Abstract
Liver cancer is one of the most lethal malignancies with very poor prognosis once diagnosed. The most common form of liver cancer is hepatocellular carcinoma (HCC). The WW domain-containing oxidoreductase (WWOX) is a large gene that is often perturbed in a wide variety of tumors, including HCC. WWOX has been shown to act as a tumor suppressor modulating cellular metabolism via regulating hypoxia-inducible factor 1α (HIF-1α) levels and function. Given that WWOX is commonly inactivated in HCC, we set to determine whether specific targeted deletion of murine Wwox affects liver biology and HCC development. WWOX liver-specific knockout mice (WwoxΔHep) showed more potent liver regeneration potential and enhanced proliferation as compared with their control littermates. Moreover, WWOX deficiency in hepatocytes combined with diethylnitrosamine treatment increased the tumor burden, which was associated with increased HIF1α levels and target gene transactivation. Inhibition of HIF1α by systemic treatment with digoxin significantly delayed HCC formation. Our work suggests that WWOX inactivation has a central role in promoting HCC through rewiring of cellular metabolism and modulating proliferation.
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, representing the fifth type of commonly diagnosed cancer worldwide and third mortality cause among other cancer malignances1. HCC prevalence has been dramatically increasing in the last decay because of the expansion of HCC risk factors, including hepatitis infection and obesity2. Therapeutic options are limited and survival after diagnosis is still poor leading to high mortality. Therefore, better understanding of the molecular basis of HCC is urgently needed.
WW domain-containing oxidoreducatase (WWOX) gene resides in one of the most common fragile sites known as FRA16D, a region that is altered in many types of cancer3–5. Frequent homozygous deletions were reported at this region in aflatoxin B1 exposed HCC6, suggesting that it might harbor a tumor suppressor. In particular, WWOX expression is absent or reduced in most of the derived liver cancer cell lines7. The gene encodes a 46 kDa protein comprising of two N-terminal WW domains, known to mediate protein–protein interactions and a short-chain dehydrogenase/reductase domain whose specific function is unknown yet8–10. Moreover, WWOX was suggested as a modulator of β-catenin protein activity in some HCC cells lines11,12. In addition to its genomic re-arrangement and hypermethylation of its regulatory region, WWOX is inactivated by other proteins or microRNAs in HCC cell lines13–15. However, no direct in vivo evidence linking WWOX tumor suppressor function with HCC development is known so far.
WWOX is commonly reported as a tumor suppressor not only owing to its common loss in many human malignancies but also due to its anti-tumorigenic effect when overexpressed and susceptibility of tumor formation in Wwox-mutant mice8,16–19. Wwox null mice die by the age of 3–4 weeks owing to severe metabolic disorders, mainly lethal hypoglycemia20 precluding studying implications of WWOX loss in adult mice. To overcome this limitation, we have recently generated a conditional mouse model in which somatic deletion of Wwox is achieved using a specific Cre recombinase21, mimicking the alterations frequently observed in human cancers and allowing study of human cancer intervention, development and progression, and assessing therapeutic strategies22,23. In this study, we utilized a Cre recombinase driven by the promoter of albumin (Alb-Cre), which results in somatic deletion of WWOX in hepatocytes; WwoxΔHep mice. Interestingly, Iatan et al. have recently demonstrated that WWOX deletion in a murine mouse model modulates levels of lipoproteins, however, these mice did not spontaneously develop HCC24. One of the most widely used and accepted models for HCC development in animal models is the use of N-nitrosodiethylamine (DEN), a known carcinogen, which alkylates DNA bases25 and results in HCC tumor formation in a defined kinetic manner23.
WWOX anti-tumorigenic functions have been shown to affect genome integrity26–28, apoptosis9,29–31, cell growth and extracellular matrix signaling32, and glucose metabolism33,34. WWOX, through its first WW domain, interacts with wide variety of proteins regulating their functions and affecting cellular outcome9,35. Previously, WWOX have been shown to physically interact with hypoxia-inducible factor 1α (HIF-1α) and inhibit its activity33. Ablation of HIF1α expression in RAS-transformed Wwox-deficient mouse embryonic fibroblasts significantly reduced tumourigenicity33, suggesting that HIF1α mediates the tumorigenic phenotype of Wwox-depelted cells. Nevertheless, the functional association of WWOX-HIF1α has not been demonstrated in a cancer mouse model. Here, we studied the effect of treating WwoxΔHep mice with DEN and followed HCC development and progression. We demonstrated that WWOX dysregulation accelerates HCC development through control of HIF1α and other master proliferation gene networks implicated in hepatocarcinogenesis. We also demonstrate that WWOX ablation modulates liver regeneration.
Results
WWOX expression is commonly lost in liver cancer
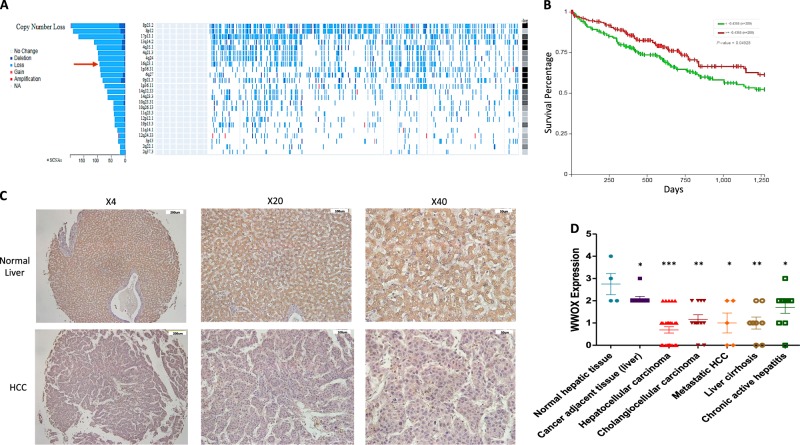
Given that WWOX expression is altered in many human malignancies36, we set to examine its genomic and expression status using different available tools and resources. Analysis of 438 liver cancer samples in the TCGA database using fire browser (www.firebrowse.org) revealed that the WWOX locus, spanning chromosomal region of 16q23.1, is one of the most significant regions harboring copy number loss in HCC patients (Fig. 1a). Using Xena browser (www.xenabrowser.net) we next evaluated the prognostic value of WWOX expression in HCC and found that cases harboring reduced WWOX mRNA levels tend to present with worse survival outcome compared to those having high WWOX expression (Fig. 1b). Consistent with these observations we further found that WWOX levels are absent or reduced in different liver pathologies, particularly in HCC, as assessed by immunohistochemical staining of a commercial tissue microarray (Fig. 1c, d). Notably, a tendency of reduced WWOX expression was observed in cancer adjacent liver tissue (P < 0.05), suggesting that loss of WWOX could be an early event in liver carcinogenesis.
Fig. 1. Loss of WWOX expression in liver cancer.
a TCGA data analysis showing that WWOX genomic region spanning long arm of chromosome 16 at 16q23.1 (red arrow) is frequently lost in HCC patients (n = 438 cases) with single copy number alteration (SCNA) equal to 96.84%. b Prognostic value of WWOX mRNA in HCC patients (n = 438) showing that patients with low expression had poorer survival compared with those with high WWOX expression. P value < 0.05. c Representative images of a liver cancer tissue microarray (TMA) (LV8011, US Biomax) showing immunohistochemical staining of WWOX in normal and HCC samples. d Quantification of WWOX protein levels from c. * indicates P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
Hepatocyte-specific WWOX ablation accelerates HCC development
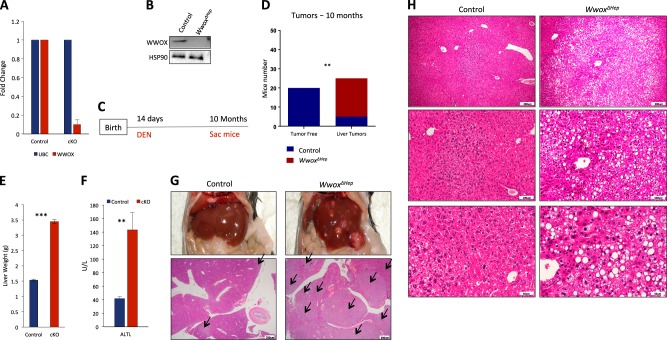
The fact that WWOX expression is reduced in early liver cancer lesions suggests that WWOX may have a role in suppressing liver carcinogenesis. To determine whether WWOX loss could contribute to HCC development or progression, we generated a hepatocyte-specific Wwox knockout mouse model and followed liver tumor formation. Wwox-floxed mice (Wwoxf/f)21 were bred with Albumin-Cre transgenic mice22 to generate Wwoxf/f;Albumin-Cre (WwoxΔHep) mice on the C57Bl6/J;SVJ129 mixed genetic background. Successful ablation of WWOX was validated using quantitative real-time (qRT)-PCR and western blot analyses (Fig. 2a, b). Follow up of WwoxΔHep and controlled littermate mice for up to 2-years did not reveal spontaneous tumor development, consistent with previous observations24. We therefore decided to examine the effect of WWOX ablation upon N-nitrosodiethylamine (DEN) treatment, a widely used chemical carcinogen for studying liver carcinogenesis22. Cohorts of WwoxΔHep mice and control littermates were intraperitoneally (IP) injected with 5 mg/Kg single injection of DEN at the age of 14 days and monitored for HCC development as a function of time (Fig. 2c). DEN treatment using this protocol is known to lead to 85% HCC development in C57BL6/J strain by the age ranging from 10 to 18 months37, whereas 129SVJ strain develop HCC later38. As shown in Fig. 2d, DEN-treated WwoxΔHep mice developed significantly higher incidence of HCC as compared with control mice (P = 0.0025). By the age of 10 months, the penetrance of tumor development was 100% in WwoxΔHep mice whereas just 20% of the control littermates’ mice developed macroscopic tumors (Fig. 2d). Furthermore, tumor load, as assessed by liver weight relative to body weight, was significantly higher in WwoxΔHep mice as compared with control mice (Fig. 2e). Interestingly, WWOX mRNA and protein was decreased in the tumors of control mice as revealed by qRT-PCR (Figure S1A) and immunohistochemistry (Figure S1B). In addition, WwoxΔHep mice had higher levels of serum alanine transaminase, indicating liver dysfunction as a result of tumor formation (Fig. 2f). Histological characterization of liver tissues revealed aggressive, poorly differentiated and highly proliferative tumors resembling HCC (Fig. 2g, h). Altogether, these findings indicate that WWOX loss accelerates HCC development.
Fig. 2. Hepatocyte-specific WWOX ablation accelerates HCC development.
a Validation of WWOX depletion in WwoxΔHep (cKO: conditional knockout) mice using qRT-PCR; n = 3 for each mouse group. Error bars indicate ± SEM. b Validation of WWOX protein ablation by immunoblotting using WWOX antibody; HSP90 was used as loading control. c DEN treatment plan: DEN was IP injected to control and WwoxΔHep mice at the age of 14 days and mice were analyzed at the age of 10 months. d χ2 analysis of macroscopic tumor incidence in control mice versus WwoxΔHep mice treated with DEN. e Tumor load analysis represented by liver weight of control mice (n = 15) versus WwoxΔHep cKO mice (n = 14) after 10 months of DEN treatment. f Serum ALTL levels of control mice (n = 6) versus WwoxΔHep mice (n = 6) after 10 months of DEN injection. g Representative pictures of control and WwoxΔHep livers of two male littermates (upper). Histological images showing liver sections of 10 months DEN-treated control and WwoxΔHep mice (lower panel) h Histological images (× 10, × 20, and × 40) of 10 months DEN-treated mice. * indicates P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
Hepatocyte-specific WWOX ablation is associated with increased proliferation
Our findings so far suggest that hepatocyte-specific WWOX deletion promoted HCC development. On one hand, WWOX overexpression was previously shown to induce apoptosis and suppress proliferation, whereas its loss is associated with enhanced survival in a hepatoma cell line39. On the other hand, DEN treatment is known to induce acute hepatic injury followed by compensatory proliferation40. We therefore set to determine whether the phenotype observed in DEN-treated WwoxΔHep mice is a consequence of impaired proliferation control. To this end, we analyzed liver tissues of DEN-treated mice at different time points starting from 1 to 10 months. Hematoxylin and Eosin (H&E) evaluation revealed that WwoxΔHep livers display hepatocyte morphological changes starting from 6-month post DEN treatment, which progressed to HCC at the age of 10 months, as shown earlier (Fig. 3a, d). No histological abnormalities were observed in the groups of 1 and 3-month post DEN treatment of WwoxΔHep mice nor in the control mice. To further support our findings, we immunostained for Ki67, a surrogate marker of proliferation. Digital quantification of Ki67-positive nuclei demonstrated that WwoxΔHep liver sections had significantly higher number of Ki67-positive cells in the groups of 1, 3, 6, and 10 months WwoxΔHep mice relative to the control littermate groups (Fig. 3e). In addition, qRT-PCR analysis of proliferative genes implicated in liver cancer, including c-Myc, c-Jun, c-Fos, and Axin, display higher levels in the WwoxΔHep mouse groups from as early as 1-month post DEN treatment (Fig. 3f). Although, DEN-free WwoxΔHep mice display no tumorigenic phenotype, levels of the previous mentioned proliferative genes were also elevated (Figure S2A). Consistent with these results, a significant increase in CTGF levels, one of the main effectors of the Hippo pathway41, was also noted in livers of WwoxΔHep mice with a slight increase in liver weight in the pre-tumorigenic phase (Figure S2B and S2C). These findings suggest that WWOX has a critical role in inhibiting proliferation of DEN-treated hepatocytes, whereas its inactivation leads to increased proliferation contributing to tumor development.
Fig. 3. Hepatocyte-specific WWOX ablation is associated with increased proliferation.
Histological images (H&E staining and Ki67 immunohistochemical staining at × 40) of DEN-treated control and WwoxΔHep liver sections at the age of 1 a, 3 b, 6 c, and 10 months d. e Quantification of positive Ki67 nuclei in 1, 3, 6, and 10 months old DEN-treated control and WwoxΔHep mice liver. (n = 5 for each group). f mRNA expression levels of proliferation genes in DEN-treated control and WwoxΔHep mice at the age of 1, 3, 6, and 10 months (n = 3 for each group). * indicates P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
Enhanced expression of glycolytic genes in livers of DEN-WwoxΔHep mice
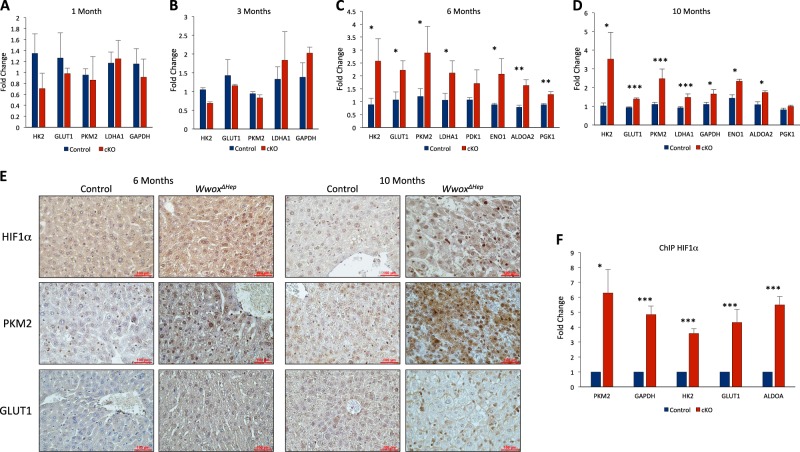
Recently, it was shown that WWOX loss is associated with Warburg effect and impaired mitochondrial respiration33,42,43. In mammalian cells, WWOX ablation is inversely correlated with increased levels and activity of hypoxia-inducible factor 1α (HIF1α-enhancing cell transformation and tumor growth33,44. In particular, Wwox-deficient cells were shown to display higher levels of glycolytic genes known to be regulated by HIF1α. Given that HIF1α has known roles in HCC development45, we next set to determine whether WWOX-targeted loss in the different DEN-treated WwoxΔHep and control mice is associated with impaired HIF1α function. Analysis of HIF1α glycolytic target genes mRNA, including Hk2, Pkm2, Glut1 and others, demonstrated no change at 1 and 3-months post treatment (Fig. 4a, b). Intriguingly, a significant upregulation of these transcripts was noted in 6 and 10 months DEN-treated WwoxΔHep mice compared with control mice (Fig. 4c, d). Interestingly, DEN-free WwoxΔHep mice on 6 months age displayed a tendency of HIF1α glycolytic target genes upregulation (Figure S3A). Moreover, nuclear HIF1α protein levels exhibited moderate and prominent increase at 6 and 10 months in DEN-treated WwoxΔHep mice, respectively (Fig. 4f). Likewise, levels of PKM2 and membranous GLUT1 protein were higher in 6 and 10 months DEN-treated WwoxΔHep mice (Fig. 4e).
Fig. 4. Enhanced expression of glycolytic genes in livers of DEN-WwoxΔHep mice.
a mRNA expression levels of HIF1α glycolytic target genes in DEN-treated control and WwoxΔHep (cKO) mice at the age of 1 a, 3 b, 6 c and 10 d months (n = 3 for each group). e Immunohistochemical staining of HIF1α, PKM2, and GLUT1 in 6 and 10 months of DEN-treated WwoxΔHep and control liver tissues. Images were taken at × 40 magnification (Bar = 100μm). f ChIP experiment with HIF1α antibody on DEN-treated control and WwoxΔHep (cKO) mice followed by qRT-PCR analysis of HIF1α target genes. * P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
Our recent studies have shown suppression of HIF1α transactivation function by WWOX29. Therefore, we next determined whether WWOX loss modulates HIF1α recruitment to promoters of these glycolytic genes. To address this, we performed a chromatin immunoprecipitation (ChIP) experiment using anti-HIF1α antibody on genomic DNA isolated from liver tissues of 10 months DEN-treated WwoxΔHep and control littermate mice. QRT-PCR analysis revealed HIF1α enrichment on promoters of target genes, including Pkm2, Hk2, Glut1, Aldh, and Gapdh, in WwoxΔHep livers (Fig. 4f). Altogether, these results imply that WWOX loss leads to HIF1α enhanced activity and, likely, reprogramming of HCC cells to glucose metabolism and Warburg effect.
Inhibition of HIF1α rescued the effect of WWOX loss
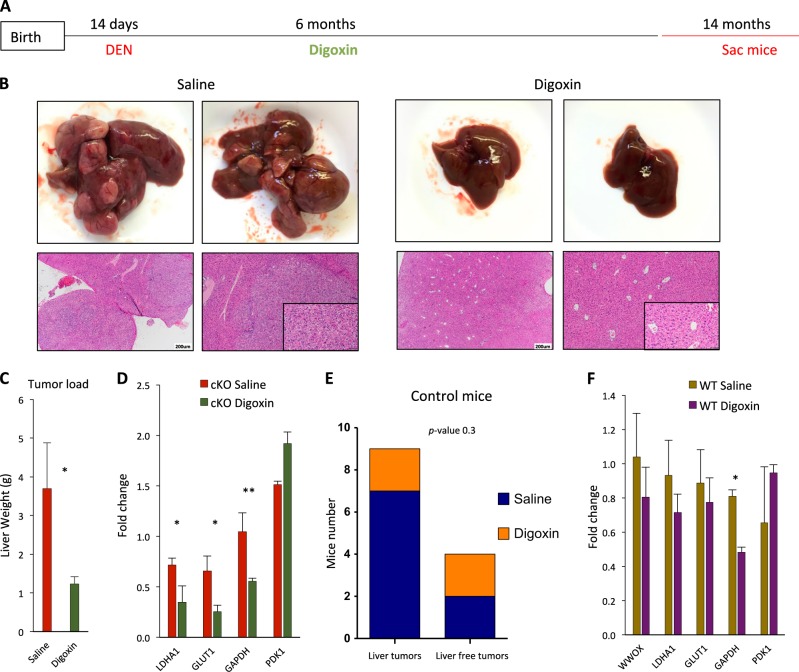
We have previously shown that digoxin, an inhibitor of HIF1α, rescued the effect of Wwox-deficient mice displaying hypoglycemia and elevated levels of glycolytic genes33. Digoxin is a cardiac glycoside, which has been shown to inhibit HIF1α transcriptional activity in vitro46 and suppress tumor growth in vivo47. We therefore set to investigate whether digoxin treatment of DEN-WwoxΔHep mice could reduce the incidence of HCC development. DEN-WwoxΔHep and control mice received digoxin (1 mg/kg) or vehicle control (saline) through intraperitoneal injections (3-times/week) for 32 weeks starting at 6 months post DEN treatment (Fig. 5a). Analysis of mice 8-months post digoxin treatment revealed marked suppression of liver tumor growth with more prominent effect in WwoxΔHep mice compared with saline-treated WwoxΔHep mice (Fig. 5b, c). In contrast, no significant difference was observed between digoxin and saline-treated DEN control mice (Fig. 5e). Consistent with these results, digoxin-treated DEN-WwoxΔHep mice showed downregulations of HIF1α targets, whereas no significant changes were observed in digoxin-treated DEN control mice (Fig. 5d, f). We conclude that WWOX loss exerts critical regulation on HIF1α function mediating rewiring of glucose metabolism and contributing to enhanced proliferation.
Fig. 5. Inhibition of HIF1α rescued the effect of WWOX loss.
a Digoxin experiment plan, DEN was IP injected to control and WwoxΔHep mice at the age of 14 days. Six-month later mice were started to be treated with digoxin (1 mg/kg) or vehicle control (saline) 3-times a week for an additional 8-months. b Representative images of saline-treated WwoxΔHep (cKO) mice versus digoxin-treated WwoxΔHep (cKO) mice livers and liver sections c Tumor load of saline-treated WwoxΔHep mice versus digoxin-treated WwoxΔHep mice livers as assessed by liver weight at the age of 14 months d mRNA expression levels of HIF1α glycolytic target genes in saline-treated WwoxΔHep (cKO) mice versus digoxin-treated WwoxΔHep (cKO) mice at the age of 14 months, (n = 3 for each group). e Χ2 analysis of macroscopic tumor incidence in saline-treated control mice versus digoxin-treated control mice at the age of 14 months. f mRNA expression levels of HIF1α glycolytic target genes in saline-treated control mice versus digoxin-treated control mice at the age of 14 months, (n = 3 for each group). * P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
High-fat diet (HFD) increased HCC incidence in WwoxΔHep mice
Obesity is known to be a major risk factor for HCC development. Indeed, combined HFD and DEN treatment have been previously shown to strongly enhance HCC development when compared with DEN mice on normal diet48. Driven by our previous observations, we decided to investigate the effect of HFD on HCC development in DEN-WwoxΔHep mice. DEN-treated cohorts of male WwoxΔHep and control mice were placed on HFD, in which 60% of calories were fat-derived, for 30 weeks. Tumors of DEN-treated WwoxΔHep HFD mice appeared 3-months earlier compared with normal chow-diet mice (Fig. 2), consistent with previous reported data showing acceleration of HCC formation in mice fed with HFD48. No difference of mice weight between the two groups of HFD-WwoxΔHep and control mice was noted (Figure S4A). Nevertheless, the number of tumors in WwoxΔHep mice was significantly higher (Fig. 6a, b) and was accompanied with higher levels of serum ALT (Figure S4B) and hepatosteatosis (Fig. 6c). Consistent with anti-proliferative role of WWOX that we showed previously (Fig. 3), DEN-treated WwoxΔHep mice fed with HFD displayed increased proliferation (Fig. 6d, e). Persistent with a prominent role of β-catenin pathway in HCC, we observed significant upregulation of Axin2 transcript in DEN-treated WwoxΔHep HFD mice (Fig. 6f). At a later stage, there was also an upregulation of HIF1α-target genes (Figure S4C). Altogether, our findings suggest that combined loss of WWOX and HFD intake accelerate HCC formation mediated by enhanced proliferation.
Fig. 6. High-fat diet (HFD) increases HCC incidence in DEN-treated WwoxΔHep mice.
a Χ2analysis of HCC incidence in DEN-treated control and WwoxΔHep mice (at age of 14 days) and fed with HFD from the age of 8 weeks. b Representative pictures of control and WwoxΔHep (cKO) livers fed with HFD at the age of 7 months. Arrows indicate macroscopic tumors. c Histological images (× 4, × 10, × 2,0 and ×40) of 7 months DEN-treated control and WwoxΔHep livers fed with HFD. d Histological images (Ki67 immunohistochemical staining) of mice from c. e Quantification of positive Ki67 nuclei of mice from d. f mRNA expression of Axin2 (control, n = 3; cKO, n = 3). * P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
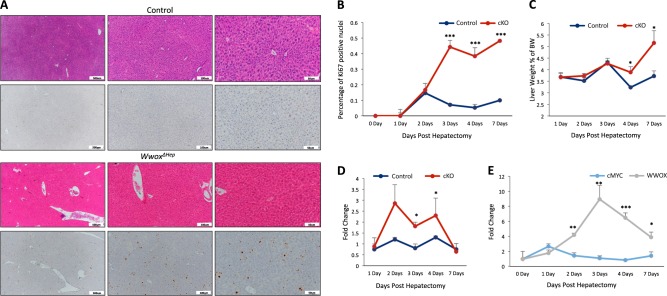
Wwox ablation is associated with increased proliferation upon partial hepatectomy
The preceding observations demonstrated that WWOX inactivation coupled with DEN treatment and HFD intake leads to enhanced proliferation and tumor formation. We next asked whether WWOX deletion affects hepatic cell proliferation upon partial hepatectomy, a process known to acutely induce hepatocyte proliferation to achieve liver regeneration. WwoxΔHep mice and their littermate controls were subjected to 30% hepatectomy and were analyzed 1, 2, 3, 4, and 7 days later for Ki67 immunohistochemical staining. As expected, an increase in the proliferation index was observed in WwoxΔHep mice compared with control mice (Fig. 7a, b). Intriguingly, whereas the proliferation index was reduced 4 and 7 days post hepatectomy in control mice, it continued rising in WwoxΔHep mice. Increased proliferation was associated with increased ratio of liver to body weight on day 7 post hepatectomy (Fig. 7c). Moreover, qRT-PCR analysis of proliferative gene implicated in liver regeneration, including c-Myc, displayed higher levels in WwoxΔHep mice group on day 2, 3, and 4 as compared with control mice (Fig. 7d). Interestingly, Wwox and c-Myc transcripts showed a trend of inverse pattern of expression in Wwox-control mice (Fig. 7e) suggesting that WWOX might negatively modulate c-Myc. These results are consistent with WWOX function to inhibit hepatocyte proliferation after liver regeneration.
Fig. 7. Wwox ablation is associated with increased proliferation upon partial hepatectomy.
a Histological images (H&E staining and Ki67 immunohistochemical staining) of control and WwoxΔHep mice liver 3-days post 50% partial hepatectomy (× 10, × 20, and × 40). b Quantification of positive Ki67 nuclei in 5 months old control (WT) and WwoxΔHep mice liver 1, 2, 3, 4, and 7 days post hepatectomy (n = 5 for each group). c Liver weight to body weight ratio of 1, 2, 3, 4, and 7 days post hepatectomy (n = 5 for each group). d mRNA expression levels of c-Myc in control and WwoxΔHep mice liver 1, 2, 3, 4, and 7 days post hepatectomy. e mRNA expression levels of Wwox in control and WwoxΔHep mice liver 1, 2, 3, 4, and 7 days post hepatectomy (n = 4 for each group). * P value < 0.05, ** P value < 0.01, *** P value < 0.001. Error bars indicate ± SEM
Discussion
We report here that WWOX loss is a common event in HCC and that its dysregulation synergizes with DEN treatment and HFD to accelerate HCC development through regulation of proliferation genes. At the molecular level, WWOX, known to act as an adapter protein through interaction via its WW1 domain, functionally associates with HIF1α and probably other transcription factors involved in proliferation, leading to inhibition of their transactivation function. For example, when WWOX is lost, HIF1α recruitment and function on its target genes are enhanced leading to increased expression of glycolytic genes and proliferation rate. We further demonstrate that pharmacological inhibition of HIF1α reduces HCC burden in DEN-treated liver-specific WWOX-deficient mice. Finally, we show that WWOX loss increases liver regeneration after hepatectomy possibly via eliminating the termination phase of liver regeneration. These findings underscore the significant role of WWOX in rewiring metabolic changes in liver cells contributing to liver cancer.
Several studies have suggested that the WWOX gene functions as a tumor suppressor in liver cancer3,4,7. Early evidence has revealed the existence of frequent homozygous deletions at chromosome 16q23, where WWOX resides, in aflatoxin B1 exposed HCC6. In another study, expression of both WWOX and FHIT, another tumor suppressor located in a fragile site, appeared to be correlated and down-regulated in liver tissues in a carcinogen-specific manner49. A subsequent study has examined the status of the WWOX gene in human HCC cell lines and found its recurrent alterations further implicating WWOX in hepatocarcinogenesis7. In our study, we further delineate the role of WWOX as a tumor suppressor in liver cancer. First, we showed that WWOX expression is reduced or absent in large cohorts of human liver pathologies, including HCC. Furthermore, WWOX low expression in HCC is correlated with decreased survival suggesting that WWOX expression has a prognostic value in HCC. Second, we provide the first in vivo evidence that WWOX loss contributes to HCC development, in two different models of murine HCC. Our data show that DEN treatment alone or in combination with HFD clearly have an advantage for HCC development upon WWOX loss. Both tumor incidence and multiplicity (load) were higher in WwoxΔHep mice implying the importance of WWOX alteration for HCC development and progression. Our findings also suggest that WWOX loss is an important contributor for HCC promotion as earlier hits are required for HCC initiation. WWOX expression could be altered by environmental cues6,49 or in consequence of microRNA dysregulation15 or even as a result of genetic variations in WWOX50. If these alterations were combined with western HFD then our findings indicate that HCC risk increases.
We also showed that WWOX anti-proliferative activity is mediated by suppression of the proto-oncogenes c-Myc, c-Fos, and c-Jun. c-Myc is known as an oncogene that promotes HCC in human51 and murine HCC models52,53. We found that c-Myc is upregulated in WwoxΔHep mice on 1, 3, 6, and 10 months continually, raising the point that WWOX partially suppresses HCC through continuous suppression of c-Myc. Previous reports have shown that WWOX suppresses the AP-1 transcriptional activity through regulating c-Jun localization54, consistent with our findings showing enhanced expression of c-Jun and c-Fos in WWOX-depleted cells. It is also likely that WWOX loss releases its inhibitory effect on a plethora of other proto-oncogenes as have been previously demonstrated9,16,55, contributing to the observed aggressive HCC phenotype. Although WWOX has been shown as an effector of the Wnt/β-catenin pathway in vitro, through interaction with DVL15,35,56, our results do not support such an effect in DEN-mediated HCC development. On the other hand, when DEN treatment was combined with HFD, WWOX loss resulted in significant increase in Axin levels, suggesting that under these conditions WWOX plays a more important role in regulating the Wnt/β-catenin pathway. It is therefore possible that other cellular pathways are also involved as WWOX function has been shown to include plethora of effectors57.
Our data further present a critical metabolic and anti-proliferative role of the tumor suppressor WWOX in suppressing HCC through regulation of HIF1α function. The fact that HIF1α protein levels and its target genes were elevated in the pre-tumor stage (6 months; Fig. 4e) implies that these metabolic changes fuel and drive HCC development in DEN-treated WwoxΔHep mice. HIF1α is significantly elevated in human HCC samples and associated with bad prognosis58,59. Moreover, HIF1α hepatocytes-specific overexpression in a murine model increases HCC-promoting M2 macrophages60, whereas HIF1α liver-specific knockout sensitizes hepatoma cells to etoposide treatment61. WWOX loss is associated with elevated HIF1α target genes starting from the age of 6 months. In addition, we show that HIF1α is enriched on its targets promoters in hepatocytes isolated from WwoxΔHep mice.
Targeting tumor suppressor genes is a major challenge. Novel approaches, including synthetic lethality and collateral vulnerability have been proposed to come over this limitation. We therefore, assessed whether targeting the proliferative/survival signals in WWOX-deficient cancer cells could help inhibit tumor progression. Several attempts to target HIF1α in HCC were reported via inhibition of its expression62, dimerization63, or activity64. In our model, we were able to rescue, at least partially, the tumor phenotype using an HIF1α inhibitor (digoxin); at the age of 14 months, DEN-WwoxΔHep mice did not show macroscopic tumors and had lower expression of glycolytic genes driven by HIF1α. These results are consistent with our previous data showing that WWOX inhibits aerobic glycolysis42–44,65,66. Rewiring of glucose metabolism in WwoxΔHep mice provides the needed building blocks for cell division and proliferation67 and hence these mice could have more proliferation compared with control mice. Future studies shall include more specific inhibitors of HIF1a and probably Myc to inhibit WWOX-mediated HCC development.
WWOX anti-proliferative function in HCC might also involve modulation of fatty acid/lipid metabolism. In fact, WWOX genetic variants in human patients68 and Wwox knockout mouse models24 display decreased serum HDL-C. Moreover, microarray analyses of Wwox liver-specific knockout mice revealed an increase in plasma triglycerides and altered lipid metabolic pathways suggesting that WWOX disruption indeed alters cellular lipid homeostasis in the liver. Whether these WWOX effects can also contribute to HCC development is unknown.
Our findings also present a vital role of WWOX in maintaining healthy liver regeneration. Remarkably, partial hepatectomy in liver-specific Wwox-deficient mice resulted in increased proliferation (Ki67) upon liver regeneration, at the later phases of this remarkably controlled homeostatic process. Interestingly, Wwox RNA levels increased 2 days post hepatectomy while c-Myc levels showed an inverse pattern. c-Myc levels are reported to be highest in 12–18 h post heptectomy and then c-Myc levels usually drop down69–71. Our results might suggest that WWOX, an anti-proliferative gene, is induced during the course of liver regeneration to allow shutting down proliferative genes, such as c-Myc, and maintaining proper organ size. Interestingly, no difference in HIF1α target genes was observed in liver regeneration of WwoxΔHep. These observations suggest that WWOX may regulate liver regeneration by HIF1α-independent mechanism.
In conclusion, our study reveals WWOX as a tumor suppressor with critical roles in HCC suppression through maintaining moderate glucose metabolism and inhibiting uncontrolled cell proliferation.
Materials and Methods
Mice and related experiments
Wwox-floxed (Wwoxfl/fl) C57BL6/J;129sv mixed genetic background mice were bred with Albumin-Cre transgenic mice to generate Wwox conditional knockout in hepatocytes (WwoxΔHep mice). Male pups of control and WwoxΔHep mice were IP injected with 5 mg/Kg DEN (Sigma Aldrich) at the age of 14 days. Partial hepatectomy (30%) of the liver was done on 4-month-aged males as described72. For digoxin treatment, digoxin (1 mg/Kg, Sigma Aldrich) was IP injected into DEN-treated mice starting at age of 6.5 months, 3-times/week, for a total of 8-months. The mice in HFD experiment were fed by 60% kcal fat (Research Diets INC, D12492) for 30 weeks. Tumor load was assessed by liver weight in grams. All experiments involving mice were approved by the Hebrew University Institutional Animal Care and Use Committee.
RNA extraction and Real-time PCR
Total RNA was prepared using TRI reagent (Sigma Aldrich) following instructions of the manufacturer. One microgram of RNA was used for complementary DNA synthesis using First-Strand cDNA Synthesis kit (Bio-Rad, Hercules, CA). QRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). All measurements were performed in triplicate and standardized to the levels of the Ubc gene. List of primers used is provided as Supplemental Table 1.
Histology and immunohistochemistry
Tissues were fixed in 4% neutral buffered-formalin and then paraffin embedded, sectioned, and stained with H&E. Immunohistochemical staining was done as previously described73. Immunohistochemical staining of WWOX (polyclonal anti-WWOX antibody, dilution, 1:4000 for 1 h) was done after antigen retrieval with 10 mM citrate buffer (pH 6.0) in a pressure cooker. Detection was done with DAB peroxidase substrate kit (Cat# SK-4100, Vector). Antibodies used were: Ki67 antibody (Cat# MA5-14520, Thermoscientific, dilution 1:200), PKM2 antibody (Cat# 38237, Abcam, dilution 1:50) and HIF1α antibody (Cat# NB100-105, Novus, dilution 1:20).
ChIP
Hepatocytes were isolated from liver tissues, and solutions were prepared for ChIP analysis according to a standard protocol74. 0.8 mg of HIF1α antibody (mouse mAB, Cat # NB100-105, Novous Biological, CO, USA) was used to precipitate HIF1α. Targeted PCR was done using list of primers (Supplemental Table 2).
Statistics
Results of the experiments were expressed as mean ± standard deviation or standard error of mean. Student’s t-test, was used to compare values of test and control samples. P < 0.05 indicates significant difference. * P-value < 0.05, ** P-value < 0.01. For the human data analysis, Single Copy Number Alteration of liver hepatocellular carcinoma (LiHC) TCGA data set (n = 434) was analyzed by the website www.firebrowse.org (developed by broad institute of MIT and Harvard). Survival curve of LiHC TCGA data set was analyzed by the web site www.xenabrowser.net (Developed by UCSC).
Electronic supplementary material
Acknowledgements
We are grateful to Chaim Preiser, Ben Cohen, and Daniel Steinberg for technical help with mice work. The Aqeilan laboratory is supported by Worldwide Cancer Research grant (grant agreement No. 14-1095) and European Research Council (ERC)-Consolidator Grant under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 682118).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. Melino
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/23/2018
Since the publication of their article, the authors reported that duplication had mistakenly occurred in Fig 3A and D - H&E panels.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41419-018-0510-4.
References
- 1.El-Serag HB. Hepatocellular carcinoma. N. Engl. J. Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J. Clin. Gastroenterol. 2013;47:S2–S6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J. Cell Physiol. 2007;212:307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- 4.Aldaz C. Marcelo, Ferguson Brent W., Abba Martin C. WWOX at the crossroads of cancer, metabolic syndrome related traits and CNS pathologies. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2014;1846(1):188–200. doi: 10.1016/j.bbcan.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baryla I, Styczen-Binkowska E, Bednarek AK. Alteration of WWOX in human cancer, a clinical view. Exp. Biol. Med. 2015;240:305–314. doi: 10.1177/1535370214561953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yakicier MC, et al. Identification of homozygous deletions at chromosome 16q23 in aflatoxin B1 exposed hepatocellular carcinoma. Oncogene. 2001;20:5232–5238. doi: 10.1038/sj.onc.1204674. [DOI] [PubMed] [Google Scholar]
- 7.Park SW, et al. Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br. J. Cancer. 2004;91:753–759. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J. Cell Biochem. 2009;108:737–745. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 9.Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6:249–259. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salah Z, Alian A, Aqeilan RI. WW domain-containing proteins: retrospectives and the future. Front. Biosci. 2012;17:331–348. doi: 10.2741/3930. [DOI] [PubMed] [Google Scholar]
- 11.Jiang QF, Tian YW, Shen Q, Xue HZ, Li K. SENP2 regulated the stability of beta-catenin through WWOX in hepatocellular carcinoma cell. Tumour Biol. 2014;35:9677–9682. doi: 10.1007/s13277-014-2239-8. [DOI] [PubMed] [Google Scholar]
- 12.Li YP, Wu CC, Chen WT, Huang YC, Chai CY. The expression and significance of WWOX and beta-catenin in hepatocellular carcinoma. APMIS. 2013;121:120–126. doi: 10.1111/j.1600-0463.2012.02947.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, et al. Upregulation of the putative oncogene COTE1 contributes to human hepatocarcinogenesis through modulation of WWOX signaling. Int. J. Oncol. 2014;45:719–731. doi: 10.3892/ijo.2014.2482. [DOI] [PubMed] [Google Scholar]
- 14.Xie B, et al. ACK1 promotes hepatocellular carcinoma progression via downregulating WWOX and activating AKT signaling. Int. J. Oncol. 2015;46:2057–2066. doi: 10.3892/ijo.2015.2910. [DOI] [PubMed] [Google Scholar]
- 15.Hua HW, Jiang F, Huang Q, Liao Z, Ding G. MicroRNA-153 promotes Wnt/beta-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget. 2015;6:3840–3847. doi: 10.18632/oncotarget.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang NS, Hsu LJ, Lin YS, Lai FJ, Sheu HM. WW domain-containing oxidoreductase: a candidate tumor suppressor. Trends Mol. Med. 2007;13:12–22. doi: 10.1016/j.molmed.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Abdeen SK, et al. Wwox inactivation enhances mammary tumorigenesis. Oncogene. 2011;30:3900–3906. doi: 10.1038/onc.2011.115. [DOI] [PubMed] [Google Scholar]
- 18.Aqeilan RI, et al. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67:5606–5610. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aqeilan RI, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc. Natl. Acad. Sci. USA. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aqeilan RI, et al. The WWOX tumor suppressor is essential for postnatal survival and normal bone metabolism. J. Biol. Chem. 2008;283:21629–21639. doi: 10.1074/jbc.M800855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdeen SK, et al. Conditional inactivation of the mouse Wwox tumor suppressor gene recapitulates the null phenotype. J. Cell Physiol. 2013;228:1377–1382. doi: 10.1002/jcp.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int J. Exp. Pathol. 2009;90:367–386. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakiri L, Wagner EF. Mouse models for liver cancer. Mol. Oncol. 2013;7:206–223. doi: 10.1016/j.molonc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iatan I, et al. The WWOX gene modulates high-density lipoprotein and lipid metabolism. Circ. Cardiovasc. Genet. 2014;7:491–504. doi: 10.1161/CIRCGENETICS.113.000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verna L, Whysner J, Williams GM. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Odeh M, Hereema NA, Aqeilan RI. WWOX modulates the ATR-mediated DNA damage checkpoint response. Oncotarget. 2016;7:4344–4355. doi: 10.18632/oncotarget.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu-Odeh M, Salah Z, Herbel C, Hofmann TG, Aqeilan RI. WWOX, the common fragile site FRA16D gene product, regulates ATM activation and the DNA damage response. Proc. Natl. Acad. Sci. USA. 2014;111:E4716–E4725. doi: 10.1073/pnas.1409252111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazan I, Hofmann TG, Aqeilan RI. Tumor suppressor genes within common fragile sites are active players in the DNA damage response. PLoS Genet. 2016;12:e1006436. doi: 10.1371/journal.pgen.1006436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aqeilan RI, et al. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004;64:8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- 30.Chang JY, et al. Signaling from membrane receptors to tumor suppressor WW domain-containing oxidoreductase. Exp. Biol. Med. 2010;235:796–804. doi: 10.1258/ebm.2010.009351. [DOI] [PubMed] [Google Scholar]
- 31.O’Keefe LV, Lee CS, Choo A, Richards RI. Tumor suppressor WWOX contributes to the elimination of tumorigenic cells in drosophila melanogaster. PloS ONE. 2015;10:e0136356. doi: 10.1371/journal.pone.0136356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gourley C, et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin alpha3. Cancer Res. 2009;69:4835–4842. doi: 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- 33.Abu-Remaileh M, Aqeilan R I. Tumor suppressor WWOX regulates glucose metabolism via HIF1α modulation. Cell Death & Differentiation. 2014;21(11):1805–1814. doi: 10.1038/cdd.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards RI, Choo A, Lee CS, Dayan S. O’Keefe L. WWOX, the chromosomal fragile site FRA16D spanning gene: Its role in metabolism and contribution to cancer. Exp. Biol. Med. 2015;240:338–344. doi: 10.1177/1535370214565990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Odeh M, et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J. Biol. Chem. 2014;289:8865–8880. doi: 10.1074/jbc.M113.506790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase’s role in myriad cancers: clinical significance and future implications. Exp. Biol. Med. 2014;239:253–263. doi: 10.1177/1535370213519213. [DOI] [PubMed] [Google Scholar]
- 37.Zimmers TA, et al. Effect of in vivo loss of GDF-15 on hepatocellular carcinogenesis. J. Cancer Res. Clin. Oncol. 2008;134:753–759. doi: 10.1007/s00432-007-0336-4. [DOI] [PubMed] [Google Scholar]
- 38.Teoh NC, et al. Defective DNA strand break repair causes chromosomal instability and accelerates liver carcinogenesis in mice. Hepatology. 2008;47:2078–2088. doi: 10.1002/hep.22194. [DOI] [PubMed] [Google Scholar]
- 39.Hu BS, et al. WWOX induces apoptosis and inhibits proliferation of human hepatoma cell line SMMC-7721. World J. Gastroenterol. 2012;18:3020–3026. doi: 10.3748/wjg.v18.i23.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolba R, Kraus T, Liedtke C, Schwarz M, Weiskirchen R. Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab. Anim. 2015;49:59–69. doi: 10.1177/0023677215570086. [DOI] [PubMed] [Google Scholar]
- 41.Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev. Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choo A, et al. Tumor suppressor WWOX moderates the mitochondrial respiratory complex. Genes Chromosomes Cancer. 2015;54:745–761. doi: 10.1002/gcc.22286. [DOI] [PubMed] [Google Scholar]
- 43.O’Keefe LV, et al. Drosophila orthologue of WWOX, the chromosomal fragile site FRA16D tumour suppressor gene, functions in aerobic metabolism and regulates reactive oxygen species. Hum. Mol. Genet. 2011;20:497–509. doi: 10.1093/hmg/ddq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Remaileh M., Seewaldt V. L., Aqeilan R. I. WWOX loss inactivates aerobic glycolysis. Mol. Cell Oncol.290, 30728–30735 (2015). [DOI] [PMC free article] [PubMed]
- 45.Wilson GK, Tennant DA, McKeating JA. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: current understanding and future directions. J. Hepatol. 2014;61:1397–1406. doi: 10.1016/j.jhep.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, et al. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc. Natl. Acad. Sci. USA. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida T, et al. Digoxin inhibits retinal ischemia-induced HIF-1alpha expression and ocular neovascularization. FASEB J. 2010;24:1759–1767. doi: 10.1096/fj.09-145664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park EJ, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iida M, et al. Unique patterns of gene expression changes in liver after treatment of mice for 2 weeks with different known carcinogens and non-carcinogens. Carcinogenesis. 2005;26:689–699. doi: 10.1093/carcin/bgi005. [DOI] [PubMed] [Google Scholar]
- 50.Lee HL, et al. Functional genetic variant of WW domain-containing oxidoreductase (WWOX) gene is associated with hepatocellular carcinoma risk. PloS ONE. 2017;12:e0176141. doi: 10.1371/journal.pone.0176141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawate S, Fukusato T, Ohwada S, Watanuki A, Morishita Y. Amplification of c-myc in hepatocellular carcinoma: correlation with clinicopathologic features, proliferative activity and p53 overexpression. Oncology. 1999;57:157–163. doi: 10.1159/000012024. [DOI] [PubMed] [Google Scholar]
- 52.Beer S, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 54.Gaudio E, et al. Physical association with WWOX suppresses c-Jun transcriptional activity. Cancer Res. 2006;66:11585–11589. doi: 10.1158/0008-5472.CAN-06-3376. [DOI] [PubMed] [Google Scholar]
- 55.Aqeilan RI, Abu-Remaileh M, Abu-Odeh M. The common fragile site FRA16D gene product WWOX: roles in tumor suppression and genomic stability. Cell Mol. Life Sci. 2014;71:4589–4599. doi: 10.1007/s00018-014-1724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouteille N, Driouch K, Hage P El, Sin S, Formstecher E, Camonis J, Lidereau R, Lallemand F. Inhibition of the Wnt/β-catenin pathway by the WWOX tumor suppressor protein. Oncogene. 2009;28(28):2569–2580. doi: 10.1038/onc.2009.120. [DOI] [PubMed] [Google Scholar]
- 57.Abu-Remaileh M, Joy-Dodson E, Schueler-Furman O, Aqeilan RI. Pleiotropic functions of tumor suppressor WWOX in normal and cancer cells. J. Biol. Chem. 2015;290:30728–30735. doi: 10.1074/jbc.R115.676346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang SL, et al. The correlation of expression levels of HIF-1alpha and HIF-2alpha in hepatocellular carcinoma with capsular invasion, portal vein tumor thrombi and patients’ clinical outcome. Jpn J. Clin. Oncol. 2014;44:159–167. doi: 10.1093/jjco/hyt194. [DOI] [PubMed] [Google Scholar]
- 59.Dai CX, et al. Hypoxia-inducible factor-1 alpha, in association with inflammation, angiogenesis and MYC, is a critical prognostic factor in patients with HCC after surgery. BMC Cancer. 2009;9:418. doi: 10.1186/1471-2407-9-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ambade A, et al. Hepatocellular carcinoma is accelerated by NASH involving M2 macrophage polarization mediated by hif-1alphainduced IL-10. Oncoimmunology. 2016;5:e1221557. doi: 10.1080/2162402X.2016.1221557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daskalow K, et al. Role of hypoxia-inducible transcription factor 1alpha for progression and chemosensitivity of murine hepatocellular carcinoma. J. Mol. Med. 2010;88:817–827. doi: 10.1007/s00109-010-0623-4. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, et al. Bufalin suppresses hepatocellular carcinoma invasion and metastasis by targeting HIF-1alpha via the PI3K/AKT/mTOR pathway. Oncotarget. 2016;7:20193–20208. doi: 10.18632/oncotarget.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee K, et al. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. USA. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Limani P, et al. Development of OXY111A, a novel hypoxia-modifier as a potential antitumor agent in patients with hepato-pancreato-biliary neoplasms - Protocol of a first Ib/IIa clinical trial. BMC Cancer. 2016;16:812. doi: 10.1186/s12885-016-2855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abu-Remaileh M, Aqeilan RI. Tumor suppressor WWOX regulates glucose metabolism via HIF1alpha modulation. Cell Death Differ. 2014;21:1805–1814. doi: 10.1038/cdd.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abu-Remaileh M, Aqeilan RI. The tumor suppressor WW domain-containing oxidoreductase modulates cell metabolism. Exp. Biol. Med. 2015;240:345–350. doi: 10.1177/1535370214561956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 68.Lee JC, et al. WW-domain-containing oxidoreductase is associated with low plasma HDL-C levels. Am. J. Hum. Genet. 2008;83:180–192. doi: 10.1016/j.ajhg.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goyette M, Petropoulos CJ, Shank PR, Fausto N. Regulated transcription of c-Ki-ras and c-myc during compensatory growth of rat liver. Mol. Cell Biol. 1984;4:1493–1498. doi: 10.1128/MCB.4.8.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson NL, et al. Sequential protooncogene expression during rat liver regeneration. Cancer Res. 1986;46:3111–3117. [PubMed] [Google Scholar]
- 71.Fausto N, et al. Proto-oncogene expression and growth factors during liver regeneration. Symp. Fundam. Cancer Res. 1986;39:69–86. [PubMed] [Google Scholar]
- 72.Hori T, et al. Simple and reproducible hepatectomy in the mouse using the clip technique. World J. Gastroenterol. 2012;18:2767–2774. doi: 10.3748/wjg.v18.i22.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aqeilan RI, et al. Loss of WWOX expression in gastric carcinoma. Clin. Cancer Res. 2004;10:3053–3058. doi: 10.1158/1078-0432.CCR-03-0594. [DOI] [PubMed] [Google Scholar]
- 74.Schmidt D, et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.