricted to fish and chickens. As a result of the discovery of this extended group of IRFs, the terms ‘IRF kingdom’ and ‘IRF world’ have been coined [6].
All members of the IRF family exhibit significant homology in their N-terminal region, which contains a DNA-binding domain (DBD) that includes a cluster of five tryptophan residues. This DBD forms a helix-turn-helix motif and recognizes the interferon-stimulated response element (ISRE) in the promoters of genes targetted by IRFs [5,6]. The C-terminal region of most IRF family members is less conserved and contains an IRF-association domain (IAD) responsible for homomeric and heteromeric interactions with other proteins, including other IRF family members and non-IRF transcription factors and cofactors (e.g. PU.1 and E47) [3,4]. Two types of IAD have been identified, namely, IAD1 and IAD2. IAD1 is present in all members of the IRF family, with the exception of IRF-1 and IRF-2, in which IAD2 is found [3]. By mediating protein–protein interactions, these IADs confer specific roles and functions upon each member of the IRF family (Figure 1).
Figure 1. Illustration of the various functional domains of IRF family members.
All IRF family members contain a DBD (blue) and a regulatory domain (light blue). In addition, most IRFs possess a type 1 (dark green) or type 2 (light green) IAD. A repression domain that functions to repress gene expression (purple) is also present in some IRF family members. Finally, a nuclear localization signal domain (orange) is found in most IRFs.
IRFs were originally recognized for their roles in innate and adaptive immunity, especially in the regulation of interferon-inducible genes in the interferon system [7]. However, recent studies have indicated their involvement in oncogenesis and other cellular responses.
IRF family members are playing a pivotal role in immune response. Amongst those, IRF-3 and IRF-7 whose antiviral role is well established. In addition, the hematopoietic factors, IRF-4 and 8. IRF-3 is expressed in all cell types and its expression is not triggered by viral infection or downstream to interferon [8]. Unlike IRF-3, IRF-7 expression is expressed downstream to interferon signaling pathway [9].
Expression of IRF-3 is up-regulated upon recognition of viral dsRNA by cellular receptor and, as a consequence of toll-like receptor-3 signaling, IRF-3 is phosphorylated and activated as a result of this post-translational modification [10]. This leads to formation of homo or heterodimers of IRF-3 and IRF-7, which translocate to nucleus and induce interferon-β, as well as, other interferon-stimulated genes (ISGs) after binding to cAMP-responsive element binding protein 1 (CREB) binding protein (CBP) [11].
Ubiquitination of IRF-3 targets its degradation by proteasome enzyme system, a process that can be triggered by propyl isomerase (Pin1) [12]. In contrast, ISG 15 (ISG-15); a ubiquitin-like protein can bind IRF-3, stabilizing it and increased its nuclear retention. This contributed to ISG-15 action in enhancing host antiviral response [13].
IRF-4 is expressed in B lymphocytes and dendritic cells and is thought to be required for B- and T-lymphocytes’ maturation and differentiation [14–16]. In this respect, an association between overexpression of IRF-4 and multiple myeloma was reported and was explained by translocation of this factor near to the locus of immunoglobulins [17]. Interestingly, IRF-4 is an endogenous antagonist of IRF-1, which is known for its tumor suppressing activity [18].
IRF-5 is another immunomodulatory factor, which has been recognized for its role as a regulator for type I interferon gene expression in response to viral infections. It has a role in regulation and development of host immune response and autoimmune responses; hence it has been recognized as a susceptibility gene for autoimmune disorders [19]. The functions of IRF-5 are extended to other disease categories including cancer, obesity, pain mediation [20], and cardiovascular disease [21]. The pleiotropic nature of IRF-5 functions could be also confirmed by its reported role as a regulator of cell growth and apoptosis, which explains its potential as tumor suppressor being down-regulated in malignant tissues [22]. In addition, metabolic activities of IRF-5 have been also reported [23].
IRF-6 is a unique member of IRF family being the only family member to be essential for embryogenesis [24]. IRF-6 is also a crucial protein for proliferation and differentiation of keratinocytes, which makes it an important element to the process of wound healing (reviewed in [25]). Defects in IRF-6 gene have been observed in patients with cleft lip and/or palate. In addition, aberrations in IRF-6 predisposes for squamous cell carcinoma and defective development of mammary gland [26]. Adding to this information, recently, a novel role of IRF-6 was reported implicating IRF-6 in development of exocrine glands as another function besides its role as a tumor suppressor [27].
Human IRF-7 gene can be induced by type I interferon and tumor necrosis factor α (TNF-α). On the other hand, regulation of type I interferon gene expression by IRF-7 has been reported, hence the relation between IRF-7 and type I interferon could be described as mutual [9,28]. This was confirmed by the finding that homozygous deletion of IRF-7 in an animal model abolished expression of type I interferon-regulated genes following activation of TLR-9 or viral infections [29]. Activation of IRF-7 is also phosphorylation dependent and is an outcome of TLR-3, -7, -8 and -9 signaling pathways [30].
IRF-8, also known as ICSBP is expressed solely in lymphoid and myeloid progenitors [31]. The function of this member depends on its interaction with other IRF members including IRF-1 and 4 [32]. IRF-1–IRF-8 heterodimer suppresses ISG-15, whereas ISG-15 is induced by IRF-4–IRF-8 complex [33]. Additionally, macrophages differentiation and activation during inflammatory response is also activated by IRF-1–IRF-8 heterodimer [34].
IRF-9, p48, or ISGF3-γ contributes to the antiviral response of interferon α, β, and γ. This role is achieved primarily by the binding of IRF-9 to interferon stimulated gene factor3, which interacts with ISRE and regulates ISGs [35,36].
This review discusses the functions of IRF-1 and IRF-2 in human cancers, with a focus on the potential contribution of IRF-1 inactivation to human carcinogenesis and the future of IRF-1 as a therapeutic target.
Antioncogenic and oncogenic potential of IRF-1 and IRF-2
The role of the IRF family in oncogenesis was first noted in 1993, when overexpression of IRF-2 was found to transform NIH 3T3 cells and enhance their tumorigenicity in nude mice, a phenotype that was shown to be reversed by IRF-1 overexpression [37]. An antioncogenic function for IRF-1 was also implied by the finding that overexpression of the Ha-ras oncogene was seen to result in transformation of IRF-1−/− but not wild-type mouse embryonic fibroblasts (MEFs) [38]. Surprisingly, ectopic expression of the N-ras oncogene in some myeloid cell lines has been shown to suppress proliferation and up-regulate the cyclin-dependent kinase (CDK) inhibitor p21WAF1/CIP1. This suppression was found to be associated with up-regulation of IRF-1, further reinforcing the notion that this IRF exerts an antioncogenic effect [39]. Moreover, overexpression of IRF-1 in a wide range of different cell types from humans, mice, and even hamsters has been reported to cause growth inhibition [40–43]. In contrast with other tumor suppressors, loss of IRF-1 function rarely induces oncogenicity; however, IRF-1 inactivation is a cofactor in increased risk of tumorigenesis mediated by p53 nullizygosity or Ha-ras oncogene overexpression [44].
The antiproliferative effect of IRF-1 has chiefly been attributed to its induction of the expression of certain target genes that down-regulate cell growth. These genes include protein kinase R (PKR) and signal transducer and activator of transcription (STAT) 1 (STAT1) in the Janus kinase (JAK)-STAT pathway [45,46]. The panel of IRF-1-induced genes also includes those encoding caspases, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), and lysyl oxidase (LOX), the latter of which has also been identified as a tumor suppressor [47]. Caspases are known for their protease activity and their activation and participation in the apoptotic cascade. IRF-1 mediates IFN-γ-induced apoptosis in ovarian cancer cell lines via induction of caspase-1 expression [48], and up-regulates caspase-8 expression in response to IFN-γ/STAT1 signaling as part of a mechanism that sensitizes cells to apoptosis [49]. IRF-1 also binds to unique sites in the p53 up-regulated modulator of apoptosis (PUMA) promoter, resulting in up-regulation of the intrinsic apoptosis pathway [50]. Cyclin D1 and survivin have also been reported as downstream targets of IRF-1, and in vitro activation of this IRF decreases cyclin D1 expression and CDK 4 (CDK4) activity [51].
The inhibitor of apoptosis, survivin, is a potential target for cancer therapy as its overexpression by tumor cells promotes their survival. Notably, overexpression of IRF-1 in breast carcinoma cells has been found to result in a 15-fold down-regulation of survivin protein levels [52], which has been attributed to the suppression of cyclin B1, CDK-1, cyclin E, E2F1, CDK2, and CDK4 expression [53]. However, survivin may also be regulated in human cancer cells by other IRF-1 signaling pathways or directly by IRF-1 itself [53]. IRF-1 also induces p21-mediated G1 cell cycle arrest in such cells [53]. IRF-1 is believed to prevent oncogenesis through initiation of apoptosis, as demonstrated by the IRF-1- and p53-mediated apoptosis, but not cell cycle arrest, of Ha-ras-overexpressing wild-type MEFs following treatment with ionizing radiation or anticancer drugs [5,6].
IRF-1 and p53, both of which are tumor suppressors, regulate DNA damage-induced apoptosis independently or co-operatively. For example, DNA damage-induced apoptosis of mature T lymphocytes relies solely on IRF-1 [54], whereas that of thymocytes depends on p53 [54,55]; thus, cell type and differentiation stage dictate the tumor suppressor that regulates this process. Furthermore, a role for IRF-1 in the DNA damage repair mechanism has been described. By using ChIP coupled to a CpG island microarray, Frontini and colleagues [56] identified 202 IRF-1 binding sites in the human genome, leading to the discovery that BRCA1-interacting protein C-terminal helicase 1 (BRIP1, also known as Fanconi anemia gene J) is up-regulated as a result of IFN-mediated induction of IRF-1 expression. BRIP1 has been implicated in breast cancer susceptibility and is known for its DNA repair function [57]. Its association with IRF-1 further supports the potential involvement of this regulatory protein in antioncogenic activity.
The concept that IRF-1 has an antioncogenic effect via cell cycle regulation is supported by change in its expression throughout the cell cycle [37]. Such changes inhibit the growth of cells with damaged DNA by inducing G1 cell cycle arrest, an effect that is dependent on ataxia telangiectasia mutated (ATM) and mediated by binding of the promoter region of p21WAF1/CIP1, which contains binding sites for both IRF-1 and p53. It has also been reported that activation of IRF-1 results in the expression of genes directly involved in various other cellular processes, including regulation of the T cell-mediated immune response to viral infection. Deletion or mutation of IRF-1 and exon skipping (a form of RNA splicing to skip faulty exons) in the corresponding mRNA are also associated with the development of various hematopoietic malignancies and syndromes [58].
In contrast with IRF-1, IRF-2 exerts a pro-oncogenic effect. IRF-2 has been reported to be up-regulated in pancreatic cancer, in which it is associated with tumor size and differentiation, tumor node metastasis stage, and survival [59]. Moreover, overexpression of the IRF-2 gene has been shown to prevent N-ras-induced growth suppression, confirming its pro-oncogenic role [60]. One study has attributed the oncogenic activity of IRF-2 to its ability to bind to the ISRE via its DNA-binding/transcription repression domain, preventing IRF-1 and other IRF family members from binding to the same DNA response element [61]. In addition, IRF-2 regulates transcription of downstream targets involved in oncogenesis, such as histone H4 [62,63]. A further mechanism underlying the oncogenic activity of IRF-2 involves its interaction with murine double minute-2 (MDM2), an enzyme that catalyzes p53 ubiquitination and degradation via the proteasome pathway [64]. Target genes of IRF-1 and IRF-2 are summarized in Table 1.
Table 1. Target genes of IRF-1 and IRF-2.
| IRF-1 Targets | Action | IRF-2 targets | Action |
|---|---|---|---|
| Cell cycle regulatory genes | DNA-binding activity | ||
| CDK inhibitor p21WAF1/CIP1 [65] | Up-regulation | ISRE [61] | Inhibition of IRF-1 transacting activity |
| Cyclin D1 [51] | Down-regulation | H4 [63] | Regulation of gene expression |
| Cyclin E [65] | Down-regulation | Anti-apoptotic | |
| CDK2 [65] | Down-regulation | MDM2 [64] | Interaction and inhibition of p53-mediated effects |
| CDK4 [65] | Down-regulation | ||
| E2F [65] | Down-regulation | ||
| Growth suppression genes | |||
| PKR [46] | Up-regulation | ||
| LOX [47] | Up-regulation | ||
| Apoptosis-inducing genes | |||
| Caspase 1 [48] | Activation | ||
| Caspase 3 [66] | Activation | ||
| Caspase 7 [66] | Activation | ||
| Caspase 8 [66] | Activation | ||
| Caspase 8 [49] | Activation | ||
| PUMA [50] | Up-regulation | ||
| BRIP1 (Fanconi Anemia gene J) [56] | Up-regulation | ||
| TRAIL [67] | Up-regulation | ||
| Immunomodulation | |||
| MHC-I [68] | Up-regulation | ||
| ISG-15 [69] | Up-regulation | ||
Gene names are italicized.
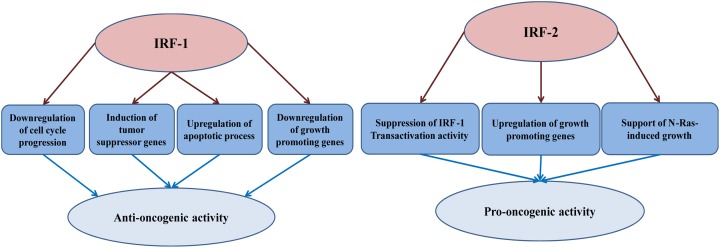
In conclusion, IRF-1 operates as a tumor suppressor whose loss, in combination with other genetic alterations, may significantly increase risk of malignancy. In the following sections, we summarize its role in different human cancers and the range of mechanisms by which this tumor suppressor loses its function (Figure 2 summarizes the anti- and pro-oncogenic potential of IRF-1 and 2).
Figure 2. The anti- and pro-oncogenic activity of IRF-1 and IRF-2.
IRF-1 anti-oncogenic activity is attributed to four main mechanisms: (I) Derailing of cell cycle; (II) Down-regulation of growth promoting genes; (III) Induction of tumor suppressor genes, and (IV) Up-regulation of apoptotic machinery. The pro-oncogenic potential of IRF-2 is owed to: (i) Suppression of IRF-1 activity; (ii) up-regulation of growth promoting genes, and (iii) supporting oncogenes-induced growth.
IRF-1 in human cancers
Human leukemia and pre-leukemic myelodysplasia
Human leukemia and pre-leukemic myelodysplastic syndrome (MDS) are characterized by a remarkable cytogenetic abnormality, namely, the loss of chromosome 5 or a deletion within its long arm (del(5q) or 5q−) [70]. This aberration accounts for 30% of MDS cases, 15% of de novo acute myelogenous leukemia (AML) cases, 50% of cases of secondary AML arising from MDS, and 2% of de novo acute lymphocytic leukemia cases [71,72]. Willman and colleagues [70] proposed that a tumor suppressor gene must be located in the common deleted segment (5q31), and eventually succeeded in mapping IRF-1 to band 31.1 of chromosome 5 using fluorescence in situ hybridization with a 19-kb IRF-1 probe. This probe only hybridized to sequences on chromosome 5q and was precisely mapped to 5q31.1 by computer-assisted fluorescence microscopy. Once the IRF-1 locus had been determined, a full-length IRF-1 cDNA probe was used to perform Southern blotting of DNA from patients with different types of leukemia and MDS associated with del(5q) to confirm IRF-1 inactivation in clinical samples. The results of the present study indicated an unusual instability in the 5q region, as deletion of one IRF-1 allele was accompanied by rearrangement or deletion of the second allele in some cases. This led to the conclusion that deletions or rearrangements are more frequent than point mutations at this locus in human leukemia and MDS, and loss of IRF-1 may be critical in the development of AML and MDS [70].
Another group has also investigated the proposal that IRF-1 loss is key in 5q− syndrome development, finding that 85.7 % of the patients included in their study exhibited loss of one allele of the IRF-1 gene, with no evidence of homozygous loss [73]. This finding is consistent with a detailed examination of the ‘critical region’ on 5q affected in MDS and AML patients, which contains crucial genes with tumor suppressing potential, such as interleukin 3 (IL-3), IL-4, IL-5, and CSF2 [74], which IRF-1 maps closely [70].
In the same context, Harada and colleagues [75] reported that a significant proportion of IRF-1 mRNA transcripts obtained from the bone marrow and peripheral mononuclear cells of patients with MDS or leukemia secondary to MDS lacks exon 2 (containing the initiation codon) and exon 3 as a result of accelerated exon skipping. The resulting IRF-1 protein lacks the ability to bind DNA and its tumor-suppressing activity is consequently lost. Thus, accelerated exon skipping comprises a second mechanism, in addition to IRF-1 loss as a result of DNA damage, by which IRF-1 is inactivated. This process may explain the development of hematopoietic malignancies in some 5q− syndrome cases in which both copies of the IRF-1 gene are retained. To validate this proposal, Green and colleagues [76] developed quantitative competitive RT-PCR assays to measure levels of full-length and exon-skipped IRF-1 transcripts (IRF-1∆2 and IRF-1∆2,3) in acute promyelocytic leukemia (APL), AML, and MDS patients, with a particular focus on those carrying an IRF-1 allele deletion. Their results showed that accelerated exon skipping is common in patients with a 5q deletion and one deleted IRF-1 allele, and occurs in most APL cases (in which IRF-1 protein expression was found to be absent). Such exon skipping thus leads to loss of IRF-1 function and increases risk of malignancy [76].
To evaluate the extent to which exon skipping-induced IRF-1 inactivation is involved in oncogenesis, investigation of the association between exon-skipped IRF-1 transcripts and other forms of leukemia was also necessary. Mutational analysis and studies of IRF-1 expression patterns have revealed a four-fold reduction in levels of full-length IRF-1 mRNA and elevated presence of abnormal splice variants in chronic myeloid leukemia (CML) patients [77]. This lends weight to the idea that production of non-functional splice variants is another mechanism underlying IRF-1 inactivation, and one that is particularly relevant to CML. Unlike the IRF-1 splice variants observed in AML, those in CML lack exons 7, 8, and 9, in addition to the AUG initiation codon in exon 2 and the DBD [77]. Loss of exons 7, 8, and 9 compromises the ability of IRF-1 to heterodimerize with cofactors and/or other IRF members, such as IRF-8, levels of which have also been reported to be reduced in CML patients [78].
Examining leukemogenesis from a different perspective, Preisler and colleagues [79] compared the IRF-1/IRF-2 gene expression ratio in AML and normal marrow, concluding that the balance between these factors, rather than the expression level of either in isolation, ultimately determines phenotype. Their study revealed that this ratio was significantly lower in AML patients as a result of low IRF-1 and high IRF-2 transcript levels, and indicated that IRF-1-responsive genes reduce AML risk. In contrast, malignant transformation related to stimulation of IRF-2 gene appears to be relatively common in leukemogenesis [79].
Human breast cancer
Following the discovery of a role for IRF-1 in human leukemia and pre-leukemic myelodysplasia, researchers began to question whether it and other IRF family members might be similarly implicated in other cancers, particularly solid tumors. In one retrospective study, IRF-1 but not IRF-2 was found to be expressed in normal breast tissue, whereas levels of the former were shown to be lower and those of the latter higher in high-grade ductal carcinoma in situ (DCIS) and lymph node-positive invasive ductal cancer [80]. This finding suggests that IRF-1 and IRF-2 protein expression profiles are altered in human breast cancer, consistent with their respective proposed roles as a tumor suppressor and oncoprotein.
In the development of previous work, Connett and colleagues [81] used immunohistochemical tissue microarrays on a larger sample of invasive breast carcinoma specimens to show that neoplastic breast tissues are less likely to maintain IRF-1 expression than adjacent normal tissues. They also demonstrated that IRF-1 expression negatively correlates with tumor size, confirming that loss of IRF-1 is associated with breast carcinogenesis [81]. No correlation was noted between IRF-2 and clinical parameters in this study; however, this may be explained at least in part by the complex nature of the regulatory relationship between IRF-1 and IRF-2 in their exertion of tumor suppressive and oncogenic effects, respectively [80,81]. Moreover, the mechanism by which dysregulation of IRF-1 and IRF-2 affects breast carcinoma cells has been proposed to involve disruption of the IRF-1/IRF-2 ratio [82], and as this ratio has been reported to change during the cell cycle, monitoring the status of IRF-1 and IRF-2 in such histological studies is highly challenging [43]. Other studies have suggested that post-translational modifications of IRF-1 and IRF-2 are more critical to the activity of these proteins than the levels at which they are present [83–85]. These observations indicate that further molecular investigations are required to establish an association between IRF-1 and IRF-2 and breast tumorigenesis.
At the genetic level, there have been no reports of point mutations that cause IRF-1 inactivation in breast cancer; however, an IRF-1 polymorphism (A4396G) has been identified in breast cancer cell lines, and has been found to be more frequent amongst African Americans [86]. It is unknown whether this variant contributes to breast oncogenesis. The nucleotide substitution involved does not change the sequence of the translated protein, and there is no evidence that this polymorphism results in the introduction of a new active splicing site, which can lead to IRF-1 inactivation in human cancers [86]. It has been hypothesized that this variant influences the binding of certain critical transcription factors with tumor-suppressing potential, including microphthalmia transcription factor (MITF), which activates INK4A, a tumor suppressor that inhibits cell cycle progression [86,87]. The identification of this polymorphism highlights the need for more thorough investigation of genetic changes in IRF-1 in patients with breast carcinoma and other tumors.
A lack of sufficient evidence concerning the exact contribution of IRF-1 to breast carcinogenesis, together with the well-documented role of this protein in hematopoietic malignancies, triggered a number of comparative genomic hybridization studies. As a result, loss of the 5q31.1 region, to which IRF-1 has been mapped, was found to be common in breast tumors [88]. In addition, 5q12-31 deletions were noted in 11% of sporadic breast cancers, and 5q31.1 loss was observed in 50% of BRCA1 mutation-positive breast tumors. Given that somatic loss of IRF-1 may be a critical event in breast oncogenesis, in 2010, Cavalli and colleagues [88] investigated its incidence in 52 patients with invasive breast tumors. Loss of heterozygosity (LOH) at the IRF-1 locus was found in 32% of cases, providing evidence of a tumor-suppressive effect of IRF-1 in breast cancer [88].
Other human cancers
The establishment of a tumor-suppressing role for IRF-1 in breast cancer and leukemia laid the foundations for further investigations exploring its function in other cancers. One such study revealed that 50% of gastric tumors exhibit LOH at the 5q region implying a critical contribution of IRF-1 to the development of stomach carcinoma [89]. In another investigation, 5q31.1 was reported to be lost in primary esophageal carcinoma, and was the smallest commonly deleted region in 57% of the specimens tested [90], implicating IRF-1 in the pathogenesis of this malignancy.
For the correct interpretation of these findings, it was important to test the so-called ‘two-hit hypothesis’ in relation to IRF-1 to confirm the role of this gene in esophageal carcinoma and stomach adenocarcinoma [91]. This hypothesis proposes that loss of function of a critical tumor suppressor gene requires allelic changes in both sister chromatids of the chromosome concerned. Sequencing of the IRF-1 gene in gastric adenocarcinoma tissues confirmed LOH at this locus, and led to the identification of a loss-of-function point mutation resulting in a methionine-to-leucine substitution at codon 8 [38]. This mutation attenuates the transcriptional activity of IRF-1 and consequently, its tumor-suppressing capability is lost. Although it is not clear how this mutation brings about this effect, it has been proposed that it may enhance the interaction of IRF-1 with other factors, impairing the function of this protein as a regulator of transcription [38]. Wang and colleagues [92] have further investigated the part played by IRFs in esophageal malignancies by measuring patterns of IRF-1 and IRF-2 protein expression in esophageal squamous cell carcinoma (ESCC) and correlating them with the clinical features of this disease. They found expression of IRF-1 to be decreased and that of IRF-2 increased in ESCCs compared with matched normal esophageal tissue. These results demonstrate that IRF-2 expression may be positively correlated with the progression of this cancer, adding to the multitude of observations supporting opposite roles for IRF-1 and IRF-2 in tumorigenesis.
A separate group has reported a correlation between IRF-1 expression in human melanoma tissue specimens and less advanced disease, although they were not able to demonstrate a clear relationship between IRF-2 expression and this malignancy in this work [93]. In an examination of cervical cancer tissues, Lee and colleagues [94] noted the presence of five different splice variants of IRF-1 mRNA lacking particular combinations of exons 7, 8, and 9. This alternative splicing results in the absence of the IRF-1 functional domain or the generation of a truncated protein with aberrant transcriptional activity that interferes with that of wild-type IRF-1. Thus, alternative splicing affecting exons 7, 8, and 9 may be another critical mechanism negatively regulating IRF-1 in cervical cancer.
Recently, it has also been reported that high IRF-1 expression in hepatocellular carcinoma (HCC) is associated with better outcome in terms of frequency of recurrence following surgical resection. In contrast, overexpression of IRF-2 is associated with increased probability of recurrence [95]. In addition, a high IRF-2/IRF-1 protein ratio positively correlates with tumor invasion and metastatic ability in human HCC cell lines [95]. The involvement of IRF-1 and IRF-2 in the progression of human pancreatic cancer has also been reported [96]. Whereas IRF-1 expression is reduced in pancreatic cancer specimens compared with adjacent normal tissues, IRF-2 gene expression is up-regulated. In the same work, it was also found that up-regulation of IRF-1, but not IRF-2, leads to better tumor differentiation, enhanced lymphocyte infiltration, smaller tumor mass, and longer survival [96]. Subsequent research using an in vitro model of pancreatic cancer has confirmed these clinical observations and thus, the tumor-suppressing and -promoting potentials of IRF-1 and IRF-2, respectively [96].
Kuroboshi and colleagues [97] have expressed doubt concerning the nature of altered IRF-1 expression in uterine endometrial carcinoma compared with pre- and postmenopausal endometrial tissue, suggesting that the modified levels of this protein could be either an outcome or a cause of the development of this malignancy. Nevertheless, considered together with other findings, whereas IRF-2 may be classified as an oncogene, these results are consistent with IRF-1 having tumor-suppressing potential. Table 2 shows a timeline of the first recognition of the influence of IRF-1 in different human cancers.
Table 2. Timeline represent the year of IRF-1 first involvement in different cancer types.
| Cancer type | Year first reported | References |
|---|---|---|
| Leukemia and pre-leukemia myelodysplasia | 1993 | [70,73,76,77,79] |
| Stomach carcinoma | 1996 | [89] |
| Esophageal carcinoma | 1996 | [90,92] |
| Gastric adenocarcinoma | 1998 | [38] |
| Breast cancer | 1998 | [80,81,86,88,98] |
| Skin melanoma | 1999 | [93] |
| Uterine endometrial carcinoma | 2003 | [97] |
| Cervical cancer | 2006 | [94] |
| HCC | 2013 | [95] |
| Pancreatic cancer | 2014 | [96] |
Inactivation of IRF-1 in human cancers
Several mechanisms responsible for attenuated IRF-1 transcriptional activity in human malignancies have been reported. The following section describes the different routes by which the function of IRF-1 and, thereby, its tumor-suppressing role may be lost in human cancers.
Genetic modulation of IRF-1 activity
Alterations in the IRF-1 gene have been reported in both hematologic malignancies and solid tumors. For instance, inactivating rearrangements or deletions in IRF-1 have been reported in AML [70], and LOH of this gene has been observed in gastric and esopharyngeal cancers and renal cell carcinoma [99]. In addition, a missense mutation in exon 2 of IRF-1 has been identified in stomach cancer. This alteration was accompanied by loss of IRF-1 transcriptional and, consequently, tumor-suppressing activity [38]. We previously mentioned in this review that genetic alterations in IRF-1 have also been documented in breast cancer [88]. Green and colleagues [76] have also described a single IRF-1 allele deletion in AML and MDS patients.
Transcriptional modulation of IRF-1 activity
IRF-1 mRNA is subject to several alterations that ultimately lead to loss of function. For example, Lee and colleagues [94] have identified splice variants of IRF-1 mRNA missing combinations of exons 7, 8, and 9. These variants are highly expressed in cervical cancer tissue and associated with attenuated IRF-1 transcriptional activity. miR-23a, which binds to the 3′-UTR of IRF-1 mRNA, is overexpressed in gastric adenocarcinoma, resulting in IRF-1 down-regulation and loss of transcriptional activity, in turn enhancing pro-proliferative and anti-apoptotic conditions [100]. Skipped exons in IRF-1 transcripts have also been reported. Skipping of exon 2 in mutant IRF-1 is associated with an absent DBD and loss of the tumor-suppressing action of the encoded protein [101]. Moreover, faulty IRF-1 mRNA can result from accelerated exon skipping, with affected transcripts lacking a translation initiation site. This constitutes one of the mechanisms leading to IRF-1 inactivation in hematopoietic cancers [76].
Proteomic modulation of IRF-1 activity
SUMOylation
SUMOylation is a post-translational modification in which lysine residues are modified by attachment of a SUMO group [102]. SUMOylation of IRF-1 stabilizes this protein and protects it from degradation, but also leads to loss of its transcriptional activity, inhibiting IRF-1-mediated apoptosis and tumor-suppressing activity; therefore, levels of IRF-1 SUMOylation are increased in tumor cells [103]. Indeed, SUMO-IRF-1 induces transformation of NIH 3T3 cells in a dose-dependent manner, implying that following SUMOylation, IRF-1 loses its antioncogenic activity and mimics the oncogenic factor IRF-2 [104].
Oncoproteins
Viruses have evolved various strategies to survive immune responses. Human papillomavirus (HPV), which is associated with increased risk of cervical cancer [105], counteracts interferon signaling to evade the human immune defense system. It has been reported that HPV oncoproteins can bind and inhibit the transcriptional activity of interferon-regulatory proteins including IRF-1 [106]. For example, HPV E6 inhibits IRF-3 activity, and HPV E7 binds to and inactivates IRF-1 and IRF-9 [107]. An early report from Park and colleagues [108] indicated a physical interaction between HPV E7 and IRF-1 leading to loss of transcriptional activity, thought to be mediated by histone deacetylation, and abrogation of IFN-α signaling.
Nucleophosmin
Nucleophosmin (NPM) predominantly acts as a nuclear shuttling protein; however, translocation of the NPM gene is associated with mislocalization of this protein to the cytoplasm, which has been implicated in loss of the function and antioncogenic effects of IRF-1. Cytosolic NPM has been observed in clinical AML specimens [109]. Moreover, overexpression of this protein has been noted in leukemia cell lines, and high levels correlate with transformation of NIH 3T3 cells [110]. Anti-apoptotic proteins of the GAGE family (a group of highly related tumor antigens) can bind and stabilize NPM/B23, which is indirectly involved in loss of IRF-1 function. In addition, interaction between GAGE proteins and IRF-1 has been reported in cancer cells, possibly explaining GAGE-induced cell survival and resistance to IFN-γ treatment [111].
Therapeutic targetting of IRF-1 in human cancers
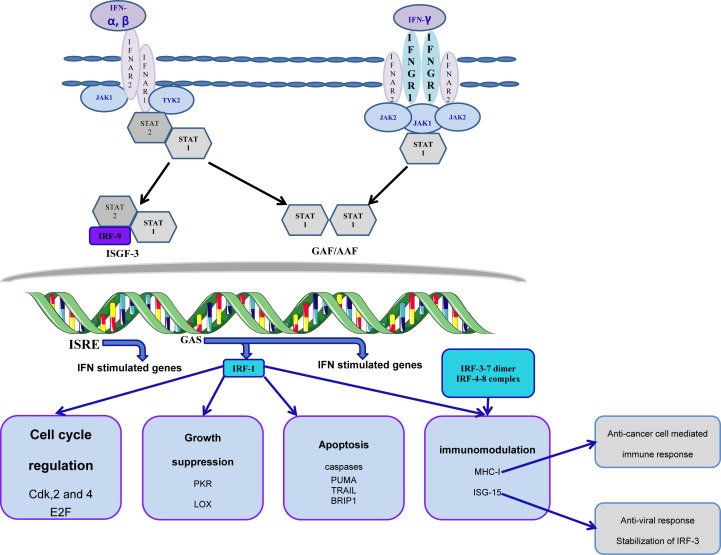
Cancer models have provided evidence on the antitumor potential of interferon and importance of signaling through type I interferon receptor in mediation of anticancer immune response [112]. Importantly, IRF-1 is induced downstream to IFN-γ-type I interferon receptor signal transduction cascade (Figure 3). In turn, IRF-1 contributes to modulation of immune response besides suppression of cell growth and transformation. Amongst targets of IRF-1 genes is the major histocompatibility class I (MHCI) gene. Induction of MHCI contributes to the long-known antitumor potential of IRF-1 enhancing tumor antigen presentation that improves the efficacy of the cell-mediated antitumor immune response. IFN-γ also triggers clonal expansion and activation of T cells and production of MHCII [113]. Therefore, functional type I IFN signaling and local expression of IFN-stimulated genes by tumors are cofactors for better disease prognosis and outcomes [114].
Figure 3. Signal transduction pathway and cross-talk in the interferon system.
Stimulation of type I and II interferon receptor recruits Janus and tyrosine kinases that phosphorylates and activates STAT. STAT proteins trigger nuclear translocation of IRF members where they interact with ISRE to regulate expression of ISG resulting in various physiological responses.
That IRF-1 is a potential target for new therapies has been highlighted by correlations between its inactivation and several types of human cancers. However, the exact IRF-1-related mechanisms that might be targetted to benefit therapy of human malignancies are not yet fully clear. It has been reported that an abnormally low IRF-1/IRF-2 ratio due to defective transcription is a basic characteristic of leukemogenesis, but that administration of certain cytokines (e.g. IL-4) can return this ratio to normal levels [79]. Similarly, Yoshino and colleagues [115] have suggested that this ratio has prognostic value with respect to diffusely infiltrating astrocytomas, and consider both IRFs to represent targets of future therapies. Consistent with this, it has been proposed that increasing the IRF-1/IRF-2 ratio may serve as a novel therapeutic strategy for pancreatic cancer [96]. Interestingly, a recent study has pointed out that down-regulation of IRF-1 may overcome resistance to anti-angiogenic drugs in glioblastoma [116]. There is also evidence that IRF-1 regulates both second mitochondria-derived activator of caspases (SMAC) and the pro-inflammatory response, suggesting that its up-regulation in human cancers could favor apoptosis [117]. With respect to breast cancer, IRF-1 has been identified as an effector that restores the sensitivity of estrogen receptor-positive metastatic breast carcinoma to the anti-estrogenic drug fulvestrant [118].
A recent study [119] has implicated the inactivation of type I interferon receptor chain (IFNAR1) in colorectal cancer (CRC) development and patients’ overall poor prognosis. Authors have reported that genetic or pharmacological stabilization of IFNAR1 can improve CRC patients’ response to chimeric antigen receptor treatment and inhibition of programmed cell death protein 1 (PD-1), which, in turn increases the efficacy of tumor immunotherapy by augmenting the activity and the number of cytotoxic T cells in the tumor niche [120]. Down-regulation of IFN signaling is associated with reduced expression of IFN-induced genes; this is indicated by lower nuclear levels of p-STAT2 in malignant colorectal tissues as compared with normal tissue specimens [119]. It has been reported that colorectal tumor microenvironment associated stress (hypoxia, for example) results in production of lower levels of IFNAR1 in CRC models [121]. In this context, significant higher levels of IFNAR1 were observed in colorectal normal tissues when compared with CRC tissues [119].
Conclusion
Up-regulation of IRF-1 in cancerous lesions is regarded as an approach that could improve prognosis and alleviate resistance to immunotherapy [95,122]. In addition, measuring IRF-1 expression within malignancies is considerably helpful in determining prognosis and predicting response to immunotherapy. The various mechanisms involved in IRF-1 inactivation in human cancers involve processes at DNA, RNA, and protein levels. Concerning DNA, LOH along with monosomy and mutation play a major role in limiting the functionality of IRF-1, ultimately resulting in oncogenesis. From the transcriptional perspective, exon skipping and alternative splicing are the two key events leading to reduced IRF-1 activity. Finally, SUMOylation, oncoproteins, and NPM have a significant impact on the IRF-1 protein and oncogenesis (Table 3). The data summarized in this review suggest that more research should be conducted to explore the potential utility of the IRF-1 as a therapeutic target in cancer.
Table 3. Mechanisms involved in IRF-1 inactivation at various molecular levels.
| Level | Inactivation mechanisms | ||
|---|---|---|---|
| DNA | LOH | Monosomy | Mutations |
| mRNA | Exon skipping | Faulty splicing | |
| Protein | SUMOylation | Oncoprotein | |
Abbreviations
- AML
acute myelogenous leukemia
- APL
acute promyelocytic leukemia
- BRCA1
breast cancer susceptibility gene 1
- BRIP1
BRCA1-interacting protein C-terminal helicase 1
- CDK
cyclin-dependent kinase
- CML
chronic myeloid leukemia
- CRC
colorectal cancer
- DBD
DNA-binding domain
- ESCC
esophageal squamous cell carcinoma
- HCC
hepatocellular carcinoma
- HPV
human papillomavirus
- IAD
IRF-association domain
- ICSBP
interferon consensus sequence binding protein
- IFN
interferon
- IFNAR1
type I interferon receptor chain
- IRF-1
interferon regulatory factor 1
- ISG
interferon-stimulated gene
- ISGF-3
interferon-stimulated gene factor 3
- ISRE
interferon-stimulated response element
- LOH
loss of heterozygosity
- MEF
mouse embryonic fibroblast
- MDS
myelodysplastic syndrome
- MHCI
major histocompatibility class I
- NPM
nucleophosmin
- STAT
signal transducer and activator of transcription
- SUMO
small ubiquitin-like modifier
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Miyamoto M., et al. (1988) Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54, 903–913 10.1016/S0092-8674(88)91307-4 [DOI] [PubMed] [Google Scholar]
- 2.Harada H., et al. (1989) Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58, 729–739 10.1016/0092-8674(89)90107-4 [DOI] [PubMed] [Google Scholar]
- 3.Meraro D., et al. (1999) Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J. Immunol. 163, 6468–6478 [PubMed] [Google Scholar]
- 4.Taniguchi T., et al. (2001) IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- 5.Yanai H., Negishi H. and Taniguchi T. (2012) The IRF family of transcription factors: Inception, impact and implications in oncogenesis. Oncoimmunology 1, 1376–1386 10.4161/onci.22475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F.F., et al. (2013) Function and mechanism by which interferon regulatory factor-1 inhibits oncogenesis. Oncol. Lett. 5, 417–423 10.3892/ol.2012.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden E.C., et al. (2007) Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6, 975–990 10.1038/nrd2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Au W.C., et al. (1995) Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. U.S.A. 92, 11657–11661 10.1073/pnas.92.25.11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marie I., Durbin J.E. and Levy D.E. (1998) Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17, 6660–6669 10.1093/emboj/17.22.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., et al. (2009) Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J. Immunol. 183, 6989–6997 10.4049/jimmunol.0901386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama M., et al. (1998) Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17, 1087–1095 10.1093/emboj/17.4.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitoh T., et al. (2006) Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7, 598–605 10.1038/ni1347 [DOI] [PubMed] [Google Scholar]
- 13.Lu G., et al. (2006) ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell. Mol. Biol. (Noisy-le-grand) 52, 29–41 [PubMed] [Google Scholar]
- 14.Eisenbeis C.F., Singh H. and Storb U. (1995) Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9, 1377–1387 10.1101/gad.9.11.1377 [DOI] [PubMed] [Google Scholar]
- 15.Lu R., et al. (2003) IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 17, 1703–1708 10.1101/gad.1104803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittrucker H.W., et al. (1997) Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275, 540–543 10.1126/science.275.5299.540 [DOI] [PubMed] [Google Scholar]
- 17.Iida S., et al. (1997) Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat. Genet. 17, 226–230 10.1038/ng1097-226 [DOI] [PubMed] [Google Scholar]
- 18.Yoshida K., et al. (2005) Active repression of IFN regulatory factor-1-mediated transactivation by IFN regulatory factor-4. Int. Immunol. 17, 1463–1471 10.1093/intimm/dxh324 [DOI] [PubMed] [Google Scholar]
- 19.Eames H.L., Corbin A.L. and Udalova I.A. (2016) Interferon regulatory factor 5 in human autoimmunity and murine models of autoimmune disease. Transl. Res. 167, 167–182 10.1016/j.trsl.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 20.Masuda T., et al. (2014) Transcription factor IRF5 drives P2X4R+-reactive microglia gating neuropathic pain. Nat. Commun. 5, 3771 10.1038/ncomms4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D., et al. (2016) Specific detection of interferon regulatory factor 5 (IRF5): a case of antibody inequality. Sci. Rep. 6, 31002 10.1038/srep31002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fresquet V., et al. (2012) High-throughput sequencing analysis of the chromosome 7q32 deletion reveals IRF5 as a potential tumour suppressor in splenic marginal-zone lymphoma. Br. J. Haematol. 158, 712–726 10.1111/j.1365-2141.2012.09226.x [DOI] [PubMed] [Google Scholar]
- 23.Dalmas E., et al. (2015) Irf5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nat. Med. 21, 610–618 10.1038/nm.3829 [DOI] [PubMed] [Google Scholar]
- 24.Ingraham C.R., et al. (2006) Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet. 38, 1335–1340 10.1038/ng1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biggs L.C., et al. (2014) Interferon regulatory factor 6 regulates keratinocyte migration. J. Cell Sci. 127, 2840–2848 10.1242/jcs.139246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith A.L., et al. (2017) Generation and characterization of a conditional allele of interferon regulatory factor 6. Genesis 55, 10.1002/dvg.23038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metwalli K.A., et al. (2018) Interferon regulatory factor 6 is necessary for salivary glands and pancreas development. J. Dent. Res. 97, 226–236 10.1177/0022034517729803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L. and Pagano J.S. (1997) IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17, 5748–5757 10.1128/MCB.17.10.5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda K., et al. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- 30.tenOever B.R., et al. (2004) Activation of TBK1 and IKKvarepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78, 10636–10649 10.1128/JVI.78.19.10636-10649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Driggers P.H., et al. (1990) An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. U.S.A. 87, 3743–3747 10.1073/pnas.87.10.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Politis A.D., et al. (1992) Modulation of interferon consensus sequence binding protein mRNA in murine peritoneal macrophages. Induction by IFN-gamma and down-regulation by IFN-alpha, dexamethasone, and protein kinase inhibitors. J. Immunol. 148, 801–807 [PubMed] [Google Scholar]
- 33.Meraro D., et al. (2002) IFN-stimulated gene 15 is synergistically activated through interactions between the myelocyte/lymphocyte-specific transcription factors, PU.1, IFN regulatory factor-8/IFN consensus sequence binding protein, and IFN regulatory factor-4: characterization of a new subtype of IFN-stimulated response element. J. Immunol. 168, 6224–6231 10.4049/jimmunol.168.12.6224 [DOI] [PubMed] [Google Scholar]
- 34.Liu J., et al. (2004) Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J. Biol. Chem. 279, 55609–55617 10.1074/jbc.M406565200 [DOI] [PubMed] [Google Scholar]
- 35.Kraus T.A., et al. (2003) A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J. Biol. Chem. 278, 13033–13038 10.1074/jbc.M212972200 [DOI] [PubMed] [Google Scholar]
- 36.Veals S.A., et al. (1992) Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12, 3315–3324 10.1128/MCB.12.8.3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada H., et al. (1993) Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science 259, 971–974 10.1126/science.8438157 [DOI] [PubMed] [Google Scholar]
- 38.Nozawa H., et al. (1998) Functionally inactivating point mutation in the tumor-suppressor IRF-1 gene identified in human gastric cancer. Int. J. Cancer 77, 522–527 10.1002/(SICI)1097-0215(19980812)77:4%3c522::AID-IJC8%3e3.0.CO;2-W [DOI] [PubMed] [Google Scholar]
- 39.Passioura T., et al. (2005) N-ras-induced growth suppression of myeloid cells is mediated by IRF-1. Cancer Res. 65, 797–804 [PubMed] [Google Scholar]
- 40.Kirchhoff S., Schaper F. and Hauser H. (1993) Interferon regulatory factor 1 (IRF-1) mediates cell growth inhibition by transactivation of downstream target genes. Nucleic Acids Res. 21, 2881–2889 10.1093/nar/21.12.2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka N., et al. (1996) Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature 382, 816–818 10.1038/382816a0 [DOI] [PubMed] [Google Scholar]
- 42.Kirchhoff S., et al. (1996) Regulation of cell growth by IRF-1 in BHK-21 cells. Cytotechnology 22, 147–156 10.1007/BF00353934 [DOI] [PubMed] [Google Scholar]
- 43.Coccia E.M., et al. (1999) Activation and repression of the 2-5A synthetase and p21 gene promoters by IRF-1 and IRF-2. Oncogene 18, 2129–2137 10.1038/sj.onc.1202536 [DOI] [PubMed] [Google Scholar]
- 44.Nozawa H., et al. (1999) Loss of transcription factor IRF-1 affects tumor susceptibility in mice carrying the Ha-ras transgene or nullizygosity for p53. Genes Dev. 13, 1240–1245 10.1101/gad.13.10.1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen H., Lin R. and Hiscott J. (1997) Activation of multiple growth regulatory genes following inducible expression of IRF-1 or IRF/RelA fusion proteins. Oncogene 15, 1425–1435 10.1038/sj.onc.1201318 [DOI] [PubMed] [Google Scholar]
- 46.Beretta L., et al. (1996) Expression of the protein kinase PKR in modulated by IRF-1 and is reduced in 5q- associated leukemias. Oncogene 12, 1593–1596 [PubMed] [Google Scholar]
- 47.Tan R.S., Taniguchi T. and Harada H. (1996) Identification of the lysyl oxidase gene as target of the antioncogenic transcription factor, IRF-1, and its possible role in tumor suppression. Cancer Res. 56, 2417–2421 [PubMed] [Google Scholar]
- 48.Kim E.J., et al. (2002) Interferon regulatory factor-1 mediates interferon-gamma-induced apoptosis in ovarian carcinoma cells. J. Cell. Biochem. 85, 369–380 10.1002/jcb.10142 [DOI] [PubMed] [Google Scholar]
- 49.Fulda S. and Debatin K.M. (2002) IFNgamma sensitizes for apoptosis by upregulating caspase-8 expression through the Stat1 pathway. Oncogene 21, 2295–2308 10.1038/sj.onc.1205255 [DOI] [PubMed] [Google Scholar]
- 50.Gao J., et al. (2010) IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ. 17, 699–709 10.1038/cdd.2009.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kroger A., et al. (2007) Tumor suppression by IFN regulatory factor-1 is mediated by transcriptional down-regulation of cyclin D1. Cancer Res. 67, 2972–2981 10.1158/0008-5472.CAN-06-3564 [DOI] [PubMed] [Google Scholar]
- 52.Pizzoferrato E., et al. (2004) Ectopic expression of interferon regulatory factor-1 promotes human breast cancer cell death and results in reduced expression of survivin. Cancer Res. 64, 8381–8388 10.1158/0008-5472.CAN-04-2223 [DOI] [PubMed] [Google Scholar]
- 53.Armstrong M.J., et al. (2012) Interferon Regulatory Factor 1 (IRF-1) induces p21(WAF1/CIP1) dependent cell cycle arrest and p21(WAF1/CIP1) independent modulation of survivin in cancer cells. Cancer Lett. 319, 56–65 10.1016/j.canlet.2011.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura T., et al. (1995) An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature 376, 596–599 10.1038/376596a0 [DOI] [PubMed] [Google Scholar]
- 55.Tamura T., et al. (1997) DNA damage-induced apoptosis and Ice gene induction in mitogenically activated T lymphocytes require IRF-1. Leukemia 11, 439–440 [PubMed] [Google Scholar]
- 56.Frontini M., et al. (2009) A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res. 37, 1073–1085 10.1093/nar/gkn1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seal S., et al. (2006) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 38, 1239–1241 10.1038/ng1902 [DOI] [PubMed] [Google Scholar]
- 58.Pamment J., et al. (2002) Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene 21, 7776–7785 10.1038/sj.onc.1205981 [DOI] [PubMed] [Google Scholar]
- 59.Cui L., et al. (2012) IRF-2 is over-expressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumour Biol. 33, 247–255 10.1007/s13277-011-0273-3 [DOI] [PubMed] [Google Scholar]
- 60.Passioura T., et al. (2005) A retroviral library genetic screen identifies IRF-2 as an inhibitor of N-ras-induced growth suppression in leukemic cells. Oncogene 24, 7327–7336 10.1038/sj.onc.1208877 [DOI] [PubMed] [Google Scholar]
- 61.Nguyen H., et al. (1995) Transcription factor IRF-2 exerts its oncogenic phenotype through the DNA binding/transcription repression domain. Oncogene 11, 537–544 [PubMed] [Google Scholar]
- 62.Vaughan P.S., et al. (1995) Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377, 362–365 10.1038/377362a0 [DOI] [PubMed] [Google Scholar]
- 63.Vaughan P.S., et al. (1998) Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF-2. J. Biol. Chem. 273, 194–199 10.1074/jbc.273.1.194 [DOI] [PubMed] [Google Scholar]
- 64.Pettersson S., et al. (2009) Role of Mdm2 acid domain interactions in recognition and ubiquitination of the transcription factor IRF-2. Biochem. J. 418, 575–585 10.1042/BJ20082087 [DOI] [PubMed] [Google Scholar]
- 65.Armstrong M.J., et al. (2012) Interferon regulatory factor 1 (IRF-1) induces p21(WAF1/CIP1) dependent cell cycle arrest and p21(WAF1/CIP1) independent modulation of survivin in cancer cells. Cancer Lett. 319, 56–65 10.1016/j.canlet.2011.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stang M.T., et al. (2007) Interferon regulatory factor-1-induced apoptosis mediated by a ligand-independent fas-associated death domain pathway in breast cancer cells. Oncogene 26, 6420–6430 10.1038/sj.onc.1210470 [DOI] [PubMed] [Google Scholar]
- 67.Clarke N., et al. (2004) Tumor suppressor IRF-1 mediates retinoid and interferon anticancer signaling to death ligand TRAIL. EMBO J. 23, 3051–3060 10.1038/sj.emboj.7600302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lorenzi S., et al. (2012) IRF1 and NF-kB restore MHC class I-restricted tumor antigen processing and presentation to cytotoxic T cells in aggressive neuroblastoma. PLoS ONE 7, e46928 10.1371/journal.pone.0046928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X.Q., et al. (2018) IRF1 up-regulates isg15 gene expression in dsRNA stimulation or CSFV infection by targeting nucleotides -487 to -325 in the 5' flanking region. Mol. Immunol. 94, 153–165 10.1016/j.molimm.2017.12.025 [DOI] [PubMed] [Google Scholar]
- 70.Willman C.L., et al. (1993) Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemic myelodysplasia. Science 259, 968–971 10.1126/science.8438156 [DOI] [PubMed] [Google Scholar]
- 71.Kerim S., et al. (1990) 5q- anomaly in lymphoid disorders. Leukemia 4, 12–15 [PubMed] [Google Scholar]
- 72.Le Beau M.M., et al. (1986) Clinical and cytogenetic correlations in 63 patients with therapy-related myelodysplastic syndromes and acute nonlymphocytic leukemia: further evidence for characteristic abnormalities of chromosomes no. 5 and 7. J. Clin. Oncol. 4, 325–345 10.1200/JCO.1986.4.3.325 [DOI] [PubMed] [Google Scholar]
- 73.Boultwood J., et al. (1993) Allelic loss of IRF1 in myelodysplasia and acute myeloid leukemia: retention of IRF1 on the 5q- chromosome in some patients with the 5q- syndrome. Blood 82, 2611–2616 [PubMed] [Google Scholar]
- 74.Le Beau M.M., et al. (1993) Cytogenetic and molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases. Proc. Natl. Acad. Sci. U.S.A. 90, 5484–5488 10.1073/pnas.90.12.5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harada H., et al. (1994) Accelerated exon skipping of IRF-1 mRNA in human myelodysplasia/leukemia; a possible mechanism of tumor suppressor inactivation. Oncogene 9, 3313–3320 [PubMed] [Google Scholar]
- 76.Green W.B., et al. (1999) Lack of IRF-1 expression in acute promyelocytic leukemia and in a subset of acute myeloid leukemias with del(5)(q31). Leukemia 13, 1960–1971 10.1038/sj.leu.2401596 [DOI] [PubMed] [Google Scholar]
- 77.Tzoanopoulos D., et al. (2002) Low expression of interferon regulatory factor-1 and identification of novel exons skipping in patients with chronic myeloid leukaemia. Br. J. Haematol. 119, 46–53 10.1046/j.1365-2141.2002.03829.x [DOI] [PubMed] [Google Scholar]
- 78.Schmidt M., et al. (1998) Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 91, 22–29 [PubMed] [Google Scholar]
- 79.Preisler H.D., et al. (2001) Alterations in IRF1/IRF2 expression in acute myelogenous leukemia. Am. J. Hematol. 68, 23–31 10.1002/ajh.1144 [DOI] [PubMed] [Google Scholar]
- 80.Doherty G.M., et al. (2001) Interferon regulatory factor expression in human breast cancer. Ann. Surg. 233, 623–629 10.1097/00000658-200105000-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Connett J.M., et al. (2005) Interferon regulatory factor 1 (IRF-1) and IRF-2 expression in breast cancer tissue microarrays. J. Interferon Cytokine Res. 25, 587–594 10.1089/jir.2005.25.587 [DOI] [PubMed] [Google Scholar]
- 82.Yim J.H., et al. (2003) The role of interferon regulatory factor-1 and interferon regulatory factor-2 in IFN-gamma growth inhibition of human breast carcinoma cell lines. J. Interferon Cytokine Res. 23, 501–511 10.1089/10799900360708623 [DOI] [PubMed] [Google Scholar]
- 83.Masumi A. and Ozato K. (2001) Coactivator p300 acetylates the interferon regulatory factor-2 in U937 cells following phorbol ester treatment. J. Biol. Chem. 276, 20973–20980 10.1074/jbc.M101707200 [DOI] [PubMed] [Google Scholar]
- 84.Masumi A., et al. (2003) Interferon regulatory factor-2 regulates cell growth through its acetylation. J. Biol. Chem. 278, 25401–25407 10.1074/jbc.M213037200 [DOI] [PubMed] [Google Scholar]
- 85.Watanabe N., et al. (1991) Activation of IFN-beta element by IRF-1 requires a posttranslational event in addition to IRF-1 synthesis. Nucleic Acids Res. 19, 4421–4428 10.1093/nar/19.16.4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bouker K.B., et al. (2007) The A4396G polymorphism in interferon regulatory factor 1 is frequently expressed in breast cancer cell lines. Cancer Genet. Cytogenet. 175, 61–64 10.1016/j.cancergencyto.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 87.Loercher A.E., et al. (2005) MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 168, 35–40 10.1083/jcb.200410115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cavalli L.R., et al. (2010) Frequent loss of heterozygosity at the interferon regulatory factor-1 gene locus in breast cancer. Breast Cancer Res. Treat. 121, 227–231 10.1007/s10549-009-0509-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamura G., et al. (1996) Allelotype of adenoma and differentiated adenocarcinoma of the stomach. J. Pathol. 180, 371–377 10.1002/(SICI)1096-9896(199612)180:4%3c371::AID-PATH704%3e3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- 90.Ogasawara S., et al. (1996) Common deleted region on the long arm of chromosome 5 in esophageal carcinoma. Gastroenterology 110, 52–57 10.1053/gast.1996.v110.pm8536888 [DOI] [PubMed] [Google Scholar]
- 91.Knudson A.G., Jr (1977) Mutation and cancer in man. Cancer 39, 1882–1886 10.1002/1097-0142(197704)39:4+%3c1882::AID-CNCR2820390821%3e3.0.CO;2- [DOI] [PubMed] [Google Scholar]
- 92.Wang Y., et al. (2007) Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 67, 2535–2543 10.1158/0008-5472.CAN-06-3530 [DOI] [PubMed] [Google Scholar]
- 93.Lowney J.K., et al. (1999) Interferon regulatory factor-1 and -2 expression in human melanoma specimens. Ann. Surg. Oncol. 6, 604–608 10.1007/s10434-999-0604-4 [DOI] [PubMed] [Google Scholar]
- 94.Lee E.J., et al. (2006) Alternative splicing variants of IRF-1 lacking exons 7, 8, and 9 in cervical cancer. Biochem. Biophys. Res. Commun. 347, 882–888 10.1016/j.bbrc.2006.06.145 [DOI] [PubMed] [Google Scholar]
- 95.Yi Y., et al. (2013) Interferon regulatory factor (IRF)-1 and IRF-2 are associated with prognosis and tumor invasion in HCC. Ann. Surg. Oncol. 20, 267–276 10.1245/s10434-012-2487-z [DOI] [PubMed] [Google Scholar]
- 96.Sakai T., et al. (2014) The roles of interferon regulatory factors 1 and 2 in the progression of human pancreatic cancer. Pancreas 43, 909–916 10.1097/MPA.0000000000000116 [DOI] [PubMed] [Google Scholar]
- 97.Kuroboshi H., et al. (2003) Interferon regulatory factor-1 expression in human uterine endometrial carcinoma. Gynecol. Oncol. 91, 354–358 10.1016/S0090-8258(03)00515-8 [DOI] [PubMed] [Google Scholar]
- 98.Tirkkonen M., et al. (1998) Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Canc. 21, 177–184 10.1002/(SICI)1098-2264(199803)21:3%3c177::AID-GCC1%3e3.0.CO;2-X [DOI] [PubMed] [Google Scholar]
- 99.Sugimura J., et al. (1997) Allelic loss on chromosomes 3p, 5q and 17p in renal cell carcinomas. Pathol. Int. 47, 79–83 10.1111/j.1440-1827.1997.tb03724.x [DOI] [PubMed] [Google Scholar]
- 100.Liu X., et al. (2013) miR-23a targets interferon regulatory factor 1 and modulates cellular proliferation and paclitaxel-induced apoptosis in gastric adenocarcinoma cells. PLoS ONE 8, e64707 10.1371/journal.pone.0064707 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Fragale A., Marsili G. and Battistini A. (2013) Genetic and epigenetic regulation of interferon regulatory factor expression: implications in human malignancies. Genet. Syndr. Gene Ther. 4, 205–219 [Google Scholar]
- 102.Seeler J.S. and Dejean A. (2003) Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4, 690–699 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- 103.Park J., et al. (2007) Elevated level of SUMOylated IRF-1 in tumor cells interferes with IRF-1-mediated apoptosis. Proc. Natl. Acad. Sci. U.S.A. 104, 17028–17033 10.1073/pnas.0609852104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park S.M., et al. (2010) SUMOylated IRF-1 shows oncogenic potential by mimicking IRF-2. Biochem. Biophys. Res. Commun. 391, 926–930 10.1016/j.bbrc.2009.11.166 [DOI] [PubMed] [Google Scholar]
- 105.Burd E.M. (2003) Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16, 1–17 10.1128/CMR.16.1.1-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lace M.J., et al. (2010) Interferon regulatory factor (IRF)-2 activates the HPV-16 E6-E7 promoter in keratinocytes. Virology 399, 270–279 10.1016/j.virol.2009.12.025 [DOI] [PubMed] [Google Scholar]
- 107.Cordano P., et al. (2008) The E6E7 oncoproteins of cutaneous human papillomavirus type 38 interfere with the interferon pathway. Virology 377, 408–418 10.1016/j.virol.2008.04.036 [DOI] [PubMed] [Google Scholar]
- 108.Park J.S., et al. (2000) Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 275, 6764–6769 10.1074/jbc.275.10.6764 [DOI] [PubMed] [Google Scholar]
- 109.Falini B., et al. (2005) Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352, 254–266 10.1056/NEJMoa041974 [DOI] [PubMed] [Google Scholar]
- 110.Kondo T., et al. (1997) Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene 15, 1275–1281 10.1038/sj.onc.1201286 [DOI] [PubMed] [Google Scholar]
- 111.Kular R.K., et al. (2009) GAGE, an antiapoptotic protein binds and modulates the expression of nucleophosmin/B23 and interferon regulatory factor 1. J. Interferon Cytokine Res. 29, 645–655 10.1089/jir.2008.0099 [DOI] [PubMed] [Google Scholar]
- 112.Diamond M.S., et al. (2011) Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 10.1084/jem.20101158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Andrea M., et al. (2002) The interferon system: an overview. Eur. J. Paediatr. Neurol. 6, A41–A46, 10.1053/ejpn.2002.0573 [DOI] [PubMed] [Google Scholar]
- 114.Zitvogel L., et al. (2015) Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 10.1038/nri3845 [DOI] [PubMed] [Google Scholar]
- 115.Yoshino A., et al. (2005) Therapeutic implications of interferon regulatory factor (IRF)-1 and IRF-2 in diffusely infiltrating astrocytomas (DIA): response to interferon (IFN)-beta in glioblastoma cells and prognostic value for DIA. J. Neurooncol. 74, 249–260 10.1007/s11060-004-7316-1 [DOI] [PubMed] [Google Scholar]
- 116.Liang J., et al. (2015) Interferon-regulatory factor-1 (IRF1) regulates bevacizumab induced autophagy. Oncotarget 6, 31479–31492 10.18632/oncotarget.5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eckhardt I., Weigert A. and Fulda S. (2014) Identification of IRF1 as critical dual regulator of Smac mimetic-induced apoptosis and inflammatory cytokine response. Cell Death Dis. 5, e1562 10.1038/cddis.2014.498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ning Y., et al. (2010) IFNgamma restores breast cancer sensitivity to fulvestrant by regulating STAT1, IFN regulatory factor 1, NF-kappaB, BCL2 family members, and signaling to caspase-dependent apoptosis. Mol. Cancer Ther. 9, 1274–1285 10.1158/1535-7163.MCT-09-1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Katlinski K.V., et al. (2017) Inactivation of interferon receptor promotes the establishment of immune privileged tumor microenvironment. Cancer Cell 31, 194–207 10.1016/j.ccell.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rosenberg S.A. and Restifo N.P. (2015) Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62 10.1126/science.aaa4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhattacharya S., et al. (2013) Anti-tumorigenic effects of type 1 interferon are subdued by integrated stress responses. Oncogene 32, 4214–4221 10.1038/onc.2012.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Murtas D., et al. (2013) IRF-1 responsiveness to IFN-gamma predicts different cancer immune phenotypes. Br. J. Cancer 109, 76–82 10.1038/bjc.2013.335 [DOI] [PMC free article] [PubMed] [Google Scholar]