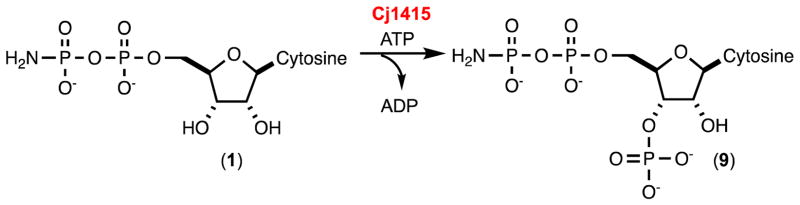
Abstract
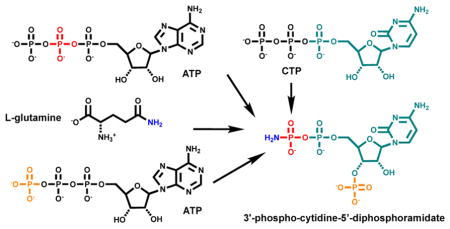
Campylobacter jejuni , a leading cause of gastroenteritis, produces a capsular polysaccharide that is derivatized with a unique O-methyl phosphoramidate (MeOPN) modification. This modification contributes to serum resistance and invasion of epithelial cells. Previously, the first three biosynthetic steps for the formation of MeOPN have been elucidated. The first step is catalyzed by a novel glutamine kinase (Cj1418), which catalyzes the ATP-dependent phosphorylation of the amide nitrogen of L-glutamine. L-Glutamine phosphate is used by CTP:phosphoglutamine cytidylyltransferase (Cj1416) to displace pyrophosphate from CTP to generate CDP-L-glutamine, which is then hydrolyzed by γ-glutamyl-CDP-amidate hydrolase (Cj1417) to form cytidine diphosphoramidate (CDP-NH2). Here we show that Cj1415 catalyzes the ATP-dependent phosphorylation of CDP-NH2 to form 3′-phospho-cytidine-5′-diphosphoramidate. Cj1415 will also catalyze the phosphorylation of adenosine diphosphoramidate (ADP-NH2) and uridine diphosphoamidate (UDP-NH2) but at significantly reduced rates. It is proposed that Cj1415 be named cytidine diphosphoramidate kinase.
Graphical Abstract
INTRODUCTION
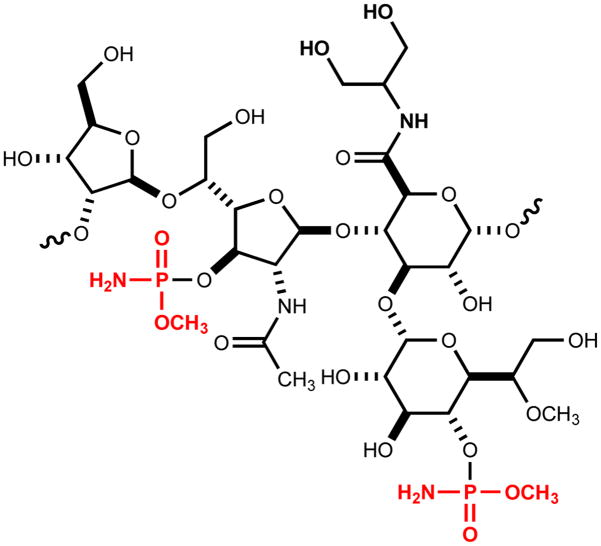
Campylobacter jejuni, a Gram-negative pathogenic bacterium, is a leading cause of gastroenteritis worldwide (1,2). C. jejuni, while pathogenic in humans, is commensal in chickens, and thus contaminated poultry have become a common source of human infections (3). While most C. jejuni infections result in gastroenteritis, rare cases are linked to the occurrence of the autoimmune disease, Guillain-Barré syndrome (4). Like many other organisms, C. jejuni produces a capsular polysaccharide (CPS) that aids in host colonization, evasion of immune responses, and serum resistance (5,6). Over 34 CPS gene clusters have been published, and it is believed that each strain produces a unique CPS (7). Currently, the structures of at least 10 C. jejuni capsular polysaccharides have been physically characterized (8). The CPS of C. jejuni NCTC 11168 consists of a four-carbohydrate repeating unit that is decorated with a unique O-methyl phosphoramidate (MeOPN) modification (9,10). Approximately 70% of all C. jejuni strains contain the MeOPN moiety attached to their CPS. In the CPS of C. jejuni NCTC 11168, the 2-acetamido-2-deoxy-β-D-galactofuranose and D-glycero-α-L-gluco-heptopyranose sugars are derivatized with the methyl phosphoramidate group (Scheme 1).
Scheme 1.
A cluster of 35 genes has been identified in C. jejuni NCTC 11168 that is primarily responsible for the biosynthesis and export of the CPS (11). Eight genes in this cluster (Cj1415 through Cj1422) have been predicted to be responsible for the biosynthesis and transfer of the MeOPN modification to the sugar backbone (10). Enzymes with the locus tags Cj1421 and Cj1422 have been implicated in the transfer of the phosphoramidate moiety to their respective sugar targets (10). Cj1422 is required for the attachment of the MeOPN group to D-glycero-α-L-gluco-heptopyranose whereas Cj1421 is needed for modification to 2-acetamido-2-deoxy-β-D-galactofuranose (10). Cj1419 and Cj1420 are annotated as SAM-dependent methyltransferases and are believed to be responsible for methylation of the phosphoramidate group (10). The other four enzymes, Cj1415, Cj1416, Cj1417, and Cj1418 are required for the biosynthesis of the phosphoramidate group of MeOPN, and if any of the four genes for these enzymes are deleted, all of the MeOPN modifications are lost (10).
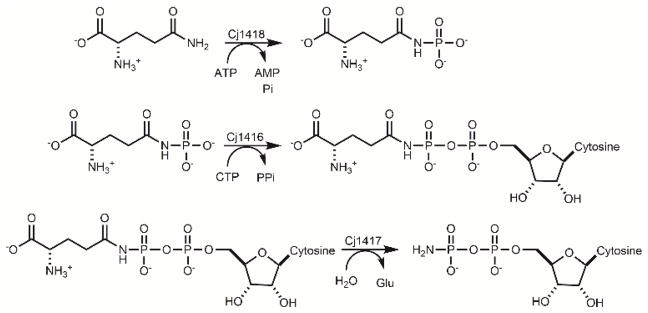
Recently, we have functionally characterized the first three enzymes in the biosynthesis of the phosphoramidate modification in C. jejuni (12,13). In the first reaction, Cj1418 catalyzes the ATP-dependent phosphorylation of L-glutamine on the amide nitrogen to make L-glutamine phosphate (12). This product is utilized by Cj1416 to displace pyrophosphate from MgCTP to form CDP-L-glutamine (13). Cj1417 then catalyzes the hydrolysis of CDP-L-glutamine to produce L-glutamate and cytidine diphosphoramidate (CDP-NH2). These reactions are summarized in Scheme 2.
Scheme 2.
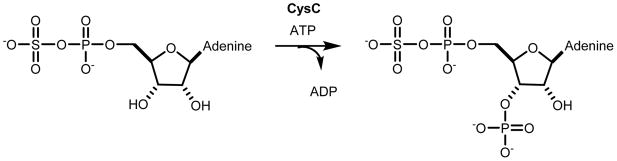
Cj1415 is currently annotated as an adenylyl sulfate kinase from cog0529 (14). While most members of this family of enzymes are bifunctional adenylyl sulfate synthases/adenylyl sulfate kinases, the closest functionally characterized homolog to Cj1415 is the monofunctional CysC from E. coli (14). CysC catalyzes the ATP-dependent phosphorylation of the 3′-hydroxyl group of adenylyl sulfate (APS) forming 3′-phosphoadenosine 5′-phosphosulfate (PAPS), a molecule that is ultimately used in the transfer of sulfate to various acceptors (Scheme 3). Since CysC is the closest functionally characterized homolog to Cj1415, we postulate that Cj1415 will phosphorylate the 3′-hydroxyl group of cytidine diphosphoramidate (1) to synthesize a new cofactor for the transfer of phosphoramidate groups to various acceptors.
Scheme 3.
MATERIALS and METHODS
Materials
All chemicals and buffers were purchased from Sigma-Aldrich, unless otherwise specified. CDP, UDP and CTP were purchased from Alfa Aesar. ADP was purchased from TCI Chemicals. DpnI was obtained from New England Biolabs. Primestar HS Polymerase was purchased from Takara Industries. The plasmid used for the expression of Cj1415 from Campylobacter jejuni NCTC 11168 was obtained from Professor Christine Szymanski of the University of Georgia.
Gene Expression and Enzyme Purification
The plasmid for the expression of Cj1415 (UniProt: Q0P8J9) with a C-terminal polyhistidine purification tag was used to transform Rosetta (DE3) E. coli cells by electroporation. Five-mL cultures of LB medium supplemented with 50 μg/mL kanamycin and 25 μg/mL chloramphenicol were inoculated with a single colony and grown overnight at 37 °C. These cultures were used to inoculate 1 L of LB medium (50 μg/mL kanamycin and 25 μg/mL chloramphenicol) and then incubated at 30 °C until an OD600 of ~0.6–0.8 was reached. The cells were induced with 1.0 mM isopropyl β-thiogalactoside (IPTG), grown for 16 hours at 16 °C and then harvested by centrifugation at 6000 rpm at 4 °C. The resulting cell pellet was resuspended into loading buffer (50 mM HEPES/K+, 300 mM KCl, 20 mM imidazole, pH 8.0) and lysed by sonication. The total cell lysate was passed through a 0.45 μm filter before being loaded onto a prepacked a 5-mL HisTrap HP (GE Healthcare) nickel affinity column. Protein was eluted with 50 mM HEPES/K+, pH 8.0, 300 mM KCl, and 400 mM imidazole over a gradient of 30 column volumes. Excess imidazole was removed by exchanging the buffers against 50 mM HEPES/K+, pH 8.0, 100 mM KCl using a 20 mL (10 kDa molecular weight cutoff) concentrator (GE Healthcare). The purified protein was flash frozen and stored at −80 °C. Approximately 30 mg of purified protein was obtained per liter of cell culture.
Synthesis of Substrates
Cytidine diphosphoramidate (CDP-NH2, 1), uridine diphosphoramidate (UDP-NH2, 6), and adenosine diphosphoramidate (ADP-NH2, 7) were prepared chemically as previously described (15). 2′-Deoxy cytidine diphosphoramidate (2′-deoxy-CDP-NH2, 2) was made enzymatically with 2′-deoxy CTP (Roche) and L-glutamine phosphate (12), using Cj1416 and Cj1417 as catalysts (13). 2′-Deoxy CTP (10 mM), L-glutamine phosphate (10 mM) and MgCl2 (15 mM) were incubated with Cj1416 (10 μM) and Cj1417 (10 μM) for three hours at room temperature (100 mM HEPES pH 8.0). The enzymes were removed by filtration and the concentration of 2′-deoxy-CDP-NH2 was determined by UV-visible spectroscopy.
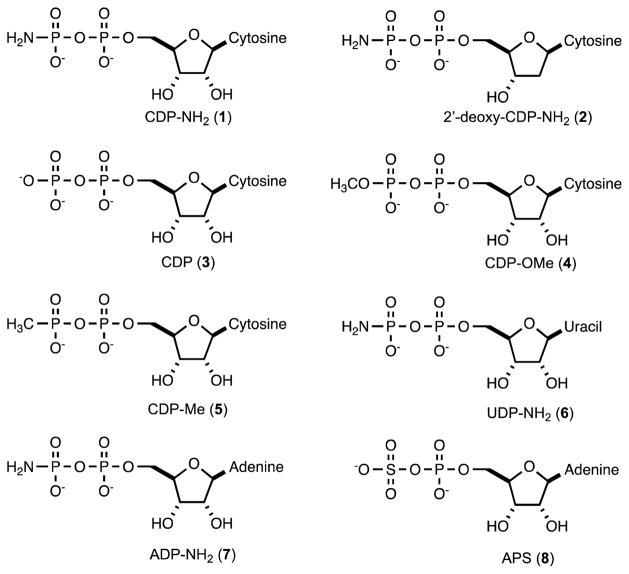
CDP-methyl phosphate (CDP-OMe, 4) and CDP-methyl phosphonate (CDP-Me, 5) were made enzymatically using Cj1416 and MnCTP (13). In these reactions methyl phosphate (15 mM) or methyl phosphonate (15 mM) was incubated with Cj1416 (15 μM) and CTP (15 mM) with MgCl2 (15 mM) and MnCl2 (4.0 mM) in 100 mM HEPES/K+, pH 8.0, 100 mM KCl. After 16 hours, the reactions were quenched and the target compounds purified by anion exchange column chromatography (DEAE Sephadex) using a gradient of 10–1000 mM triethylamonium bicarbonate (pH 8.0). The elution of the products from the column was monitored at 260 nm. The fractions were pooled and the structures of the desired compounds confirmed by 31P NMR spectroscopy and ESI (negative) mass spectrometry. CDP-Me (5) was obtained in an isolated yield of ~30% and CDP-OMe (4) was obtained with a yield of ~20%, based on the starting concentration of CTP. The 31P NMR spectrum of CDP-Me (5) showed two doublets at 17.92 (β-P) and −10.79 ppm (α-P) whereas the 31P NMR spectrum for CDP-OMe (4) showed two doublets at −9.06 (β-P), and −10.82 ppm (α-P). The ESI (negative) mass spectrum for CDP-Me (5) indicated an m/z of 400.03 (expected m/z for M-H = 400.03 for C10H17N3O10P2) whereas the ESI (negative) mass spectrum for CDP-OMe (4) had an m/z of 416.02 (expected m/z for M-H = 416.02 for C10H17N3O11P2). The structures of the compounds used in this investigation are presented in Scheme 4
Scheme 4.
Determination of Kinetic Constants
The kinetic constants for wild type Cj1415 were determined using a pyruvate kinase/lactate dehydrogenase coupled enzyme assay, following the oxidation of NADH at 340 nm at 25 °C with a SpectraMax340 UV-visible spectrophotometer (16). Assays were performed in 100 mM HEPES/K+, pH 8.0, and 100 mM KCl. The coupling system contained 2.0 mM phosphoenol pyruvate, 0.3 mM NADH, 8 units/mL lactate dehydrogenase, and 8 units/mL pyruvate kinase. When determining the kinetic constants for CDP-NH2 (20–200 μM), 2′-deoxy CDP-NH2 (50 – 500 μM), ADP-NH2 (1 – 10 mM), UDP-NH2 (2.6–26 mM), CDP-OMe (200 – 2000 μM), and CDP-Me (360 – 3600 μM), a fixed concentration of 10 mM ATP was used. The kinetic constants for ATP (200 – 7000 μM) were determined with a fixed concentration of 400 μM CDP-NH2. For these experiments, MgCl2 was added in a 4.0 mM excess of the total nucleoside concentration. Kinetic constants for the consumption of CDP (0.5 – 15 mM) were determined using 31P NMR spectroscopy because CDP interfered with the coupled enzyme assay. For these assays, each reaction mixture contained 10 mM ATP and a 4 mM excess of MgCl2. The reactions were followed for 30 minutes, collecting a 31P NMR spectrum every 5 minutes. With the S85A mutant of Cj1415 the kinetic constants for CDP-NH2 (0.20 – 10 mM) were determined at a fixed concentration of ATP (10 mM). The kinetic constants for ATP (1 – 8 mM) were determined at a fixed concentration of CDP-NH2 (15 mM). Kinetic constants for ATP, CDP-NH2, dCDP-NH2, CDP-OMe, and CDP-Me were determined using 200 nM Cj1415, adenylyl sulfate, ADP-NH2 and UDP-NH2 used 1, 2.1 and 10 μM of enzyme, respectively.
The kinetic parameters were determined by fitting the initial rates to eqn. 1 using GraFit 5, where v is the initial velocity of the reaction, Et is the enzyme concentration, kcat is the turnover number, [A] is the substrate concentration, and Km is the Michaelis constant.
| (1) |
31P NMR Spectroscopy
The product of the ATP-dependent phosphorylation of each substrate by Cj1415 was analyzed by 31P NMR spectroscopy. Each reaction was conducted in 100 mM HEPES/K+, pH 8.0, and 100 mM KCl at 25 °C and initially contained 5.0 mM ATP, 5.0 mM substrate, 14 mM MgCl2, and 5 μM Cj1415. All reactions were quenched after 60 minutes with 15 mM EDTA, except for UDP-NH2 (6) and ADP-NH2 (7), which were quenched after 3 hours. After the reactions were quenched, the pH was adjusted to 9.0 before the spectra were collected.
Cj1415 S85A Mutant
Ser85 in Cj1415 was determined to be a residue of interest based on previous reports that the homologous residue in CysC from E. coli is phosphorylated during catalysis (17). The pET 30b plasmid for the expression of wild type Cj1415 with a C-terminal hexahistidine tag was used as a template for the construction of a plasmid for the expression of the S85A mutant. The forward primer (5′-GTATGATGGTTATTGTCACTACGATTGCAATGTTTAATGAGATTTATG-3′) and reverse primer (5′-CATAAATCTCATTAAACATTGCAATCGTAGTGACAATAACCATCATAC-3′) were used to convert the serine codon (TCA) to alanine (GCA). Primestar HS polymerase was used to amplify the gene. The reaction used a three-step thermal cycle (98 °C for 10 sec, 63 °C for 40 sec, and 72 °C for 10 minutes) and continued through thirty cycles. After amplification, DpnI was used to digest the template DNA for 2 hours at 37 °C. Following DpnI digestion, PCR cleanup (Qiagen) was performed and the plasmid transformed into E. coli BL21 (DE3) cells. Single colonies were selected and the DNA sequence of the mutant gene was confirmed.
Cog0529 Sequence Similarity Network
The identification codes for cog0529 (1160 total sequences) were downloaded from Uniprot. The collected identifiers were used to generate a network using the Enzyme Function Initiative- Enzyme Similarity Tool through Option-D (18,19). The full network (each sequence is a node) was downloaded and used to generate a sequence similarity network using Cytoscape (20). Edges below a 50% identity threshold were removed, and each cluster was assigned an arbitrary number and color.
RESULTS
Determination of the Catalytic Activity of Cj1415
CysC in E. coli catalyzes the final step in the biosynthesis of PAPS, the cofactor used for sulfuryl transfer in biological systems (14). Since the closest functionally characterized homolog to Cj1415 is CysC, adenylyl sulfate (APS, 8) was tested as a substrate for this enzyme using ATP as the phosphoryl donor. Within the limits of the coupled enzyme assay, no activity could be detected using Cj1415 to catalyze the phosphorylation of APS. Because of its presumptive connection to the biosynthesis of the O-methyl phosphonate moiety in C. jejuni, Cj1415 was tested with cytidine diphosphoamidate (1) as a substrate since this compound has been demonstrated to be the final product in the collective reactions catalyzed by Cj1416, Cj1417, and Cj1418 (Scheme 2) (12,13). CDP-NH2 (1) is an excellent substrate for Cj1415 with a turnover number of 2.2 s−1
Characterization of the Cj1415 Catalyzed Reaction Product
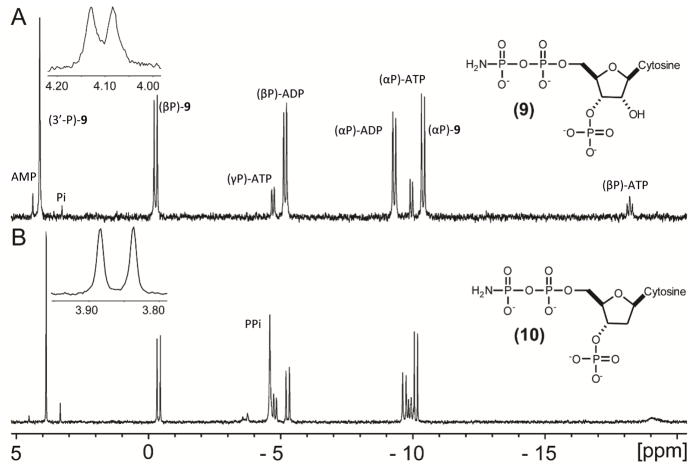
The reaction between CDP-NH2 and MgATP, as catalyzed by Cj1415, was analyzed by 31P NMR spectroscopy. After an incubation period of approximately one hour, the 31P NMR spectrum shows that most of the ATP was converted to ADP, and that a new resonance was observed at ~4.1 ppm (Figure 1a). In the 1H-coupled spectrum this resonance is a doublet with a coupling constant of 7.3 Hz, indicating that this phosphorus is coupled to a non-exchangeable proton. The molecular weight of the new product (9) was determined by ESI (negative ion) mass spectroscopy and shown to be 480.99. This value is consistent with the expected m/z of 480.99 for C9H17N4O13P3. Based on the molecular structure of CDP-NH2, the site of phosphorylation can only correspond to the 2′- or 3′-hydroxyl group of the ribose ring.
Figure 1.
31P NMR spectra of the products in the reaction catalyzed by Cj1415. (A) Cj1415 (5 μM) was mixed with 5.0 mM MgATP and 5.0 mM CDP-NH2 for 60 minutes at pH 8.0 before the reaction was quenched with 15 mM EDTA. (B) Cj1415 (5 μM) was mixed with 5.0 mM MgATP and 5.0 mM 2′-deoxy-CDP-NH2 for 60 minutes before the reaction was quenched with 15 mM EDTA. The insets show the 1H-coupled spectra for the resonances at 4.1 ppm (A) and 3.85 ppm (B). Additional details are available in the text.
The 2′-deoxy derivative of CDP-NH2 (2) was synthesized enzymatically and tested as a potential substrate for Cj1415. In the presence of MgATP, 2′-deoxy-CDP-NH2 (2) was phosphorylated with a kcat of 2.2 s−1 (Table 1). The 31P NMR spectrum of the phosphorylated reaction product (Figure 1b) shows the presence of a new resonance at ~3.85 ppm that is a doublet, with a coupling constant of 7.8 Hz, in the absence of proton decoupling. The molecular weight of this product (10) was determined by negative ion ESI-MS and shown to be 465.00. This value is consistent with the expected m/z of 464.98 for C9H17N4O12P3 Since the kinetic constants for the phosphorylation of CDP-NH2 and 2′-deoxy-CDP-NH2 are essentially identical, the site of phosphorylation is at the hydroxyl group attached to C3′ of the ribose ring. This result is fully consistent with the site of phosphorylation catalyzed by CysC. The reaction catalyzed by Cj1415 is summarized in Scheme 5.
Table 1.
Kinetic Constants of Cj1415 at 25 °C, pH 8.0
| substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) | |
|---|---|---|---|---|
| CDP-NH2 | (1) | 2.2 ± 0.3 | 0.11 ± 0.01 | 20000 ± 3300 |
| dCDP-NH2 | (2) | 2.2 ± 0.2 | 0.39 ± 0.07 | 5600 ± 1100 |
| CDP | (3) | 0.64 ± 0.04 | 4.2 ± 0.7 | 150 ± 30 |
| CDP-OMe | (4) | 0.65 ± 0.06 | 1.4 ± 0.2 | 480 ± 90 |
| CDP-Me | (5) | 1.4 ± 0.1 | 0.8 ± 0.2 | 1700 ± 400 |
| UDP-NH2 | (6) | 0.023 ± 0.002 | 21 ± 3 | 1.1 ± 0.2 |
| ADP-NH2 | (7) | 0.08 ± 0.01 | 5.6 ± 0.8 | 14 ± 3 |
| APS | (8) | < 0.01 | ||
| CDP-NH2a | (1) | 1.6 ± 0.2 | 4.4 ± 0.4 | 360 ± 60 |
| ATP | 2.2 ± 0.3 | 1.5 ± 0.1 | 1500 ± 200 | |
| ATPa | 1.6 ± 0.2 | 0.7 ± 0.1 | 2300 ± 500 |
S85A mutant of Cj1415
Scheme 5.
Substrate Specificity of Cj1415
The range of substrates accepted by Cj1415 was tested with five additional compounds. To determine if Cj1415 is selective for the cytosine base, uridine diphosphoramidate (6) and adenosine diphosphoramidate (7) were tested as substrates (Table 1). The values of kcat/Km for these two compounds are 3–4 orders of magnitude poorer that the value for CDP-NH2 and thus Cj1415 greatly prefers the cytosine base. Two other cytidine diphosphoramidate analogs were synthesized and assayed with Cj1415. Similar to CDP-NH2, these analogues possess a single negative charge on the β-phosphorus group. CDP-methyl phosphate (4) and CDP-methyl phosphonate (5) were both found to be substrates for Cj1415. The values of kcat are 30 to 60% as good as for CDP-NH2, but the Km values are about an order of magnitude greater than for CDP-NH2. CDP can be phosphorylated by Cj1415, but the value of kcat/Km is approximately 2 orders of magnitude poorer than for CDP-NH2. For all seven substrates discovered for Cj1415, the negative ion ESI mass spectra of the phosphorylated reaction products are fully consistent with the expected molecular structure.
Mutation of Serine-85
The closest functionally characterized homolog to Cj1415 is CysC from E. coli (14). Previous investigations with CysC have indicated the requirement for the phosphorylation of a serine residue as an obligatory reaction intermediate (17). The phosphorylated enzyme was isolated and shown to phosphorylate APS in the absence of added ATP (17). This serine residue is conserved in close CysC homologs and in Cj1415. To determine if the corresponding residue in Cj1415 is required for catalytic activity, Ser-85 was mutated to an alanine residue (S85A). The value of kcat for the S85A mutant is 73% of the value of the wild type enzyme, but the value of Km is 40 times greater, suggesting that this residue is not phosphorylated during catalysis but is perhaps interacting with the substrate in the active site (Table 1).
DISCUSSION
Substrate Specificity
In this study we have established the substrate profile of Cj1415, an enzyme that functions in the biosynthesis of the cofactor used for the transfer of phosphoramidate to acceptor carbohydrates in C. jejuni. As anticipated, the product of the reaction catalyzed by Cj1417, cytidine diphosphoramidate (1), is the best substrate of those tested for Cj1415 and is phosphorylated on the C3′-hydroxyl group (Table 1). We also examined the catalytic activity with the corresponding adenine and uridine diphosphoramidate derivatives, and confirmed that cytidine is the nucleotide preferred by Cj1415. CDP is a relatively poor substrate for Cj1415 and this result is likely to be a reflection of the two negative charges on the β-phosphoryl group, where CDP-NH2 has only one. In addition to CDP-NH2, the enzyme will also accept the substitution of –CH3 or –OCH3 for the –NH2 functional group on the β-phosphorus substituent. We propose that this enzyme be called cytidine diphosphoramidate kinase (CDK).
Comparison with Adenylyl Sulfate Kinase
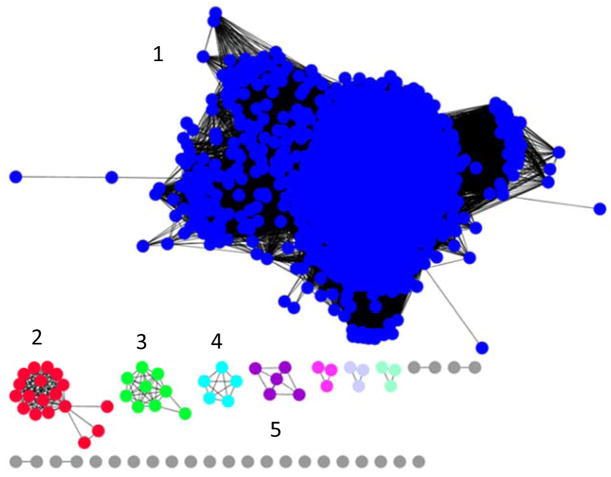
Cj1415 is a member of cog0529 and the sequence similarity network (SSN) is presented in Figure 2 at a percent identity cutoff of 50% (18). The only known catalytic activities from cog0529 are adenylyl sulfate kinase or a bifunctional adenylyl sulfate synthase and adenylyl sulfate kinase, which represent arbitrary Group-1 (Figure 2). At a 50% identity cutoff, Cj1415 is Group-2, which contains no functionally characterized enzymes. This cluster contains enzymes primarily from various Campylobacter and Helicobacter species. Groups 3–5 are not similar to adenylyl sulfate kinase or Cj1415 and thus likely represent other unique catalytic activities. Group-3 contains sequences only from Desulfovibrio species, indicating this group of enzymes may be specific to this bacterium.
Figure 2.
Sequence similarity network for proteins of cog0529 at a percent identity cutoff of 50%. All protein sequences for cog0529 obtained from Uniprot were used to create the network. Each node (colored sphere) represents a unique protein sequence and each edge (black line) represents a connection between two sequences at the given percent identity cutoff. Group 1 contains known adenylyl sulfate kinases or bifunctional adenylyl sulfate kinase/adenylyl sulfate synthases. Group 2 contains Cj1415 and homologs from several Campylobacter and Helicobacter species. Groups 3–5 represent enzymes that are dissimilar from adenylyl sulfate kinases and Cj1415, and likely represent unique activities.
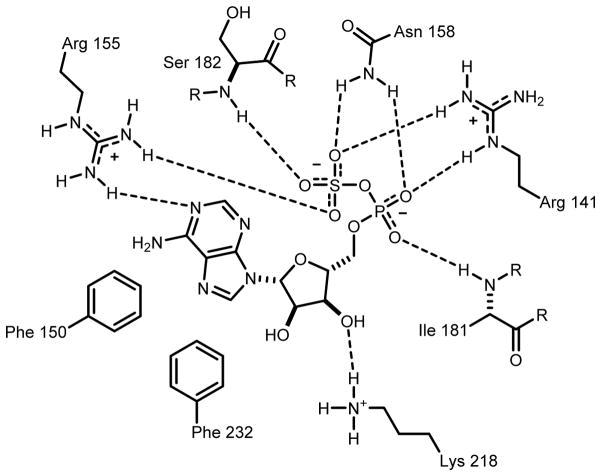
The closest functionally characterized homolog to Cj1415 is CysC from E. coli with a 26% identity. Two other monofunctional adenylyl sulfate kinases from Arabidopsis thaliana (UniProt Q43295) and Penicillium chrysogenum (UniProt Q12657) also share a 26% identity to Cj1415. A sequence alignment of these four proteins is presented in Figure 3. The three-dimensional structure of the A. thaliana enzyme has previously been solved, and the adenylyl sulfate binding residues have been identified (Figure 4). Comparing the residues in the active site of adenylyl sulfate kinase to Cj1415, there are three notable differences in residues that interact with the nucleoside base and the sulfate moiety of the bound adenylyl sulfate. In adenylyl sulfate kinase, there are two phenylalanine residues that interact with the adenine base, however, in Cj1415 these two residues are replaced by tyrosine and valine. These two residues may help to explain the specificity for cytosine over adenine (21). The sulfate of adenylyl sulfate in CysC is stabilized by an asparagine residue, which donates two hydrogen bonds to the substrate. In Cj1415 this residue is a methionine, a hydrogen bond acceptor. Methionine commonly forms hydrogen bonds with the backbone amide in a polypeptide chain (22). This change from a hydrogen bond donor to an acceptor in Cj1415 may help to explain why the amidate substrate is preferred over sulfate.
Figure 3.
Sequence alignment of CysC homologs. Sequence alignment of E. coli CysC (UniProt P0A6J1), Arabadopsis CysC (UniProt Q43295), Pennicillian CysC (UniProt Q12657) and Cj1415 (UniProt Q0P8J9). The annotated adenylyl sulfate binding residues are highlighted in red boxes (residues based on Arabidopsis crystal structure PDB id: 3UIE) (25).
Figure 4.
Active site of Arabidopsis adenylyl sulfate kinase (PDB id: 3UIE) (25). This image was adapted from the ligand interactions found within the Protein Data Bank for this structure. Potential hydrogen bonds are indicated by the dashed lines where the distances between the two heteroatoms are ≤3.1Å.
Previously, the E. coli CysC enzyme has been reported to require a phosphoserine enzyme intermediate (17). However, this residue has never been mutated and a crystal structure is not available for the E. coli enzyme. Mutation of the A. thaliana and P. chrysogenum CysC homologs show that when this serine is mutated there is no loss of catalytic activity (23,24). In the crystal structure of A. thaliana (Figure 4) this serine residue is in the active site, but is removed from the 3′-hydroxyl group of adenylyl sulfate (25). While this residue is not forming a phosphorylated intermediate in Cj1415 its involvement in substrate binding can be seen in the increased Km of CDP-NH2 (Table 1).
We have now demonstrated that Cj1415, Cj1416, Cj1417, and Cj1418 collectively catalyze the biosynthesis of 3′-phosphocytidine-5′-diphosphoamidate (9) using L-glutamine, CTP and 2 molecules of ATP. The β-phosphoramidate group of 9 must ultimately be methylated and transferred to a specific carbohydrate that is incorporated into the growing capsular polysaccharide chain. However, at this point it is unclear as to the timing of the methylation and phosphoryl transfer reactions in the biosynthesis of the capsular polysaccharide chain. From the appropriate gene cluster contained within C. jejuni 11168 it is highly likely that the enzymes Cj1419 and Cj1420 will be responsible for the transfer of the methyl group of SAM to the appropriate substrate and that Cj1421 and Cj1422 will catalyze the transfer of the phosphoramidate or the methylated phosphoramidate group to either an NDP-functionalized sugar or the growing capsular polysaccharide itself. Experiments are currently underway to unravel the substrate profiles for Cj1419, Cj1420, Cj1421, and Cj1422.
Acknowledgments
This work was supported by grants from the Robert A. Welch Foundation (A-840) and the National Institutes of Health (GM 122825). We thank Professor Hazel M. Holden of the University of Wisconsin for many fruitful discussions during the course of this investigation.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 2.Guerry P, Poly F, Riddle M, Maue AC, Chen YH, Monteiro MA. Campylobacter Polysaccharide Capsules: Virulence and Vaccines. Front Cell Infect Microbiol. 2012;2:1–7. doi: 10.3389/fcimb.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee MD, Newell DG. Campylobacter in Poultry: Filling an Ecological Niche. Avian Dis. 2006;50:1–9. doi: 10.1637/7474-111605R.1. [DOI] [PubMed] [Google Scholar]
- 4.Poropatich KO, Walker CLF, Black RE. Quantifying the Association between Campylobacter Infection and Guillain-Barré Syndrome: A Systematic Review. J Health Popul Nutr. 2010;28:545–552. doi: 10.3329/jhpn.v28i6.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 6.Bacon DJ, Szymanski CM, Burr DH, Silver RP, Alm RA, Guerry P. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81–176. Mol Microbiol. 2001;40:769–777. doi: 10.1046/j.1365-2958.2001.02431.x. [DOI] [PubMed] [Google Scholar]
- 7.Liang H, Zhang A, Gu Y, You Y, Zhang J, Zhang M. Genetic Characteristics and Multiple-PCR Development for Capsular Identification of Specific Serotypes of Campylobacter jejuni. PLoS One. 2016;11:e0165159. doi: 10.1371/journal.pone.0165159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poly F, Serichatalergs O, Schulman M, Ju J, Cates CN, Kanipes M, Mason C, Guerry P. Discrimination of Major Capsular Types of Campylobacter jejuni by Multiplex PCR. J Clin Microbiol. 2011;49:1750–1757. doi: 10.1128/JCM.02348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michael FS, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson JR, Monteiro MA. The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur J Biochem. 2002;269:5119–5136. doi: 10.1046/j.1432-1033.2002.03201.x. [DOI] [PubMed] [Google Scholar]
- 10.McNally DJ, Lamoureux MP, Karlyshev AV, Fiori LM, Li J, Thacker G, Coleman RA, Khieu NH, Wren BW, Brisson JR, Jarrell HC, Szymanski CM. Commonality and Biosynthesis of the O-Methyl Phosphoramidate Capsule Modification in Campylobacter jejuni. J Biol Chem. 2007;282:28566–28576. doi: 10.1074/jbc.M704413200. [DOI] [PubMed] [Google Scholar]
- 11.Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol. 2000;35:529–541. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- 12.Taylor ZW, Brown HA, Narindoshvili T, Wenzel CQ, Szymanski CM, Holden HM, Raushel FM. Discovery of a Glutamine Kinase Required for the Biosynthesis of the O-Methyl Phosphoramidate Modifications Found in the Capsular Polysaccharides of Campylobacter jejuni. J Am Chem Soc. 2017;139:9463–9466. doi: 10.1021/jacs.7b04824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor ZW, Brown HA, Holden HM, Raushel FM. Biosynthesis of Nucleoside Diphosphoramidates in Campylobacter jejuni. Biochemistry. 2017;56:6079–6082. doi: 10.1021/acs.biochem.7b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leyh TS, Taylor JC, Markham GD. The sulfate activation locus of Escherichia coli K12: cloning, genetic, and enzymatic characterization. J Biol Chem. 1988;263:2409–2416. [PubMed] [Google Scholar]
- 15.Wehrli W, Verheyden D, Moffatt J. Dismutation Reactions of Nucleoside Polyphosphates. II Specific Chemical Syntheses of α-, β-, and γ-P32-Nucleoside 5′-Triphosphates1. J Am Chem Soc. 1965;87:2265–2277. doi: 10.1021/ja01088a028. [DOI] [PubMed] [Google Scholar]
- 16.Technikova-Dobrova Z, Sardanelli AM, Papa S. Spectrophotometric determination of functional characteristics of protein kinases with coupled enzymatic assay. FEBS Lett. 1991;292:69–72. doi: 10.1016/0014-5793(91)80836-r. [DOI] [PubMed] [Google Scholar]
- 17.Satishchandran C, Hickman YN, Markham GD. Characterization of the phosphorylated enzyme intermediate formed in the adenosine 5′-phosphosulfate kinase reaction. Biochemistry. 1992;31:11684–11688. doi: 10.1021/bi00162a003. [DOI] [PubMed] [Google Scholar]
- 18.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using Sequence Similarity Networks for Visualization of Relationships Across Diverse Protein Superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copeland KL, Anderson JA, Farley AR, Cox JR, Tschumper GS. Probing Phenylalanine/Adenine π-Stacking Interactions in Protein Complexes with Explicitly Correlated and CCSD(T) Computations. The Journal of Physical Chemistry B. 2008;112:14291–14295. doi: 10.1021/jp805528v. [DOI] [PubMed] [Google Scholar]
- 22.Biswal HS, Gloaguen E, Loquais Y, Tardivel B, Mons M. Strength of NH···S Hydrogen Bonds in Methionine Residues Revealed by Gas-Phase IR/UV Spectroscopy. The Journal of Physical Chemistry Letters. 2012;3:755–759. doi: 10.1021/jz300207k. [DOI] [PubMed] [Google Scholar]
- 23.Lillig CH, Schiffmann S, Berndt C, Berken A, Tischka R, Schwenn JD. Molecular and Catalytic Properties of Arabidopsis thaliana Adenylyl Sulfate (APS)-Kinase. Arch Biochem Biophys. 2001;392:303–310. doi: 10.1006/abbi.2001.2453. [DOI] [PubMed] [Google Scholar]
- 24.MacRae IJ, Segel IH, Fisher AJ. Crystal Structure of Adenosine 5′-Phosphosulfate Kinase from Penicillium chrysogenum. Biochemistry. 2000;39:1613–1621. doi: 10.1021/bi9924157. [DOI] [PubMed] [Google Scholar]
- 25.Ravilious GE, Nguyen A, Francois JA, Jez JM. Structural Basis and Evolution of Redox Regulation in Plant Asenosine-5′-phosphosulfate Kinase. PNAS. 2012;109:309–314. doi: 10.1073/pnas.1115772108. [DOI] [PMC free article] [PubMed] [Google Scholar]