Abstract
In bacteria and yeast, glutamine synthetase (GS) expression is tightly regulated by the metabolic status of the cell, both at the transcriptional and posttranscriptional levels. We discuss the relative contributions of light and metabolic cues on the regulation of members of the GS gene family (chloroplastic GS2 and cytosolic GS1) in Arabidopsis. These studies reveal that the dramatic induction of mRNA for chloroplastic GS2 by light is mediated in part by phytochrome and in part by light-induced changes in sucrose (Suc) levels. In contrast, the modest induction of mRNA for cytosolic GS1 by light is primarily mediated by changes in the levels of carbon metabolites. Suc induction of mRNA for GS2 and GS1 occurs in a time- and dose-dependent manner. Suc-induced changes in GS mRNA levels were also observed at the level of GS enzyme activity. In contrast, amino acids were shown to antagonize the Suc induction of GS, both at the level of mRNA accumulation and that of enzyme activity. For GS2, the gene whose expression was the most dramatically regulated by metabolites, we used a GS2 promoter-β-glucuronidase fusion to demonstrate that transcriptional control is involved in this metabolic regulation. Our results suggest that the metabolic regulation of GS expression in plants is controlled by the relative abundance of carbon skeletons versus amino acids. This would allow nitrogen assimilation into glutamine to proceed (or not) according to the metabolic status and biosynthetic needs of the plant. This type of GS gene regulation is reminiscent of the nitrogen regulatory system in bacteria, and suggests an evolutionary link between metabolic sensing and signaling in bacteria and plants.
The assimilation of inorganic nitrogen into amino acids is a biochemical process that is critical for plant growth and has marked effects on plant productivity and crop yield (Lawlor et al., 1989; Mattsson et al., 1991). The enzyme Gln synthetase (GS) (EC 6.3.1.2) is key in this nitrogen assimilatory process, as it catalyzes the first step in the conversion of inorganic nitrogen (ammonium) into its organic form (Gln). Distinct isoenzymes of GS exist in the chloroplast (GS2) and cytosol (GS1) of many plant species (Mann et al., 1979; Hirel and Gadal, 1980; McNally et al., 1983; Lam et al., 1996; Oliveira et al., 1997). These distinct GS isoenzymes are encoded by distinct nuclear genes in all higher plants studied. Expression studies showing that the distinct GS genes display organ-specific, cell-specific, developmental, and temporal patterns of gene expression suggest that the chloroplastic GS2 and cytosolic GS1 isoforms perform distinct functions in vivo (Edwards and Coruzzi, 1989; Sakamoto et al., 1990; Cock et al., 1991; Sakakibara et al., 1992; Li et al., 1993).
Despite its small genome, Arabidopsis, like all other higher plants examined, has a family of GS genes: a single nuclear gene for chloroplastic GS2 and multiple genes (three identified to date) for cytosolic GS1. These GS genes have been shown to display organ-specific patterns of mRNA expression (Peterman and Goodman, 1991; Bernhard and Matile, 1994). We have furthered the study of GS gene regulation in Arabidopsis by testing the effects of light, carbon, and organic nitrogen supplementation on the expression of genes for chloroplastic GS2 or cytosolic GS1. These studies include measurements of changes in GS transcription, levels of steady-state mRNA, and levels of GS enzyme activity. The experiments were performed in planta and analyzed within a time frame compatible with a normal day/night cycle, thus addressing the possible physiological significance of such regulation.
Our findings reveal that levels of mRNA for the chloroplastic GS2 or the cytosolic GS1 are each induced by light or by carbon metabolites in a time frame compatible with a normal day/night cycle. The dramatic light induction of mRNA for GS2 is mediated in part by phytochrome and in part by light-induced changes in levels of Suc. In contrast, the modest light induction of mRNA for GS1 is primarily mediated by metabolic cues. We further demonstrate that organic nitrogen in the form of amino acids has an antagonistic effect on Suc induction of mRNA for both GS2 and GS1. These effects appear to be mediated transcriptionally, as amino acids are shown to antagonize the Suc induction of a GS2 promoter-GUS gene construct. Additionally, we show that regulation of GS expression by carbon and amino acids is reflected in changes in the levels of GS enzyme activity. Thus, Suc and amino acids appear to have reciprocal effects on GS expression observed at the transcriptional, posttranscriptional, and enzyme activity levels. The similarities between the metabolic control of GS in Arabidopsis and mechanisms described in microorganisms are discussed.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The plant tissues used in all experiments were from the Columbia ecotype of Arabidopsis; for the determination of RFLPs for the GS genes the Landsberg ecotype was also used. Arabidopsis recombinant inbred (RI) lines used for mapping purposes were from the Arabidopsis Stock Center at Ohio State University (Lister and Dean, 1993). For genomic DNA isolation, plants were grown in soil in a growth chamber (Environmental Growth Chamber, Chagrin Falls, OH) at an average irradiance of 60 mmol photons m−2 s−1 on a 16 h/8 h light/dark cycle until bolting (approximately 30 d). For protein and RNA extraction, plants were grown semihydroponically. The semihydroponic system for plant culture consisted of growing plants on nylon nets (pore size 250 mm, Tetko, Briarcliff Manor, NY) suspended on a semisolid Murashige and Skoog (MS) medium (0.6% [w/v] agar) in Phytatrays (Sigma).
Except when noted otherwise, plants were grown in an average irradiance of 60 mmol photons m−2 s−1 on a 16 h/8 h light/dark cycle in a medium consisting of ammonium-free/nitrate-free MS medium (Sigma, catalog no. M–9911, or Life Technologies, Long Island, NY, catalog no. 97–5068) supplemented with a total of 4 mm nitrate and 2 mm ammonium (as KNO3 and NH4NO3), and 0.5% (w/v) Suc until the first pair of true leaves were fully expanded (14–18 d). The plants were then transferred to fresh MS medium containing 4 mm nitrate/2 mm ammonium in the absence of carbon, and dark-adapted for 48 h. Thereafter, the plants were transferred to fresh MS medium containing the indicated supplementations and incubated as described in the legend for each figure. The semihydroponic system permits the growth of a large number of plants under identical experimental conditions. Moreover, due to its low agar concentration, this system also allows the easy transfer of plants to different culture media simultaneously and with negligible root damage by just lifting the nylon net and transferring the plants to fresh medium.
Mapping of the GS Genes Using RI Arabidopsis Lines
Genomic DNA was isolated as previously described (Ausubel et al., 1987). Restriction enzyme digests of genomic DNA (1 mg) from either the Landsberg or Columbia ecotype of Arabidopsis were compared to identify a RFLP (Botstein et al., 1980) for each of the Arabidopsis GS genes (Peterman and Goodman, 1991). Distinct restriction patterns were produced between the two ecotypes of Arabidopsis for the following combinations of GS genes/enzymes: gln2/BamHI, gln1;1/EcoRI, gln1;2/HhaI, and gln1;3/EcoNI. To comply with international rules of nomenclature (Price et al., 1996), we adopted the name gln2 for the gene encoding the chloroplastic GS2 isoenzyme (GSL1, Peterman and Goodman, 1991) and gln1;1, gln1;2, and gln1;3 for the genes encoding the cytosolic GS1 isoenzymes (GSR1, GSR2, and GSKB, respectively, Peterman and Goodman, 1991).
The resulting RFLPs were used to perform genomic Southern-blot analysis from 30 different RI lines (Lister and Dean, 1993). Specificity for all RFLPs was ensured by cross-hybridization with each of the GS probes. The GS 3′-specific antisense probes were generated by PCR (Myerson, 1991) using the cDNAs for the Arabidopsis GS genes gln2, gln1;1, gln1;2, and gln1;3 as templates. The probes were labeled with a digoxigenin DNA-labeling kit, following the protocol provided by the manufacturer (Boehringer Mannheim). PCR amplification was performed using a forward oligonucleotide internal to each GS cDNA (CCAGTTCTCATGGGGCGTGG spanning base positions 1,151–1,149 of the gln2 cDNA; CGATAAATTGGGACTGAGACAC spanning base positions 865–1,351 of the gln1;1 cDNA; GCGTCGTCTCACGGGACACC spanning base positions 937–1,458 of the gln1;2 cDNA; and GCGATAGGGAAGCTTCAGC spanning base positions 801–1,267 of the gln1;3 cDNA) and a second, reverse oligonucleotide (CACAGGAAACAGCTATGACC for gln2 and CGTCGTTTTACAACGTCGTG for gln1;1, gln1;2, and gln1;3), hybridizing to the vector plasmid pBlueScript (Stratagene). Base positions for the GS cDNAs are as described by Peterman and Goodman (1991). The GS genes were mapped relative to 742 markers by Mary Anderson (John Innes Centre, Norwich, UK).
Northern-Blot Analysis
Total RNA extraction and northern-blot analyses were performed as described previously (Ausubel et al., 1987). Each experimental point represented a pool of 200 to 400 semihydroponically grown Arabidopsis plants. In addition to the GS gene-specific probes described above, we used a probe for the β-ATPase cDNA identified in an EST library from Arabidopsis (EST clone no. 107I9T7; accession no. T22836) (Newman et al., 1994) as a control. Gel loading was monitored by probing the blots for the 16S rRNA subunit (rRNA cDNA probe provided by Ben Scheres, University of Utrecht, The Netherlands). Incorporation of digoxigenin label in the probes was estimated according to the protocol provided by the manufacturer (Boehringer Mannheim). Prehybridization, hybridization, washing (0.5× SSC, 65°C), and detection of the probes were performed as indicated by the manufacturer (Boehringer Mannheim). After detection, the blots were exposed to x-ray film and the signals were quantified by densitometry using an imaging software (NIH Image version 1.41, National Institutes of Health, Bethesda, MD). Each northern-blot experiment was repeated at least three times with similar results. Results from a representative experiment are shown in each figure.
Analysis of Phytochrome-Mediated Gene Activation
Arabidopsis plants were grown in the dark (etiolated) for 5 d in nitrogen-free MS medium supplemented with a total of 4 mm nitrate and 2 mm ammonium (as KNO3 and NH4NO3) and no carbon source. Red light was provided by exposing the etiolated seedlings to red fluorescent light bulbs (58,000 μmol photons m−2 in total). A subsequent far-red light pulse (800 μmol photons m−2 in total) was given by exposing Arabidopsis seedlings to incandescent light bulbs fitted with a plexiglass filter (FRF 700, AIN Plastics, Mount Vernon, NY).
Measurement of GS and GUS Activity
A sample equivalent to 100 to 200 μL of frozen, ground tissue was collected from the same pool of plants (200–400 individuals) used for RNA extraction. The samples were extracted in 200 μL of buffer (50 mm Tris-HCl, pH 8.0, 10 mm imidazole, and 0.5% [w/v] β-mercaptoethanol) and kept on ice. GS activity was measured with the transferase method (Shapiro and Stadtman, 1971). A volume of 50 μL of plant extract was mixed with 500 μL of GS assay buffer and incubated for 30 to 60 min at 37°C. The reaction was stopped and the A540 was read in a spectrophotometer. The results were compared with a standard curve using l-Glu γ-monohydroxamate as the standard. GUS activity was determined fluorimetrically as described previously (Jefferson, 1989). The results were normalized to total protein as determined by the method of Bradford (1976) (Bio-Rad) using BSA as a standard.
HPLC Analysis
A sample equivalent to 100 to 200 μL of frozen, ground tissue was collected from the same pool of plants (200–400 individuals) used for RNA extraction and GS enzyme activity. HPLC analysis was performed as previously described (Brears et al., 1993) with minor modifications. The samples were ground in a buffer containing 50 mm Tris-HCl, pH 8.0, 10 mm imidazole, and 0.5% (w/v) β-mercaptoethanol, followed by extraction with 200 mL of methanol:chloroform (6:2.5). The aqueous phase was vacuum-dried, resuspended in 400 μL of water, and filtered at 0.22 μm. HPLC analysis of amino acids was performed using a reverse-phase analytical column (25 cm × 4.6 mm, i.d., particle size 5 mm; model Supelcosyl LC-18, Supelco, Bellefonte, PA). The mobile phase consisted of a gradient of 26 mm phosphate buffer, pH 7.5 (buffer A), with increasing concentrations of 72% (v/v) methanol in water (buffer B). The column eluate was read by a luminescence spectrometer (model LS30, Perkin-Elmer) and recorded in an integrator (ChromJet, ThermoSeparations, Bergenfield, NJ). The amino acid analog norvaline (25 nm/sample, Sigma) was added to the plant extracts immediately before the organic extraction of amino acids and used as an internal standard.
RESULTS
GS Genes from Arabidopsis: Map Positions and Gene-Specific Probes
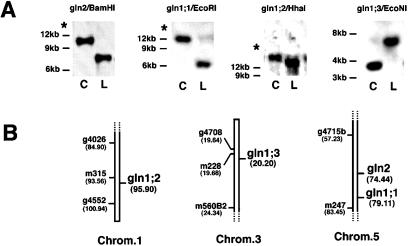
The GS isoenzymes in Arabidopsis are encoded by a gene family including a nuclear gene for a chloroplastic GS2 isoenzyme (gln2) and at least three genes encoding cytosolic GS1 isoenzymes (gln1;1, gln1;2, and gln1;3) (Peterman and Goodman, 1991). Gene-specific probes for chloroplastic GS2 (gln2) and the cytosolic GS1 isoenzymes (gln1;1, gln1;2, and gln1;3) were used to map the GS genes to distinct locations on the Arabidopsis chromosomes (Fig. 1). These results provided finer map positions for some GS genes (gln2, gln1;1, and gln1;3) (Nam et al., 1989) and positioned the unmapped gln1;2 gene on chromosome 1.
Figure 1.
Mapping of GS genes to Arabidopsis chromosomes. Genomic DNA of Arabidopsis was digested with the indicated restriction enzymes and analyzed by Southern blot. A, RFLP produced after restriction digestion of genomic DNA from Arabidopsis ecotypes Columbia (C) and Landsberg (L) with the enzymes BamHI (gln2), EcoRI, (gln1;1), HhaI (gln1;2), and EcoNI (gln1;3), respectively. RFLPs were used to map each gene using DNA from RI lines (Lister and Dean, 1993). Blotting, prehybridization, hybridization, and washing conditions were as described in Methods. The positions of the size marker bands are indicated on the left. The star indicates the migration position of undigested DNA. B, Enlargement of regions of chromosomes (Chrom.) 1, 3, and 5 showing the results of mapping of the GS genes. Markers for the RI mapping are indicated to the left of each chromosome figure. The numbers in parentheses are the positions of the markers (in cM) according to the RI line map of Lister and Dean (1993) as of May, 1998.
Light Affects Levels of GS mRNA in Arabidopsis
GS gene expression is known to be modulated by light in a number of species (Edwards and Coruzzi, 1989; Sakamoto et al., 1990; Cock et al., 1991; Peterman and Goodman, 1991; Sakakibara et al., 1992). We designed experiments to determine the relative contribution of phytochrome versus carbon metabolites on light induction of GS gene expression in Arabidopsis. We also compared the kinetics of light induction of mRNAs for both chloroplastic GS2 and cytosolic GS1 to determine whether this induction might occur in a short, physiological time frame of up to 16 h (Fig. 2A). GS gene-specific probes developed for the mapping experiments described above were used in all northern-blot experiments.
Figure 2.
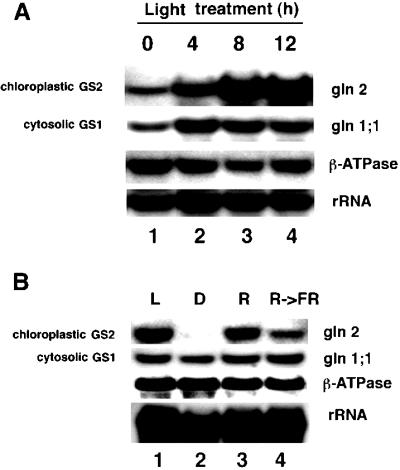
Light induction of GS mRNA accumulation. A, Kinetics of light induction. Arabidopsis plants were grown semihydroponically as described in Methods. The dark-adapted Arabidopsis plants were transferred to light and samples were collected at 4, 8, and 12 h (lanes 2–4). Control plants were collected immediately before transfer to light (lane 1). B, Involvement of phytochrome. Arabidopsis plants were grown in the dark (etiolated) for 5 d in nitrogen-free MS medium with no carbon source supplementation. Thereafter, plants were left in the dark (lane 2), exposed to red light (lane 3), or exposed to red light followed by a pulse of far-red light (lane 4), and re-incubated in the dark for 5 h. Control plants were transferred to light for 5 h at the beginning of the treatments (lane 1). RNA extraction and northern-blot analysis were performed as described in Methods. Northern blots were probed with gene-specific cDNA probes for the Arabidopsis chloroplastic (GS2) isoenzyme gln2 and the cytosolic GS1 isoenzyme gln1;1 as described in Methods. The β-ATPase gene was used as a control gene, and the 16S subunit of the rRNA was used to monitor gel loading.
Dark-adapted Arabidopsis plants were grown and exposed to the light regimens described in Methods. Light exposure of dark-adapted plants induced a progressive accumulation of mRNA for chloroplastic GS2 beginning at 4 h (4-fold induction) and peaking at 12 h (8-fold induction, Fig. 2A, lanes 2–4). The maximal induction of GS2 mRNA ranged from 8- to 19-fold in replicate experiments (Figs. 3, 4, and 5A, compare lanes 1 and 2). In contrast, the levels of mRNA for a representative cytosolic GS1 gene (gln1;1) showed a lower but reproducible induction by light (2- to 3-fold), peaking after only 4 h of light exposure (Fig. 2A, lanes 2–4). The relatively rapid kinetics of light induction of mRNAs for either chloroplastic GS2 or cytosolic GS1 demonstrates that these changes occur in the time course of a normal day. Measurement of GS enzyme activity in leaves of these plants revealed that light has similar effects on the levels of GS enzyme activity (data not shown).
Figure 3.
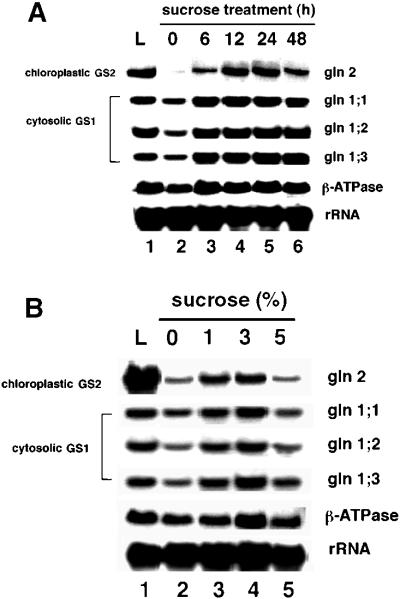
Suc induction of GS mRNA accumulation. Arabidopsis plants were grown semihydroponically and dark-adapted as described in Methods. A, Dark-adapted Arabidopsis plants were transferred in the dark to a low-nitrogen MS medium (2 mm ammonium and 4 mm nitrate) supplemented with 3% (w/v) Suc, and samples were collected after 0, 6, 12, 24, or 48 h of incubation in the dark (lanes 2–6). Control plants (no carbon supplementation) were incubated in constant light for 12 h (lane 1). B, Dark-adapted Arabidopsis plants grown as above and transferred in the dark to a low-nitrogen MS medium (2 mm ammonium and 4 mm nitrate) with no carbon source supplementation (lane 2) or supplemented with 1%, 3%, or 5% Suc and incubated in the dark for 12 h (lanes 3–5). Control plants (no carbon supplementation) were incubated in constant light for 12 h (lane 1). RNA extraction and northern-blot analysis were performed as described in Methods. Probes were as in the legend for Figure 2 except that probes for the cytosolic forms gln1;2 and gln1;3 were also used.
Figure 4.
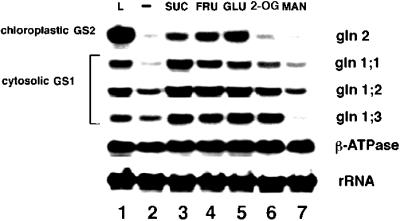
Effects of different carbon metabolites on GS mRNA accumulation. Arabidopsis plants were grown semihydroponically and dark-adapted as described in Methods. The dark-adapted Arabidopsis plants were transferred in the dark to a low-nitrogen MS medium with either no carbon source (lane 2) or supplemented with 90 mm of the following carbon sources: Suc (lane 3), Fru (lane 4), Glc (lane 5), 2-oxoglutarate (2-OG, lane 6), or mannitol (MAN, lane 7). After transfer to these treatments the plants were further incubated for 12 h in the dark (lanes 2–7). Control plants were transferred in the dark to fresh low-nitrogen MS medium with no carbon supplementation and incubated in constant light for 12 h (lane 1). RNA extraction and northern-blot analysis were performed as described in Methods. Probes were as in the legend for Figure 3.
Studies of GS gene expression in other species have revealed that the light induction of mRNA for chloroplastic GS2 is mediated at least in part by phytochrome (Edwards and Coruzzi, 1989; Sakamoto et al., 1990). We sought to determine the participation of phytochrome in the light induction of mRNAs for chloroplastic GS2 or cytosolic GS1 in Arabidopsis (Fig. 2B). Levels of mRNA for chloroplastic GS2 (gln2) were induced by red light (Fig. 2B, lane 3), and this induction was reversed by a subsequent pulse of far-red light (Fig. 2B, lane 4) in a typical phytochrome-dependent response. In contrast, no phytochrome-mediated effect on the expression of mRNA for cytosolic GS1 was seen (gln1;1, Fig. 2B). Levels of mRNA for the control gene β-ATPase were not significantly affected by light exposure (1.1- to 1.4-fold, as shown by densitometry). We conclude that light induction of mRNA for chloroplastic GS2 is at least partially the result of a direct effect of light via phytochrome. In contrast, the low-level induction of mRNA for cytosolic GS1 by light involves a mechanism other than phytochrome activation (see below).
Levels of GS mRNA and GS Enzyme Activity Are Induced by Suc Treatment in the Absence of Light
Light can trigger the direct modulation of gene expression through phytochrome activation (see Fig. 2A). Light has also been shown to affect the expression of genes indirectly via the activation of photosynthesis and increases in levels of carbon metabolites (Sheen 1990; Vincentz et al., 1993; Lam et al., 1994; Chevalier et al., 1996; Jang and Sheen, 1997). We therefore investigated whether Suc could induce levels of mRNA for chloroplastic GS2 or cytosolic GS1 mRNA in the absence of light. The effects of exogenously supplied Suc were monitored in both a time-dependent (Fig. 3A) and dose-dependent (Fig. 3B) manner. To determine the kinetics of Suc induction of GS mRNA, plants were grown semihydroponically in a normal day/night cycle prior to treatment (see Methods).
Dark-adapted plants were transferred in the dark to MS medium supplemented with 3% (90 mm) Suc, and samples were collected at 0, 6, 12, 24, and 48 h (Fig. 3A, lanes 2–6). Control plants were grown in the light without Suc for 48 h (Fig. 3A, lane 1). The Suc induction of mRNA for chloroplastic GS2 began as early as 6 h (8-fold increase), increased steadily with a peak at 12 to 24 h (16- to 17-fold), and decreased by 48 h (8-fold, Fig. 3A, lanes 2–6). Suc induction of GS2 mRNA accumulation paralleled light induction in magnitude and kinetics (14- to 19-fold induction at 12- to 24-h time points) in the same set of experiments. Suc treatment could also induce levels of GS enzyme activity in these dark-adapted plants (Fig. 5). The parallel kinetics of induction of GS2 mRNA by light or Suc in the absence of light suggests that Suc can at least partially mimic the effects of light. Moreover, these results indicate that maximal expression of the GS2 gene may require both an environmental component (light via phytochrome) and a metabolic component (light induction of carbon metabolites).
Figure 5.
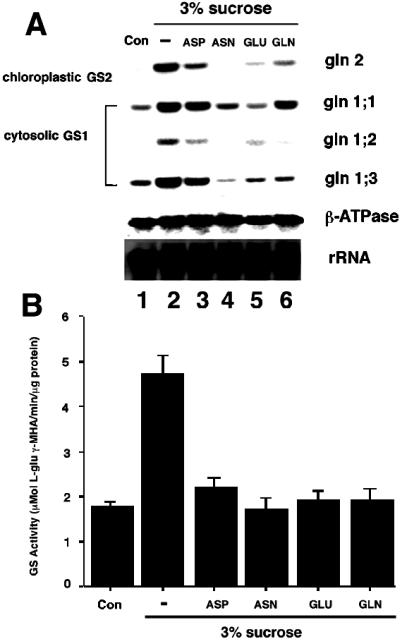
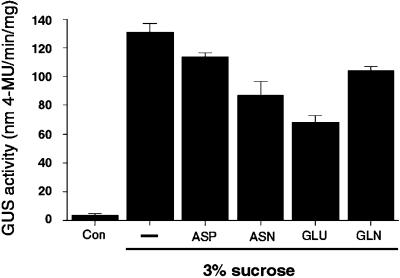
Amino acids and carbon reciprocally regulate GS mRNA accumulation and enzyme levels. Arabidopsis plants were grown semihydroponically and dark-adapted as described in Methods. The dark-adapted Arabidopsis plants were transferred in the dark to fresh low-nitrogen MS medium with no carbon supplementation (Con, lane 1), to fresh low-nitrogen MS medium with 3% Suc (lane 2), or to fresh low-nitrogen MS medium with 3% Suc in addition of either 10 mm Asp (lane 3), 10 mm Asn (lane 4), 10 mm Glu (lane 5), or 10 mm Gln (lane 6). After transfer, the plants were further incubated for 12 h in the dark. Probes were as in the legend for Figure 3. B, Same as A except that an aliquot of each sample was collected for determination of total GS activity. A representative experiment of two repetitions is shown (results are GS activity per milligram of total protein ± se of three independent determinations). Protein extraction and measurement of GS activity were performed as described in Methods.
Levels of mRNA for the cytosolic GS1 isoenzymes showed a moderate induction (2- to 3-fold) when treated with Suc, peaking after only 6 h of treatment (Fig. 3A, compare lanes 2 and 3). This induction and the kinetics of induction of cytosolic GS1 by Suc also mimicked the effects of light. Dose-response studies showed that treatment with 3% Suc (90 mm) resulted in the maximum induction of mRNAs for GS2 or GS1 during the 12-h period tested. Significant (albeit lower) levels of induction were also seen with 1% Suc (Fig. 3B); 5% Suc was not effective in inducing GS mRNA within the 12-h time point. This may reflect a difference in the kinetics of Suc induction of GS mRNA caused by an increased concentration of Suc. The levels of mRNA for the control gene β-ATPase were either not affected or were only marginally affected (1.1- to 1.4-fold, as shown by densitometry) by the Suc treatments (compare Fig. 3A and Figs. 4 and 5 below). Together, these in planta experiments provide evidence supporting a physiologically significant role for carbon in the metabolic regulation of GS gene expression and levels of GS enzyme activity in Arabidopsis.
Levels of GS mRNA Are Differentially Regulated by Distinct Carbon Metabolites
The best-studied example of a putative carbon-mediated sensing mechanism in plants involves the enzyme hexokinase (Jang and Sheen, 1997), which has been proposed as a sensor for Suc and Suc-derived monosaccharides in plants and yeast (Jang et al., 1997). However, hexokinase-mediated carbon sensing alone cannot account for all of the carbon-sensing mechanisms in plants. Additional mechanisms have therefore been invoked to explain the differential sensing of hexoses and the less-complex downstream metabolites of the glycolytic pathway (Jang and Sheen, 1997). The effect of Suc on GS gene expression prompted us to test the effects of different carbon sources on the expression of GS genes. We tested the hexoses Fru and Glc, as well as a representative non-hexose, the tricarboxylic acid (TCA) cycle intermediate 2-oxoglutarate. Mannitol, a non-metabolizable carbon source, was used as a control. To determine the effects of different carbon sources on the induction of GS mRNA accumulation, dark-adapted Arabidopsis plants (see Methods) were transferred in the dark to MS medium supplemented with different carbon sources (Fig. 4, lanes 3–7) or to fresh MS medium lacking any carbon source, and incubated either in the light (Fig. 4, lane 1) or in the dark (Fig. 4, lanes 2–7) for 12 h. The concentration used for all carbon sources tested (90 mm) was identical to the optimal concentration of Suc for GS gene expression in Arabidopsis (equivalent to 3% Suc; data not shown; Fig. 3). These semihydroponic culture conditions permitted us to grow plants to full maturation before transferring them to specific metabolic treatments for short periods of time. Short exposure to the various metabolites tested diminishes potential toxic and/or nonspecific effects caused by long-term exposure to exogenously provided metabolites.
Our results demonstrate that Suc, Fru, and Glc could all induce accumulation of mRNA for chloroplastic GS2 or cytosolic GS1 to a similar extent (Fig. 4, lanes 2–5). In contrast, treatment of Arabidopsis plants with the TCA cycle intermediate 2-oxoglutarate led to a specific induction (2- to 5-fold) of mRNA for genes encoding cytosolic GS1 (gln1;1, gln1;2, and gln1;3; Fig. 4, compare lanes 2 and 6). The inductive effect of 2-oxoglutarate was specific for cytosolic GS1 mRNA and did not affect the levels of mRNA for chloroplastic GS2 (1.4-fold, Fig. 4, lane 6) or the control gene β-ATPase (1.1- to 1.3-fold induction). Mannitol had no significant effect on GS mRNA accumulation (Fig. 4, lane 7), except for a reduction in gln1;3. These results support the notion that levels of carbon metabolites may play an important role in the regulation of mRNA for specific GS isoenzymes in Arabidopsis. These changes in levels of GS mRNA by carbon metabolites are reflected by corresponding changes in GS enzyme activity (see below).
GS Expression Is Reciprocally Regulated by Organic Nitrogen and Carbon in Arabidopsis
The effects of light and the metabolic status of the cell are two critical factors controlling the conversion of inorganic nitrogen into amino acids in plants (Ratajczak et al., 1981; Vincentz et al., 1993; Lam et al., 1994). We therefore investigated whether exogenously supplied amino acids have any effect on the levels of GS mRNA (Fig. 5A) or on GS enzyme activity (Fig. 5B). Plants were grown semihydroponically and dark-adapted for 48 h, as described in Methods. Thereafter, plants were maintained in the dark and transferred to MS medium supplemented with 3% (90 mm) Suc or 3% Suc plus Asp, Asn, Glu, or Gln (10 mm each), and incubated for 12 h in the dark. Control plants were transferred to fresh MS medium with no carbon or amino acid supplementation, and incubated for 12 h in the dark. Samples were collected from the same set of plants for both northern-blot analysis (Fig. 5A) and GS enzyme activity assays (Fig. 5B).
Northern-blot analysis showed that the low levels of mRNA for all GS genes could be induced by Suc treatment in the absence of light (Fig. 5A, compare lanes 1 and 2). This induction was reflected by the increase in levels of GS enzyme activity in Suc-treated plants (Fig. 5B, column 2). Under these experimental conditions, the amino acids Asp, Asn, Glu, and Gln all had antagonistic effects on the Suc-induction of levels of GS mRNA, albeit reproducibly affecting the expression of the GS genes to different extents (2.4- to 16-fold inhibition, Fig. 5A, lanes 2–6). Levels of GS enzyme activity were also reduced in samples treated with Suc plus amino acids compared with those given Suc alone (Fig. 5B, columns 2–6). We conclude that amino acids have a negative effect on the Suc induction of GS, and that this can be observed both at the level of mRNA accumulation and that of GS enzyme activity. This inhibitory effect of amino acids may be physiologically significant, as it occurs within a 12-h treatment period. Moreover, the negative effect of amino acids on the levels of GS mRNA was not due to a general toxic and/or inhibitory effect, as the expression of a gene coding for the β-subunit of mitochondrial ATPase was not significantly affected by the same treatment (0.9- to 1.3-fold inhibition, Fig. 5A).
We conducted HPLC analysis to verify that the amino acids supplied in the medium were taken up in the semihydroponic growth system (Table I) using an aliquot from the same Arabidopsis plants used to perform northern-blot analysis and GS enzyme activity (Fig. 4). Table I shows that treatment of plants with 10 mm of each amino acid for 12 h led to a nontoxic (μm range) but physiologically effective (Fig. 5) internal accumulation of each amino acid used. Furthermore, exogenous feeding with a single amino acid had an overall effect on the accumulation of the other amino acids to a significant extent (Table I). The correlation between the levels of amino acid accumulation in planta and the observed down-regulation of GS mRNA accumulation support the notion that organic nitrogen antagonizes the Suc induction of GS mRNA and enzyme levels.
Table I.
Determination of free amino acid levels in Arabidopsis after incubation of plants with different carbon and nitrogen combinations
| Treatment | Asp | Asn | Glu | Gln |
|---|---|---|---|---|
| pmol mg−1 total protein | ||||
| Suc | 13.5 ± 0.6 | 288.6 ± 105.3 | 43.9 ± 1.3 | 114.4 ± 4.0 |
| Suc + Asp | 1,410.7 ± 84.5 | 1,575.2 ± 59.2 | 130.9 ± 54.3 | 660.0 ± 26.2 |
| Suc + Asn | 92.7 ± 26.7 | 7,105.2 ± 33.8 | 272.2 ± 14.4 | 731.0 ± 25.3 |
| Suc + Glu | 354.6 ± 5.8 | 6,809.3 ± 125.2 | 6,211.3 ± 63.9 | 2,017.0 ± 19.6 |
| Suc + Gln | 85.6 ± 10.0 | 1,659.4 ± 729.2 | 214.1 ± 27.6 | 2,625.4 ± 221.0 |
Results are means ± se (n = 3).
To investigate whether the inhibitory effects of amino acids on Suc induction of GS mRNA could be explained by changes in GS gene transcription, we tested the effects of amino acid and Suc treatments on the activation of a reporter gene (GUS) whose expression is driven by a GS2 promoter (Fig. 6). The GS2 gene was chosen for this study, as it exhibits the most dramatic regulation by Suc and amino acids. Arabidopsis was transformed with a construct containing a promoter for pea chloroplastic GS2 (−399/+11) placed upstream of a GUS reporter gene (line GS2/5′−399; Tjaden et al., 1995).
Figure 6.
Amino acids and carbon reciprocally regulate GS at the transcriptional level. Arabidopsis plants were grown etiolated in low-nitrogen MS medium with no carbon supplementation (Con, column 1), with 3% Suc (column 2), or with 3% Suc plus 0.5 mm Asp (column 3), 0.5 mm Asn (column 4), 3.0 mm Glu (column 5), or 3.0 mm Gln (column 6). After 6 d, samples containing 100 to 200 etiolated seedlings were collected for determination of total GUS activity. Results are GUS activity expressed in nanomoles of 4-methylumbeliferone (4-MU) produced per minute per milligram of total protein ± se of three independent determinations. Protein extraction and measurement of GUS activity were performed as described in Methods. A representative experiment of two repetitions is shown.
Because GS2 expression is induced by light and the GUS protein is very stable, metabolic treatment experiments were performed on dark-grown plants (etiolated) to avoid interference due to the light-induced expression of GS. As it was not technically feasible to transfer dark-grown plants to amino acid treatments, the plants were germinated and grown on levels of amino acids shown to have no negative effects on long-term growth, as specified below. Etiolated plants were germinated on MS medium with: (a) no carbon source (Fig. 6, column 1); (b) 3% (90 mm) Suc (Fig. 6, column 2); or (c) 3% Suc supplemented with Asp or Asn (0.5 mm each) or Glu or Gln (3.0 mm each) (Fig. 6, columns 3–6). These concentrations of amino acids have been previously shown to cause no negative effects on the long-term growth of Arabidopsis plants in culture (Lam et al., 1994; Schultz et al., 1998). Control plants were germinated and grown in the dark on MS medium with no carbon or amino acid supplementation (Fig. 6, column 1).
Measurement of GUS activity in the GS2-GUS plants showed that the low basal levels of GUS in dark-grown plants could be induced by Suc treatment in the absence of light (Fig. 6, compare columns 1 and 2). The addition of the amino acids Asp, Asn, Glu, and Gln all had antagonistic effects on the Suc-induced activation of the GS2-GUS construct to different extents (Fig. 6, columns 2–6). These GS2-GUS studies suggest that reduction in GS mRNA by amino acids observed at the northern-blot level can be at least partially explained by metabolic control of GS gene transcription. Together, these results suggest that organic nitrogen can lead to a down-regulation of Suc induction of GS gene transcription, levels of GS mRNA, and GS enzyme activity in Arabidopsis.
DISCUSSION
We report the reciprocal or antagonistic effects of carbon and amino acids on the modulation of GS expression in Arabidopsis. The effects of these metabolites were monitored at the level of GS gene transcription, GS mRNA accumulation, and GS enzyme activity. Our results point to an important physiological role for carbon and organic nitrogen in the modulation of GS levels in higher plants. Light regulation of chloroplastic GS2 mRNA is mediated in part by phytochrome and in part by light-induced increases in the levels of Suc. In contrast, light induction of cytosolic GS1 mRNA can be accounted for by metabolic induction by Suc alone. Interestingly, the non-hexose carbon source 2-oxoglutarate also induced accumulation of mRNA for cytosolic GS1, but had negligible effects on the levels of mRNA for chloroplastic GS2 (Fig. 4). Thus, the expression of distinct GS isoenzymes in Arabidopsis appears to be modulated by hexose-dependent and possibly hexose-independent pathways.
As the cytosolic GS1 isoenzymes are predominantly expressed in roots, it is possible that carbon compounds such as the components of the TCA cycle work as effectors of gene expression in such non-photosynthetic organs. It is also possible that the differential effects on gene expression were due to the differential accumulation of 2-oxoglutarate levels in leaves versus roots. This potential regulatory role of 2-oxoglutarate on GS1 gene expression may be especially significant in plant roots, because TCA cycle intermediates are thought to play a pivotal metabolic role in regulating nitrogen assimilation in this organ (Oaks, 1992).
As carbon levels were shown to positively affect GS gene expression and result in increased GS enzyme activity, we next tested whether these inductive effects could be reversed by treatment of Arabidopsis with organic nitrogen in the form of amino acids. Indeed, under the experimental conditions tested, the amino acids Asp, Asn, Glu, and Gln all had a pronounced inhibitory effect on the Suc-induced accumulation of GS mRNA, albeit to different extents. This inhibitory effect of amino acids on Suc-induced GS gene expression occurred in a physiological time frame (12 h) and was not due to a general negative effect, as levels of mRNA for the control gene β-ATPase were unaffected. The ability of amino acids to inhibit Suc-induced GS mRNA accumulation operates at least partially at the transcriptional level, as judged by experiments using Arabidopsis plants transformed with a GS2 promoter-GUS construct. The ability of amino acids to antagonize Suc induction of GS was also observed at the level of GS enzyme activity. In previous studies of GS in lupine, GS enzyme activity was reported to be inhibited by a 3-d treatment with 35 mm Gln and Asn (Ratajczak et al., 1981). However, the very extreme conditions used (both the length of treatment and the dose of metabolite) clouds the actual physiological significance of their observations made in that study.
The mechanisms by which plant genes respond to metabolic signals is presently unknown. Genes involved in nitrogen assimilation have been demonstrated to be induced by Suc (and its derivative hexoses) in an effect that is at least partially mimicked by light. For instance, exogenous Suc supplementation of dark-adapted plants has been demonstrated to induce the expression of genes for nitrate reductase in Arabidopsis and tobacco (Cheng et al., 1992; Vincentz et al., 1993; Jang et al., 1997) and for nitrite reductase in tobacco (Vincentz et al., 1993). Similarly, light repression of gene expression may also be mimicked by sugars, as is the case with Asn synthetase in Arabidopsis and maize (Lam et al., 1994; Chevalier et al., 1996) and Glu dehydrogenase in Arabidopsis (Melo-Oliveira et al., 1995). There is also strong evidence for hexokinase-dependent sugar-sensing pathways regulating photosynthesis-related genes in plants (Sheen, 1990; Jang and Sheen, 1997; Jang et al., 1997).
While progress has been made in understanding sugar sensing and signaling in plants, very little is known about amino acid sensing and signaling in plants. Early work in this area showed that the amino acids Glu, Gln, or Asn could inhibit accumulation of nitrate reductase and nitrite reductase mRNA and activity in tobacco (Vincentz et al., 1993). This was later shown for Asn- or Gln-treated maize seedlings (Sivasankar et al., 1997). Previous work from our laboratory revealed that the amino acids Glu, Gln, and Asn could at least partially relieve Suc repression of a gene encoding Asn synthetase in Arabidopsis (Lam et al., 1994). These findings were later demonstrated to occur in maize as well (Chevalier et al., 1996).
A recent report described the inductive effect of Glu on GS gene expression in radish (Watanabe et al., 1997). In that study, the authors used radish protoplasts cultured in a high concentration of Glu (50 mm) for extended periods of time (5 d). These extreme conditions may have led to the induction of senescence-related genes. They also did not report whether these results were reproduced in a whole plant. We believe that the discrepancy between their observations on GS regulation by amino acids in radish protoplasts and ours on GS in whole Arabidopsis plants is due to the significant differences between the systems used. Our data showing that amino acids repress the Suc-induced GS gene expression was performed in whole plants exposed to brief treatments with metabolites, and our results are reminiscent of previous studies on nitrate reductase and nitrite reductase genes in maize and tobacco (Vincentz et al., 1993; Chevalier et al., 1996; Sivasankar et al., 1997).
We report that different amino acids antagonize the Suc induction of GS gene expression at the levels of transcription of the GS2 gene, accumulation of individual GS mRNAs, and GS enzyme activity. One possible explanation for the differences in the efficacy of the amino acids used is that each amino acid elicits its effects through different but partially overlapping pathways. Another possible explanation is that differences are due to rate of uptake or metabolism to a common “sensed” intermediate (e.g. Glu). Further investigations should help clarify the specific nature by which amino acids or their derivatives are the effectors of gene regulation in plants.
It is noteworthy that amino acid treatment reversed both the Suc induction of GS mRNA accumulation and levels of GS activity. However, the changes in levels of GS enzyme activity did not quantitatively parallel the reduction in the levels of GS mRNA accumulation (Fig. 5). The effects of amino acid treatment on the regulation of a GS2 promoter-GUS construct suggest that modulation of transcription alone is not sufficient to account for the observed decrease in the steady-state levels of Suc-induced GS mRNA accumulation by amino acids (Fig. 6). A possible explanation for these observations is that amino acid inhibition of Suc-induced GS activity could be due to a mechanism operating at the transcriptional, posttranscriptional, and posttranslational levels.
The above observations on GS regulation by carbon and nitrogen metabolites in Arabidopsis are reminiscent of a similar nitrogen regulatory (Ntr) mechanism in Escherichia coli. In E. coli, the assimilation of inorganic nitrogen into Gln by GS is negatively regulated by the Ntr system at both the transcriptional and posttranslational levels, when the internal levels of Gln are high in relation to 2-oxoglutarate (Magasanik and Neidhardt, 1987; Neidhardt, 1987). Recently, a homolog of a component of the bacterial Ntr system, PII, has been isolated from Arabidopsis and has been implicated as a component of a C:N sensing mechanism in planta (Hsieh et al., 1998). Thus, some apparent links between metabolic regulatory mechanisms in plants and microorganisms have begun to emerge (Schubert, 1986; Alderson et al., 1991; Le Guen et al., 1992; Hsieh et al., 1998). Future studies should help clarify whether common pathways leading to the regulation of gene expression by nitrogen exist in bacteria and plants. In addition, the identification of components of metabolic signaling pathways in plants may be aided by genetic selections for mutants in Arabidopsis. Understanding the mechanisms involved in metabolite-mediated signaling and gene expression in plants should have an impact on understanding the controls of plant nitrogen assimilation, growth, and development.
ACKNOWLEDGMENTS
The authors are deeply indebted to Rosana Melo-Oliveira for her important insights. We thank Eric D. Brenner and Philip M. Benfey for critical reading of the manuscript, Neofitos Stefanides, Lee Borghi, and Dimitrios Bliagos for their assistance with the mapping experiments using RI lines, and Ravi Mistri, Alexandra Clark, Paula Gonzalez, Chia-Hung Yuan, and Yana Pikman for their help in various technical aspects of this project.
Footnotes
This research was supported by the National Institutes of Health (grant no. GM32877).
LITERATURE CITED
- Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreis M, Shewry PR, Halford NG. Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast by a plant protein kinase cDNA. Proc Natl Acad Sci USA. 1991;88:8602–8605. doi: 10.1073/pnas.88.19.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seiden JG, Smith JA, Struhl K (1987) Current Protocols in Molecular Biology. Greene Publishing & John Wiley & Son, New York
- Bernhard WR, Matile P. Differential expression of glutamine synthetase genes during the senescence of Arabidopsis thaliana rosette leaves. Plant Sci. 1994;98:7–14. [Google Scholar]
- Botstein D, White RI, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brears T, Liu C, Knight T, Coruzzi G. Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol. 1993;103:1285–1290. doi: 10.1104/pp.103.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-L, Acedo GN, Cristinsin M, Conkling MA. Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA. 1992;89:1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Just D, Raymond P. Metabolic regulation of asparagine synthetase gene expression in maize root tips. Plant J. 1996;9:1–11. doi: 10.1046/j.1365-313x.1996.09010001.x. [DOI] [PubMed] [Google Scholar]
- Cock JM, Brock IW, Watson AT, Swarup R, Morby AP, Cullimore JV. Regulation of glutamine synthetase genes in leaves of Phaseolus vulgaris. Plant Mol Biol. 1991;17:761–771. doi: 10.1007/BF00037059. [DOI] [PubMed] [Google Scholar]
- Edwards JW, Coruzzi GM. Photorespiration and light act in concert to regulate the expression of the nuclear gene for chloroplast glutamine synthetase. Plant Cell. 1989;1:241–248. doi: 10.1105/tpc.1.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Gadal P. Glutamine synthetase in rice: a comparative study of the enzyme from roots and leaves. Plant Physiol. 1980;66:619–623. doi: 10.1104/pp.66.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M-H, Lam H-M, van de Loo FJ, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Sheen J. Sugar sensing in higher plants. Trends Pharmacol Sci. 1997;2:208–214. [Google Scholar]
- Jefferson RA. The GUS reporter system. Nature. 1989;342:837–838. doi: 10.1038/342837a0. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Coschigano K, Oliveira IC, Melo-Oliveira R, Coruzzi G. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:596–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Peng SS-Y, Coruzzi GM. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol. 1994;106:1347–1357. doi: 10.1104/pp.106.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW, Kontturi M, Young AT. Photosynthesis by flag leaves of wheat in relation of protein, ribulose bisphosphate carboxylase activity and nitrogen supply. J Exp Bot. 1989;40:43–52. [Google Scholar]
- Le Guen L, Thomas M, Bianchi M, Halford NG, Kreis M. Structure and expression of a gene from Arabidopsis thaliana encoding a protein related to SNF1 protein kinase. Gene. 1992;120:249–254. doi: 10.1016/0378-1119(92)90100-4. [DOI] [PubMed] [Google Scholar]
- Li M-G, Villemur R, Hussey PJ, Silflow CD, Gantt JS, Snustad DP. Differential expression of six glutamine synthetase genes in Zea mays. Plant Mol Biol. 1993;23:401–407. doi: 10.1007/BF00029015. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Magasanik B, Neidhardt FC (1987) Regulation of carbon and nitrogen utilization. In FC Neidhardt, JL Ingraham, KB Low, B Magasanik, M Schaechter, HE Umbarger, eds, Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp 1313–1325
- Mann AF, Fentem PA, Stewart GR. Identification of two forms of glutamine synthetase in barley. Biochem Biophys Res Commun. 1979;88:515–521. doi: 10.1016/0006-291x(79)92078-3. [DOI] [PubMed] [Google Scholar]
- Mattsson M, Johansson E, Lundborg T, Larsson M, Larsson CM. Nitrogen utilization in N-limited barley during vegetative and generative growth. I. Growth and nitrate uptake kinetics in vegetative cultures grown at different relative addition rates of nitrate. J Exp Bot. 1991;42:197–205. [Google Scholar]
- McNally SF, Hirel B, Gadal P, Mann AF, Stewart GR. Glutamine synthetase from higher plants: evidence for a specific isoform content related to their possible physiological role and their compartmentation within the leaf. Plant Physiol. 1983;72:22–25. doi: 10.1104/pp.72.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Oliveira R, Oliveira IC, Coruzzi GM. Proc Natl Acad Sci USA. 1995;93:4718–4723. doi: 10.1073/pnas.93.10.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myerson D. Producing single-stranded DNA probes with the Taq DNA polymerase: a high yield protocol. Biotechniques. 1991;10:35–38. [PubMed] [Google Scholar]
- Nam H-G, Giraudat J, den Boer B, Moonan F, Loos WDB, Hauge BM, Goodman HM. Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell. 1989;1:699–705. doi: 10.1105/tpc.1.7.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC (1987) Multigene systems and regulons. In FC Neidhardt, JL Ingraham, KB Low, B Magasanik, M Schaechter, HE Umbarger, eds, Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp 1313–1317
- Newman T, de Brujin FJ, Green P, Keegstra K, Kende H, Mcintosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, Retzel E, Somerville C. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks A. A re-evaluation of nitrogen assimilation in roots. BioScience. 1992;42:103–111. [Google Scholar]
- Oliveira IC, Coschigano K, Lam HM, Melo-Oliveira R, Coruzzi G. Molecular-genetic dissection and metabolic engineering of nitrogen assimilation in plants. Plant Physiol Biochem. 1997;35:185–198. [Google Scholar]
- Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;230:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Price CA, Reardon EM, Lonsdale DM. A guide to naming sequenced plant genes. Plant Mol Biol. 1996;30:225–227. doi: 10.1007/BF00020109. [DOI] [PubMed] [Google Scholar]
- Ratajczak L, Ratajczak W, Mazurowa H. The effect of different carbon and nitrogen sources on the activity of glutamine synthetase and glutamate dehydrogenase in lupine embryonic axes. Physiol Plant. 1981;51:277–280. [Google Scholar]
- Sakakibara H, Kawabata S, Takahashi H, Hase T, Sugiyama T. Molecular cloning of the family of glutamine synthetase genes from maize: expression of genes for glutamine synthetase and ferredoxin-dependent glutamate synthase in photosynthetic and non-photosynthetic tissues. Plant Cell Physiol. 1992;33:49–58. [Google Scholar]
- Sakamoto A, Takeba G, Shibata D, Tanaka K. Phytochrome-mediated activation of the gene for cytosolic glutamine-synthetase (GS1) during inbibition of photosensitive lettuce seeds. Plant Mol Biol. 1990;15:317–323. doi: 10.1007/BF00036917. [DOI] [PubMed] [Google Scholar]
- Schubert KR. Products of biological nitrogen fixation in higher plants: synthesis, transport, and metabolism. Annu Rev Plant Physiol. 1986;37:539–574. [Google Scholar]
- Schultz CJ, Hsu M, Miezak B, Coruzzi G. Arabidopsis mutants define an in vivo role for isoenzymes of the aspartate aminotransferase in plant nitrogen assimilation. Genetics. 1998;149:491–499. doi: 10.1093/genetics/149.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BM, Stadtman ER. Glutamine synthetase (Escherichia coli) Methods Enzymol. 1971;17A:910–922. doi: 10.1016/s0076-6879(85)13032-6. [DOI] [PubMed] [Google Scholar]
- Sheen J-Y. Metabolic repression of transcription in higher plants. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Rothstein S, Oaks A. Regulation of the accumulation and reduction of nitrate by nitrogen and carbon metabolites in maize seedlings. Plant Physiol. 1997;114:583–589. doi: 10.1104/pp.114.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden G, Edwards JW, Coruzzi GM. cis Elements and trans-acting factors affecting regulation of a non-photosynthetic light-regulated gene for chloroplast glutamine synthetase. Plant Physiol. 1995;108:1109–1117. doi: 10.1104/pp.108.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz M, Moureaux T, Leydecker M-T, Vaucheret H, Caboche M. Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 1993;3:315–324. doi: 10.1111/j.1365-313x.1993.tb00183.x. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Takagi N, Hayashi H, Chino M. Internal gln/glu ratio as a potential regulatory parameter for the expression of a cytosolic glutamine synthetase gene of radish in cultured cells. Plant Cell Physiol. 1997;38:1000–1006. [Google Scholar]