Abstract
Campylobacter jejuni is a pathogenic Gram-negative bacterium and a leading cause of food-borne gastroenteritis. C. jejuni produces a capsular polysaccharide (CPS) that contains a unique O-methyl phosphoramidate modification (MeOPN). Recently, the first step in the biosynthetic pathway for the assembly of the MeOPN modification to the CPS was elucidated. It was shown that the enzyme Cj1418 catalyzes the phosphorylation of the amide nitrogen of L-glutamine to form L-glutamine phosphate. In this investigation the metabolic fate of L-glutamine phosphate was determined. The enzyme Cj1416 catalyzes the displacement of pyrophosphate from MgCTP by L-glutamine phosphate to form CDP-L-glutamine. The enzyme Cj1417 subsequently catalyzes the hydrolysis of CDP-L-glutamine to generate cytidine diphosphoramidate and L-glutamate. The structures of the two novel intermediates, CDP-L-glutamine and cytidine diphosphoramidate, were confirmed by 31P NMR spectroscopy and mass spectrometry. It is proposed that the enzyme Cj1416 be named CTP: phosphoglutamine cytidylyltransferase and that the enzyme Cj1417 be named γ-glutamyl-CDP-amidate hydrolase.
TOC image
The pathogenic Gram-negative bacterium Campylobacter jejuni is a leading cause of food-borne gastroenteritis (1). While pathogenic to humans, C. jejuni is a commensal organism in chickens, and as a result, contaminated poultry serves as a common route of human infection. While most C. jejuni infections cause a case of gastroenteritis, approximately 1 in 1000 infections results in the autoimmune disease Guillain-Barré Syndrome (2, 3). Like many other organisms, C. jejuni uses a capsular polysaccharide (CPS) to improve fitness. The capsular polysaccharide of C. jejuni protects the organism from bacteriophages and shields it from the host immune response (4, 5). Over forty different strains of C. jejuni have been identified to date, and each strain is believed to produce a unique CPS variant (6, 7). In the C. jejuni strain NCTC 11168 a cluster of approximately 35 genes is responsible for the synthesis and export of the CPS (8). Moreover, C. jejuni has evolved the ability to synthesize a unique O-methyl phosphoramidate (MeOPN) modification found on the CPS that improves the pathogenicity of the bacterium and promotes evasion of the host immune response (9). The structures of the MeOPN modification to the CPS in the C. jejuni strain NCTC 11168 and the hypermotile strain 11168H are illustrated in Scheme 1.
Scheme 1.
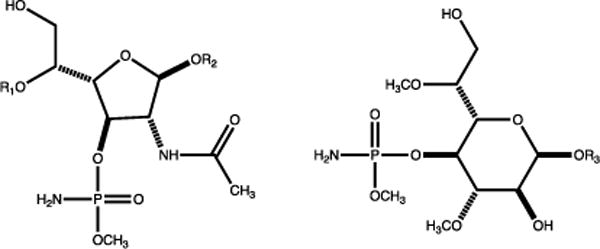
Structures of the O-methyl phosphoramidate modifications to the CPS in C. jejuni.
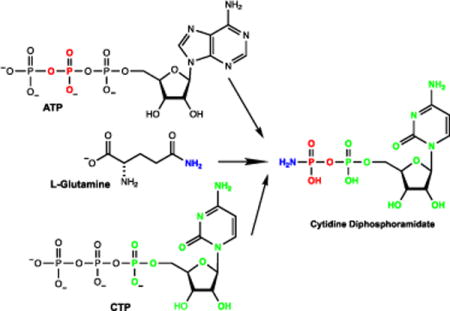
We have recently shown that the enzyme denoted with the locus tag Cj1418 from C. jejuni strain 11168H catalyzes the first committed step in the biosynthesis of the MeOPN (10). This enzyme, L-glutamine kinase, catalyzes the unprecedented ATP-dependent phosphorylation of the amide nitrogen of L-glutamine (1) to form L-glutamine phosphate (2) as shown in Scheme 2A. However, the subsequent metabolic fate of this novel enzyme intermediate has not been addressed. The primary focus of this investigation is to identify those enzymes that can harness the phosphoramidate moiety contained within L-glutamine phosphate for the ultimate construction of the O-methyl phosphoramidate modification of the CPS.
Scheme 2.
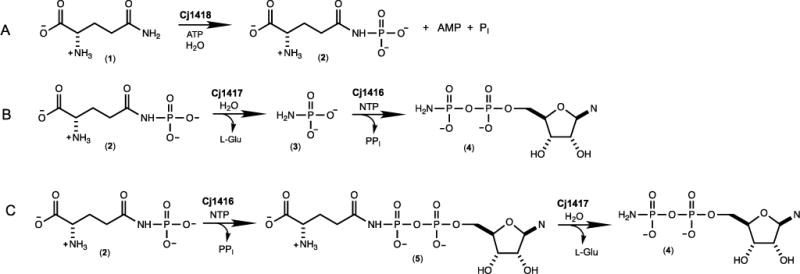
Reaction catalyzed by Cj1418 and the predicted functions of Cj1417 and Cj1416.
The two most likely enzymes of unknown function from C. jejuni to utilize L-glutamine phosphate as a substrate during the biosynthesis of the O-methyl phosphoramidate modification of the CPS are Cj1417 and Cj1416. Cj1417 is functionally annotated as a Type I amidotransferase from cog2071. The Type I amidotransferase family of enzymes typically catalyze the hydrolysis of L-glutamine or amide-substituted derivatives of this amino acid (11, 12). Currently, the closest functionally characterized homologue to Cj1417 is the enzyme PuuD (23% sequence identity) from Escherichia coli; an enzyme that catalyzes the hydrolysis of 4-(γ-L-glutamylamino)butanoate to 4-aminobutanoate and L-glutamate (12). Cj1416 is currently annotated as a nucleotidyl transferase from cog1213. The closest functionally characterized homologue of Cj1416 is CTP: phosphocholine cytidylyltransferase from Streptococcus pneumonia with a sequence identity of 28% (13). This enzyme catalyzes the formation of CDP-choline and pyrophosphate from CTP and choline phosphate. We therefore predict that the combination of Cj1417 and Cj1416 will catalyze the synthesis of a nucleoside diphosphoramidate (4) using L-glutamine phosphate (2) and a nucleoside triphosphate as substrates.
The biosynthesis of a nucleoside diphosphoramidate (4) by the consecutive reactions catalyzed by Cj1417 and Cj1416 can be envisioned to occur via one of two possible reaction schemes. In the first scenario, Cj1417 catalyzes the hydrolysis of L-glutamine phosphate (2) to L-glutamate and phosphoramidate (3). This reaction is followed by the displacement of pyrophosphate from a nucleoside triphosphate by phosphoramidate (3) to generate the nucleoside diphosphoramidate (4) in a reaction catalyzed by Cj1416 as illustrated in Scheme 2B. Alternatively, Cj1416 catalyzes the displacement of pyrophosphate from a nucleoside triphosphate by L-glutamine phosphate to form NDP-L-glutamine (5). This reaction is subsequently followed by the hydrolysis of this intermediate by Cj1417 to form L-glutamate and the nucleoside diphosphoramidate (4) as presented in Scheme 2C.
To test our initial prediction that the combination of Cj1416 and Cj1417 catalyze the formation of a nucleoside diphosphoramidate, these two enzymes were incubated together in the presence of MgCTP and an excess of L-glutamine phosphate (2) at pH 8.0. After 45 minutes, all of the CTP (retention time of 8.7 minutes, Figure 1A) was converted to a new product with a retention time of 5.9 minutes (Figure 1B). The retention time of the new reaction product formed in the presence of Cj1417 and Cj1416 is identical to that of authentic cytidine diphosphoramidate (Figure 1E) (14).
Figure 1.
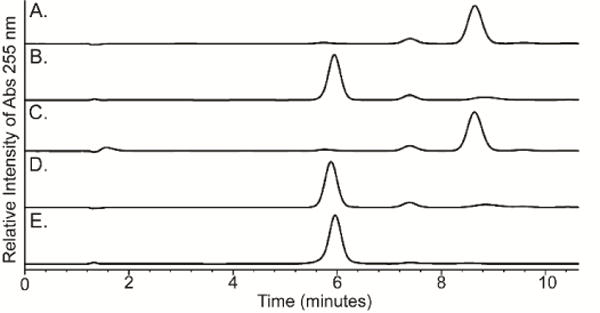
Anion exchange chromatograms of nucleotide standards and enzyme-catalyzed reaction products formed in 100 mM HEPES, pH 8.0 at 25 °C with an incubation time of 45 minutes. The elution profiles were monitored at 255 nm. The nucleotides were separated using a 0-17% gradient of 10 mM triethanolamine (pH 8) and 2 M KCl over 17 column volumes on a 1 mL resource Q column. (A) Control sample of 1.0 mM CTP and 2.0 mM MgCl2 in the absence of any added enzyme. (B) 1.0 mM CTP, 2.0 mM MgCl2, 5.0 mM L-glutamine phosphate, Cj1416 (5 μM), and Cj1417 (5 μM). (C) 1.0 mM CTP, 2.0 mM MgCl2, 5.0 mM phosphoramidate (3), 5 μM Cj1416 and 5 units/mL pyrophosphatase. (D) 1.0 mM CTP, 2.0 mM MgCl2, 5.0 mM L-glutamine phosphate, 5 μM Cj1416 and 5 units/mL pyrophosphatase. (E) Control sample of chemically synthesized CDP phosphoramidate.
The identity of the new reaction product as cytidine diphosphoramidate (4, Scheme 2C) was confirmed by 31P NMR spectroscopy. A reaction mixture containing CTP, MgCl2 and an excess of L-glutamine phosphate (2) was incubated at pH 8.0 for 90 minutes in the presence of Cj1416 and Cj1417 until the reaction was quenched by the addition of 10 mM EDTA. The 31P NMR spectrum of the control reaction (Figure 2A), obtained in the absence of Cj1416 and Cj1417, showed the expected resonances for CTP (−20.99 ppm (β-P); −10.33 ppm (α-P); and –5.51 ppm (γ-P)) and L-glutamine phosphate (−3.57 and –3.83 ppm). In the presence of Cj1416 and Cj1417, the resonances for CTP and L-glutamine phosphate (2) essentially disappear and are replaced by new resonances for pyrophosphate (−6.63 ppm) and a pair of doublets at –0.42 (β-P) and –10.23 ppm (α-P) for cytidine diphosphoramidate (Figure 2B). The 31P NMR spectrum for authentic cytidine diphosphoramidate is presented in Figure 2C. The formation of cytidine diphosphoramidate was further supported by acquisition of the negative ion ESI mass spectrum of the unfractionated reaction mixture upon incubation of MgCTP, L-glutamine phosphate, Cj1417, Cj1416 at pH 8.0 in ammonium bicarbonate buffer. A peak with an m/z of 401.03 was observed that is consistent with that expected for cytidine diphosphoramidate (Supplementary Figure S1). These experiments demonstrate that Cj1417 and Cj1416 use MgCTP and L-glutamine phosphate (2) to catalyze the formation of CDP phosphoramidate, pyrophosphate, and L-glutamate.
Figure 2.
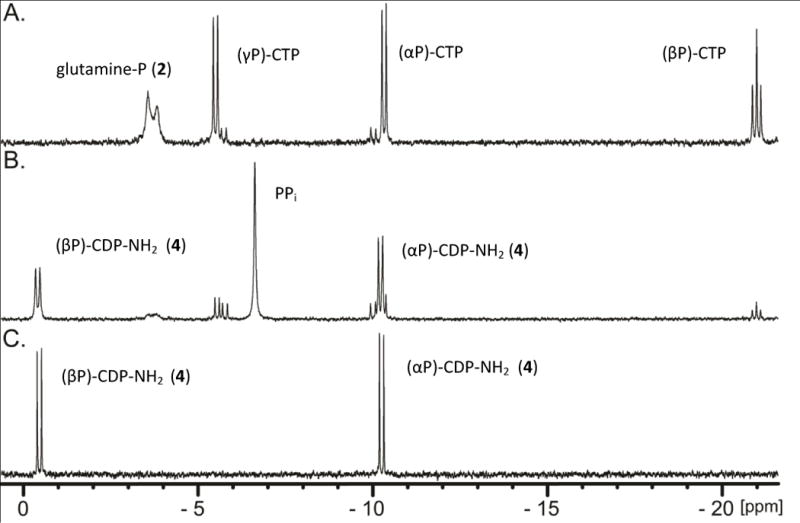
31P NMR spectra of nucleotide standards and enzyme-catalyzed reaction products formed in 100 mM HEPES, pH 8.0 at 30 °C with an incubation time of 90 minutes before being quenched with 10 mM EDTA. (A) Control sample containing 5.0 mM CTP, 5.0 mM MgCl2, and 6.0 mM L-glutamine phosphate. (B) 5.0 mM CTP, 5.0 mM MgCl2, 6.0 mM L-glutamine phosphate, 20 μM Cj1416, and 20 μM Cj1417. (C) Control sample of 5.0 mM CDP phosphoramidate.
In Schemes 2B and 2C we have proposed that either phosphoramidate (3) or L-glutamine phosphate (2) is used to displace pyrophosphate from a nucleoside triphosphate to form either a nucleoside diphosphoramidate (4) or NDP-L-glutamine (5) as an intermediate. The reactivity of Cj1416 with each of these potential substrates was tested with MgCTP as the acceptor nucleotide, and the reaction was monitored by ion exchange chromatography at 255 nm. In separate experiments, either phosphoramidate (3) or L-glutamine phosphate (2) was incubated with CTP, MgCl2 and Cj1416 at pH 8.0 for 45 minutes. Utilizing the chemically synthesized phosphoramidate (3) as a potential substrate, there was no change in the HPLC chromatogram when compared to CTP alone (Figure 1C) (15). However, when Cj1416 was incubated with L-glutamine phosphate (2) and MgCTP, all of the CTP was converted to a new product that corresponds to a molecule with a net charge of approximately −2 (Figure 1D)(10). This result demonstrates that Cj1416 is fully capable of using L-glutamine phosphate (2) to displace pyrophosphate from CTP.
To provide further spectroscopic support for the Cj1416-catalyzed formation of CDP-L-glutamine, the reaction products were analyzed by 31P NMR spectroscopy. In this experiment Cj1416 was incubated with CTP, Mg2+, L-glutamine phosphate (2) and inorganic pyrophosphatase until the reaction was quenched with EDTA. After an incubation period of 90 minutes, essentially all of the CTP and L-glutamine phosphate (2) were converted to products (Figure 3A). A new resonance is observed at 3.02 ppm for phosphate (from the hydrolysis of pyrophosphate) and two new doublets are observed at −10.98 ppm (α-P) and −16.13 ppm (β-P) for CDP-L-glutamine. The formation of CDP-L-glutamine was further supported by acquisition of the negative ion ESI mass spectrum of the unfractionated reaction mixture upon incubation of MgCTP, L-glutamine phosphate and Cj1416 at pH 8.0 in ammonium bicarbonate buffer. A peak with an m/z of 530.07 was observed that is consistent with that expected of CDP-L-glutamine (Supplementary Figure S2).
Figure 3.
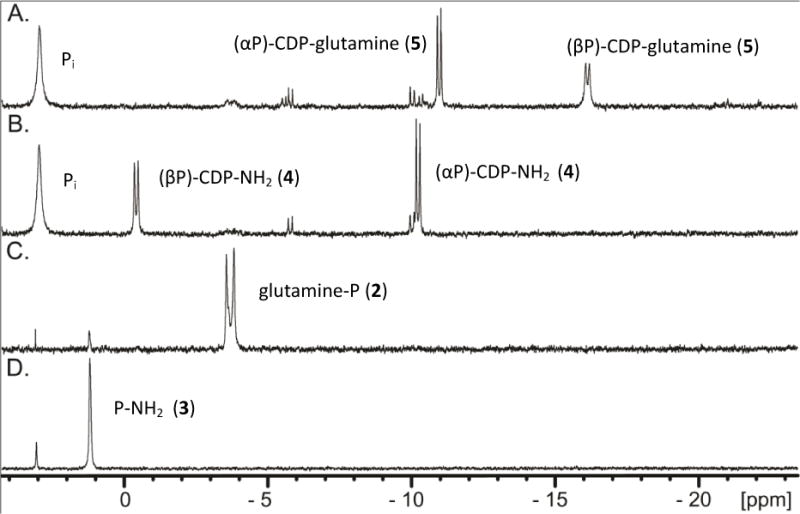
31P NMR spectra of nucleotide standards and enzyme-catalyzed reaction products formed in 100 mM HEPES, pH 8.0 at 25 °C with an incubation time of 90 minutes before being quenched with 10 mM EDTA. (A) 5.0 mM CTP, 5.0 mM MgCl2, 5 unit/mL of pyrophosphatase, and 20 μM Cj1416. (B). 20 μM Cj1417 was added to the reaction mixture shown in Figure 3A and allowed to reaction for an additional 90 minutes. (C). 20 μM Cj1417 and 5.0 mM glutamine phosphate. (D). Control sample of 5.0 mM phosphoramidate.
When Cj1417 is subsequently added to the reaction mixture containing CDP-L-glutamine, the 31P NMR resonances for CDP-L-glutamine disappear and are replaced by two new pairs of doublets at −0.43 ppm (β-P) and −10.24 ppm (α-P) that can be assigned to cytidine diphosphoramidate (Figure 3B and Figure 2C). These experiments demonstrate that Cj1416 catalyzes the formation of CDP-L-glutamine from CTP and L-glutamine phosphate (2) and that Cj1417 catalyzes the hydrolysis of CDP-L-glutamine to L-glutamate and cytidine diphosphoramidate.
The remaining issue to address for the functional characterization of Cj1417 is whether or not this enzyme is capable of catalyzing the hydrolysis of L-glutamine phosphate to L-glutamate and phosphoramidate (2). Cj1417 was therefore incubated with L-glutamine phosphate (2) at pH 8.0 for 90 minutes. The 31P NMR spectrum (Figure 3C) of the reaction mixture demonstrated that ~93% of the original L-glutamine phosphate (2) remained intact. Two other resonances are observed that are consistent with the presence of a small amount of phosphate (3.08 ppm, 1.2%) and phosphoramidate (1.21 ppm, 5.5%). The 31P NMR spectrum for chemically synthesized phosphoramidate (2) is shown in Figure 3D where a resonance is observed at 1.19 ppm. Based on these results, it is clear that the preferred pathway for the synthesis of cytidine diphosphoramidate (4) is for Cj1416 to catalyze the displacement of pyrophosphate from CTP to form CDP-L-glutamine (5) and for Cj1417 to catalyze the hydrolysis of this intermediate to generate cytidine diphosphoramidate (4) as shown in Scheme 2C.
The kinetic constants for the catalytic activity of Cj1417 and Cj1416 were determined. At a fixed concentration of either 5.0 mM MgCTP or 2.0 mM L-glutamine phosphate (2), the rates of the reaction catalyzed by Cj1416 were determined at 25 °C and pH 8.0 by monitoring the formation of products via anion exchange chromatography at 255 nm. The observed kinetic constants using L-glutamine phosphate (2) as the variable substrate are as follows: Km = 120 ± 30 μM, kcat = 57 ± 6 min−1 and kcat/Km = 4.8 (± 1.3) × 105 M−1 min−1. The observed kinetic constants using MgCTP as the variable substrate are as follows: Km = 170 ± 35 μM, kcat = 57 ± 6 min−1 and kcat/Km = 3.4 (± 0.8) × 105 M−1 min−1.
The kinetic constants for the hydrolysis of CDP-L-glutamine (5) catalyzed by Cj1417 were determined at 25 °C and pH 8.0 using a glutamate dehydrogenase coupled assay that monitors the formation of NADH at 340 nm. The kinetic constants were determined to be Km = 28 ± 3 μM, kcat = 34 ± 1.2 min−1 and a kcat/Km = 1.2 (± 0.2) × 106 M−1 min−1. In an effort to determine the upper limit of the rate constant for the hydrolysis of L-glutamine phosphate (2) by Cj1417, 10 mM L-glutamine phosphate (2) was incubated with 20 μM Cj1417, and the reaction monitored by 31P NMR for 13 hours. From this experiment, an upper limit of 1.6 hr−1 was obtained for the hydrolysis of L-glutamine phosphate by Cj1417.
Previously, we have demonstrated that the first step in the biosynthesis of the MeOPN modification to the CPS of C. jejuni is catalyzed by Cj1418, where ATP is utilized to phosphorylate the amide nitrogen of L-glutamine (10). Here we have shown that Cj1416 catalyzes the displacement of pyrophosphate from MgCTP by L-glutamine phosphate (2) yielding CDP-L-glutamine (5). We have also established that the catalytic function of Cj1417 is to hydrolyze CDP-L-glutamine to L-glutamate and cytidine diphosphoramidate (4). This investigation has thus unveiled the identities of two new nucleoside diphosphoramidate derivatives that are involved in the biosynthesis of MeOPN. It is likely that cytidine diphosphoramidate (4) will be subsequently phosphorylated at the hydroxyl group attached to the C3 carbon of the ribose ring prior to transfer of the phosphoramidate group to various carbohydrates of the CPS. Based upon sequence similarity network analysis of the enzymes of unknown function contained within C. jejuni, we predict that Cj1415 will catalyze this reaction. Cj1415 is a member of cog0529 and the closest functionally characterized enzyme is CysC, an adenylyl-sulfate kinase, from E. coli (26% identity); an enzyme that catalyzes the ATP-dependent phosphorylation of the 3′ hydroxyl of adenylyl sulfate (16). The unique biosynthetic pathway for the assembly of the phosphoramidate functionality found in the CPS of C. jejuni offers many opportunities for the development of potent inhibitors that may ultimately be useful in the therapeutic control of this pathogenic organism.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the Robert A. Welch Foundation (A-840) to F.M.R. and the NIH (GM 115921) to H.M.H. We thank Professor C. Szymanski and Mr. C. Wenzel for the plasmids that were used to express Cj1417 and Cj1416.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:
ORCID
Frank M. Raushel: 0000-0002-5918-3089
Notes
The authors declare no competing financial interest.
References
- 1.Young KT, Davis LM, Dirita VJ. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs BC, Rothbarth PH, van der Meché FGA, Herbrink P, Schmitz PIM, de Klerk MA, van Doorn PA. Neurology. 1998;51:1110–1115. doi: 10.1212/wnl.51.4.1110. [DOI] [PubMed] [Google Scholar]
- 3.Acheson D, Allos BM. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 4.Guerry P, Poly F, Riddle M, Maue A, Chen YH, Monteiro M. Front Cell Infect Microbiol. 2012;2:1–7. doi: 10.3389/fcimb.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sørensen MCH, van Alphen LB, Harboe A, Li J, Christensen BB, Szymanski CM, Brøndsted L. J Bacteriol. 2011;193:6742–6749. doi: 10.1128/JB.05276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlyshev AV, Champion OL, Churcher C, Brisson JR, Jarrell HC, Gilbert M, Brochu D, St Michael F, Li J, Wakarchuk WW, Goodhead I, Sanders M, Stevens K, White B, Parkhill J, Wren BW, Szymanski CM. Mol Microbiol. 2005;55:90–103. doi: 10.1111/j.1365-2958.2004.04374.x. [DOI] [PubMed] [Google Scholar]
- 7.Michael FS, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson JR, Monteiro MA. Eur J Biochem. 2002;269:5119–5136. doi: 10.1046/j.1432-1033.2002.03201.x. [DOI] [PubMed] [Google Scholar]
- 8.Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW. Mol Microbiol. 2000;35:529–541. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- 9.van Alphen LB, Wenzel CQ, Richards MR, Fodor C, Ashmus RA, Stahl M, Karlyshev AV, Wren BW, Stintzi A, Miller WG, Lowary TL, Szymanski CM. PLoS One. 2014;9:e87051. doi: 10.1371/journal.pone.0087051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor ZW, Brown HA, Narindoshvili T, Wenzel CQ, Szymanski CM, Holden HM, Raushel FM. J Am Chem Soc. 2017;139:9463–9466. doi: 10.1021/jacs.7b04824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massière F, Badet-Denisot MA. Cell Mol Life Sci. 1998;54:205–222. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, Suzuki H. J Biol Chem. 2005;280:4602–4608. doi: 10.1074/jbc.M411114200. [DOI] [PubMed] [Google Scholar]
- 13.Kwak BY, Zhang YM, Yun M, Heath RJ, Rock CO, Jackowski S, Park HW. J Biol Chem. 2002;277:4343–4350. doi: 10.1074/jbc.M109163200. [DOI] [PubMed] [Google Scholar]
- 14.Wehrli W, Verheyden D, Moffatt J. J Am Chem Soc. 1965;87:2265–2277. doi: 10.1021/ja01088a028. [DOI] [PubMed] [Google Scholar]
- 15.Makoto W, Shoji S, Khosaku W. Bull Chem Soc Jpn. 1990;63:1243–1245. [Google Scholar]
- 16.Leyh TS, Taylor JC, Markham GD. J Biol Chem. 1988;263:2409–2416. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


