Abstract
The initiation and development or inflammatory bowel disease (IBD) and associated colorectal cancers, have been linked to inflammation. MicroRNAs are non-coding regulators of gene expression that have gained great attention due to their capability to regulate the expression of a number of target transcripts. It is now generally admitted that microRNAs are instrumental in gut pathologies, in particular through their targeting of transcripts encoding proteins of the intestinal barrier (IB) and their regulators. Intense research is conducted to identify microRNAs susceptible to be used as biomarkers and to design new therapeutic approaches based upon using synthetic microRNA mimics and inhibitors as well as finding new drugs capable to restore or modify microRNA expression in the context of gut pathologies.
Introduction
Food digestion and nutrient absorption rest on the activity of intestinal epithelial cells (IECs) and of a plethora of microorganisms, collectively referred to as the microbiota. The greatest proportion of microbiota corresponds to commensal bacteria that both participate to the digestion of nutrients and help to keep the gut healthy by competing with pathogens for the same ecological niche. Interactions between the microbiota, IECs and immune cells modulate the renewal of the epithelium and the activation status of immune cells.
Immune cells must tolerate commensal bacteria and antigens produced from food digestion while retaining their full activation capacity to fight invading pathogens. The health of the gut thus rests on a delicate equilibrium between IECs loss and proliferation, on one hand, and immune cells activation or tolerance, on the second hand. When this equilibrium is broken, inflammatory conditions can occur and induce gut pathologies such as Inflammatory bowel disease (IBD). IBD is a group of chronic idiopathic, relapsing and remitting immune disorders of the gastrointestinal tract in genetically susceptible individuals exposed to environmental risk factors [1]. IBD present with two main forms: Crohn’s disease (CD), that may affect any part of the gastrointestinal tract, and ulcerative colitis (UC), that is confined to the colon [2].
The critical role played by the intestinal barrier (IB) on gut health and function came to light recently, although whether intestinal barrier defect is a cause or a consequence of IBD remains debated [3–5]. The semi-permeable IB established by IECs must allow the captation of nutrients while avoiding the passage of pathogens. This double function depends on the production of a luminal mucosal barrier, that restricts the access of bacteria to intestinal epithelium, and the presence of different types of IECs that concur to IB homeostasis [6,7]. Organized in a single layer, IECs are linked by three types of junctions, namely, from apical to basal, tight junctions, adherens junctions and desmosomes. Tight junctions are implicated in blocking the transit of bacteria and food while facilitating paracellular flux through the pore pathway and the leak pathway [8]. Chronic, enhanced inflammation can damage IB, allowing unwanted bacteria and molecules to cross IB and further exacerbate the immune response, depending on the quality of mucosal layer and tight junctions, the composition of the microbiota, and ultimately the genotype of each individual.
MicroRNAs are small non-coding RNAs that regulate the translation and/or stability of target transcripts. Micro-RNAs control the expression of tens to hundreds of transcripts, either directly through binding or indirectly by targeting transcripts encoding transcription factors, epigenetic regulators or effectors of signal transduction pathways. As target transcripts usually contain one short sequence partly complementary to targeting microRNAs, one can predict, with a certain risk of error, the targeting of a given transcript by a given microRNA. MicroRNAs are major regulators of cell function and homeostasis, and their aberrant expression has been found in virtually all diseases, including IBD, CD, UC and cancers. Therefore, molecules that would allow targeting of specific micro-RNAs in a specific context should improve the efficiency of existing treatments. In this review, we will emphasize results of the last 2–3 years that implicate microRNAs in IBD and inflammation-related gut cancers, and summarize the therapeutic potential of using specific micro-RNAs as biomarkers and/or therapeutic agents in these pathologies.
MicroRNAs in IBD
Frozen nondysplastic colonic mucosa collected from CD and UC patients showed differential expression of micro-RNAs, implicated in the regulation of genes encoding proteins of epithelial adhesion junctions, integrin, glycolysis and cell cycle [9••]. Altered microRNA expression profiles were found between inflamed and non-inflamed ascending colon mucosae of CD patients [10••], and microRNAs of the miR-200 family showed enhanced expression in UC dysplastic lesions [11,12]. Patients with pediatric CD showed enhanced duodenal expression of miR-146, miR-155 and miR-122 [13,14]. MicroRNAs regulated cytokine production in regulatory T cells of UC patients [15], miR-155 targeted Jarid2 transcripts in Th17 cells in an experimental dextran sulfate sodium (DSS)-induced colitis in mouse [16], and increased miR-511-3p expression in mouse macrophages has been linked to intestinal inflammation [17]. By contrast, miR-31 expression increased while miR-21, miR-155 and miR-146a levels decreased in colonic CD3+ T cells in UC remission [18]. Table 1 presents microRNAs with validated target transcripts in gut mucosa or cells of the immune system in the context of IBD or in different mouse models [16,19,20•,21••,22–32].
Table 1.
Validated target transcripts of microRNAs implicated in IBD.
| MicroRNA(s) | Target transcript(s) | Proteins | Pathology | References |
|---|---|---|---|---|
| miR-132, miR-223 | FOXO3 | Forkhead box O3 | IBD | [19] |
| miR-223 | NLRP3 | NLR family pyrin domain containing 3 | IBD | [20] |
| miR-320 | NOD2 | Nucleotide binding oligomerization domain containing 2 | IBD | [21] |
| miR-665 | XBP1/ORMDL3 | X-box binding protein 1/ORMDL sphingolipid biosynthesis regulator 3 | IBD | [22] |
| miR-31 | IL-25 | Interleukin 25 | CD | [23] |
| miR-124 | AHR | Aryl hydrocarbon receptor | CD | [24] |
| miR-29b | MCL1 | MCL1, BCL2 family apoptosis regulator | CD fibrosis | [25] |
| miR-16 | ADORA2A | Adenosine A2a receptor | UC | [26] |
| miR-206 | ADORA3 | Adenosine A3 receptor | UC | [27] |
| miR-429 | MARCKS | Myristoylated alanine rich protein kinase C substrate | UC | [28] |
| miR-155 | FOXO3 | Forkhead box O3 | UC | [29] |
| miR-155 | Ship-1/INPP5D | Inositol polyphosphate-5-phosphatase D | Spontaneous colitisa | [30] |
| miR-155 | Jarid2 | jumonji and AT-rich Interaction domain containing 2 | Induced colitisb | [16] |
| miR-193a-3p | PepT1 | Solute carrier family 15 (oligopeptide transporter), member 1 | Induced colitisb | [31] |
| miR-223 | CLDN8 | Claudin 8 | Induced colitisc | [32] |
Bacteria-induced spontaneous colitis in mouse.
Mouse model of dextran sulfate sodium (DSS)-induced colitis.
Mouse model of 2,4,6-trinitrobenzene sulphonic acid (TNBS)-induced colitis.
MicroRNAs in IB
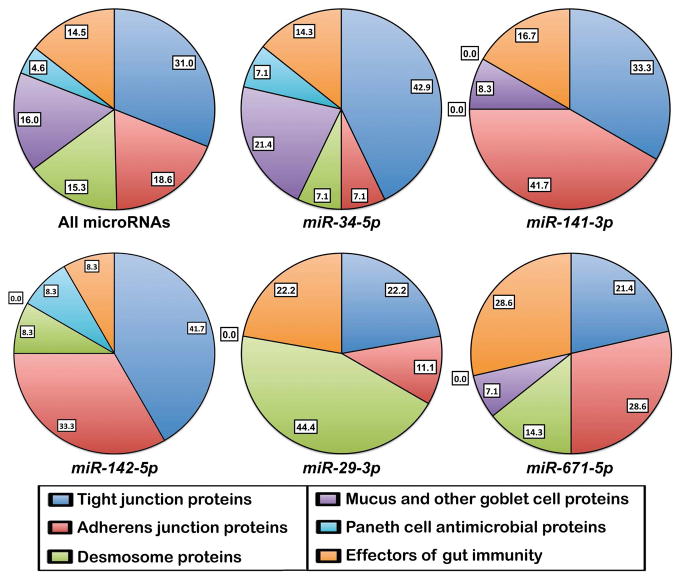
Patients with IBD and their healthy relatives have increased intestinal permeability, suggesting that barrier dysfunction might contribute to inflammation in IBD and other inflammatory-related gut pathologies [4,33,34]. IB external, mucosal layer is represented by glycosylated mucins and less abundant proteins secreted by goblet cells. Tight junctions that regulate paracellular transport require the presence of proteins such as Claudins, Occludin, MarvelD2/Tricellulin, MarvelD3, Zona occludens (Z0)-1-3 and the regulator of tight junction permeability myosin light chain kinase (MLCK) [5,35]. Paneth cells have a very active autophagy pathway and secret antibacterial peptides, including Lysozyme, RegIIIA and αDefensins such as HD5 and HD6. Abnormal expression of genes encoding barrier proteins has been linked to the development of IBD, CD, UC and other inflammatory-related diseases [5,7,34]. Several microRNAs, whose expression changed in IBD [16,19,20•,21••,22–32,36] have been shown to target transcripts encoding some of the above proteins in the context of gut inflammatory diseases. For example, by targeting Claudin 8, pro-inflammatory miR-223 was instrumental in allowing IL23 pathway to initiate trinitrobenzene sulphonic acid (TNBS)-induced colitis in mice [32]. Computer analysis using Targetscan software (http://www.targetscan.org) allowed us to predict microRNA–mRNA interactions possibly implicated in IB homeostasis and dysfunction (Table 2). Interestingly, transcripts encoding IB proteins Claudin 1, Occludin, MarvelD3, ZO-1, MYLK, Catenin delta1/p120, VEZT, and Desmocolin 3 are likely target of the highest number of microRNAs, suggesting that micro-RNA deregulation might be an initiating or aggravating factor for IBD. Taken all together, microRNAs listed in Table 2 do not show any preference between the six classes of transcripts (Figure 1). By contrast, other micro-RNAs should preferentially target specific components of tight junctions, adherens junctions, desmosomes, or factors implicated in the immune response. Thus, it is likely that deregulation of different microRNAs will have differential effects on paracellular permeability, cell–cell adhesion or the level of inflammation, and thus should be associated with different intestinal pathologies. IB is now considered a key therapeutic target [3–5], therefore any treatment susceptible to restore normal microRNA expression is likely to improve the treatment of gut diseases. Nevertheless, certain genes encoding important barrier proteins or mucins that are not listed in Table 2 have a very short 3′-untranslated region and likely are not directly regulated by microRNAs.
Table 2.
MicroRNAs whose expression changed in IBD [16,19,20•,21••,22–32] or a mouse model of colitis [36] potentiallya target transcripts encoding proteins of IB and gut immunity.
| Proteins | Target transcripts | MicroRNAs | Number of targeting microRNAs |
|---|---|---|---|
| Tight junction proteins | |||
| Claudin 1 | CLDN1 | 29a-5p; 29-3p; 34-5p; 142-5p; 144-5p; 155-5p; 194-5p; 196-5p; 449-5p; 455-5p; 483-3p; 584-5p; 1290; 4725-3p | 14 |
| Claudin 2 | CLDN2 | 29a-5p; 34-5p; 122-5p; 142-3p; 449-5p; 483-3p; 1291; 3622a-3p; 3622b-3p | 9 |
| Claudin 3 | CLDN3 | 455-3p | 1 |
| Claudin 4 | CLDN4 | 142-3p; 144-5p; 455-3p.1; 483-3p; 1291; 4793-3p | 6 |
| Claudin 5 | CLDN5 | 122-5p; 671-5p; 1291 | 3 |
| Claudin 7 | CLDN7 | 122-5p; 140-3p; 223-3p; 483-5p; 483-3p | 3 |
| Claudin 8 | CLDN8 | 21-5p; 34-5p; 155-5p; 142-5p; 143-3p; 144-5p; 181-5p; 223-3p; 449; 455-3p | 3 |
| Occludin | OCLN | 29-3p; 122-5p; 142-5p; 142-3p.1; 144-3p; 155-5p; 181-5p; 192-3p; 200b-3p; 200c-3p; 429; 455-5p; 455-3p.2; 584-5p; 3622b-5p | 15 |
| MARVEL domain containing 2/Tricellulin | MARVELD2 | 29a-5p; 141-3p; 145-5p; 194-5p; 200b-3p; 200c-3p; 223-3p; 429; 671-5p; 3622b-5p; 3622a-3p; 3622b-3p; 4793-3p | 13 |
| MARVEL domain containing 3 | MARVELD3 | 29a-5p; 34-5p; 93-5p; 106-5p; 122-5p; 141-3p; 142-3p.1; 145-5p; 155-5p; 200a-3p; 200b-3p; 200c-3p; 429; 449-5p; 455-3p.2; 483-5p; 1290; 3622a-3p; 3622b-3p | 19 |
| Tight junction protein 1/ZO-1 | TJP1 | 34-5p; 141-3p; 142-5p; 142-3p.1; 144-3p; 145-5p; 194-5p; 200a-3p; 200b-3p; 200c-3p; 429; 449-5p; 455-5p; 1290; 4725-3p | 15 |
| Myosin light chain kinase | MYLK/MLCK | 34-5p; 93-5p; 106-5p; 141-3p; 142-5p; 142-3p.1; 142-3p.2; 144-5p; 145-5p; 155-5p; 194-5p; 200b-3p; 200c-3p; 221-3p; 222-3p; 429; 449-5p; 584-5p; 671-5p; 1290; 3622b-5p; 4793-3p | 22 |
| Adherens junction proteins | |||
| Cadherin 1/E-cadherin | CDH1 | 93-5p; 98-5p; 106-5p; 140-5p; 142-5p; 146-5p; 181-5p; 221-3p; 222-3p; 483-3p; 671-5p; let-7-5p | 12 |
| Catenin alpha 1 | CTNNA1 | 140-3p.2; 141-3p; 142-3p.2; 144-3p; 181-5p; 200a-3p; 671-5p | 7 |
| Catenin beta 1 | CTNNB1 | 141-3p; 142-5p; 142-3p.2; 146-5p; 200a-3p; 483-3p.1; 483-3p.2 | 7 |
| Catenin delta 1/p120 | CTNND1 | 29-3p; 34-5p; 93-5p; 106-5p; 141-3p; 142-3p; 145-5p; 155-5p; 181-5p; 200b-3p; 200c-3p; 223-3p; 429; 449-5p; 455-5p; 671-5p; 1290 | 17 |
| Pleckstrin homology domain containing A7 | PLEKHA7 | 93-5p; 106-5p; 141-3p; 142-5p; 196-5p; 200a-3p; 455-3p; 671-5p | 8 |
| Vezatin, adherens junctions | VEZT | 29a-5p; 93-5p; 98-5p; 106-5p; 122-5p; 141-3p; 142-5p; 144-3p; 145-5p; 155-5p; 181-5p; 194-5p; 196-5p; 200a-3p; 200b-3p; | 28 |
| transmembrane protein | 200c-3p; 221-3p; 222-3p; 429; 455-3p.1; 483-5p; 483-3p.1; 584-5p; 1290; 3622b-5p; 3622a-3p; 3622b-3p; let-7-5p | ||
| Desmosome proteins | |||
| Desmoglein 2 | DSG2 | 29-3p; 93-5p; 106-5p; 122-5p; 155-5p; 223-3p; 483-3p.1; 671-5p; 4725-3p; 4793-3p | 10 |
| Desmoglein 3 | DSG3 | 29-3p; 93-5p; 98-5p; 106-5p; 144-3p; 221-3p; 222-3p; 223-3p; 483-5p; 584-5p; 671-5p; 3622a-3p; 3622b-3p; 4793-3p; let-7-5p | 15 |
| Desmoglein 4 | DSG4 | 29-3p; 93-5p; 106-5p; 142-5p; 142-3p.1; 142-3p.2; 144-3p; 192-5p; 455-5p; 4725-3p; 4793-3p | 11 |
| Desmocollin 2 | DSC2 | 29a-5p; 29-3p; 142-3p.2; 144-3p; 181-5p; 194-5p; 455-3p.2; 1290; 1291 | 9 |
| Desmocollin 3 | DSC3 | 29a-5p; 34-5p; 93-5p; 98-5p; 106-5p; 145-5p; 200b-3p; 200c-3p; 223-3p; 429; 449-5p; 455-5p; 455-3p.2; 584-5p; 4725-3p; 4793-3p; let-7-5p | 17 |
| Mucus and other goblet cell proteins | |||
| Mucin 1, cell surface associated | MUC1 | 122-5p; 145-5p; 455-3p.1; 1291; 3622a-3p; 3622b-3p | 6 |
| Anterior gradient 2, protein disulphide isomerase family member | AGR2 | 34b-5p; 194-5p; 196-5p; 431-5p; 449-5p; 483-3p.2 | 6 |
| Zymogen granule protein 16 | ZG16 | 34-5p; 93-5p; 98-5p; 106-5p; 122-5p; 142-3p.1; 145-5p; 155-5p; 181-5p; 194-5p; 196-5p; 431-5p; 449-5p; 455-3p.1; 455-3p.2; 483-3p.1; 584-5p; 1290; 4793-3p; let-7-5p | 20 |
| Cystic fibrosis transmembrane conductance regulator | CFTR | 34-5p; 142-3p.2; 144-3p; 145-5p; 200b-3p; 200c-3p; 223-3p; 429; 449-5p; 455-3p.2; 671-5p | 11 |
| Autophagy related 16 like 1 | ATG16l1 | 29a-5p; 93-5p; 98-5p; 106-5p; 141-3p; 142-3p.1; 142-3p.2; 181-5p; 200a-3p; 455-5p; 483-3p; 1291; let-7-5p | 13 |
| Autophagy related 3 | ATG3 | 29a-5p; 142-3p.2; 155-5p; 194-5p; 221-3p; 222-3p; 431-3p; 455-5p; 584-5p; 1290; 3622b-5p | 11 |
| Paneth cell antimicrobial proteins | |||
| Defensin alpha 5 | DEFA5/HD-5 | 155-5p | 1 |
| Defensin alpha 6 | DEFA6/HD-6 | 142-5p | 1 |
| Lysozyme | LYZ | 140-5p; 142-3p.1; 455-3p.2; 483-3p.1 | 4 |
| Angiogenin | ANG | 21-5p; 34-5p; 34b-5p; 93-5p; 106-5p; 122-5p; 142.5p; 144-5p; 146-5p; 155-5p; 192-5p; 455-3p.2; 483-3p.1; 584-5p | 14 |
| Effectors of gut immunity | |||
| Signal transducer and activator of transcription 3 | STAT3 | 29-3p; 34-5p; 93-5p; 98-5p; 106-5p; 122-5p; 181-5p; 192-5p; 196-5p; 221-3p; 222-3p; 449-5p; 671-5p; let-7-5p | 14 |
| Signal transducer and activator of transcription 5A | STAT5A | 29a-5p; 141-3p; 200a-3p; 223-3p; 483-3p; 3622b-5p | 6 |
| Signal transducer and activator of transcription 5A | STAT5B | 141-3p; 194-5p; 200a-3p; 221-3p; 222-3p; 455-5p; 483-5p; 671-5p; 1291; 3622b-5p | 10 |
| Nucleotide binding oligomerization domain containing 2 | NOD2 | 29a-5p; 34b-5p; 122-5p; 142-5p; 192-5p; 431-5p; 449-5p; 483-5p; 483-3p.1; 671-5p | 10 |
| C-type lectin domain containing 7A/DECTIN1 | CLEC7A | 29-3p; 144-3p; 192-5p; 200b-3p; 200c-3p; 429; 3622b-5p; 4793-3p | 8 |
| TNF receptor superfamily member 1B | TNFRSF1B/TNFR2 | 93-5p; 98-5p; 106-5p; 122-5p; 142-3p.1; 431-5p; 671-5p; 1291; let- 7-5p | 9 |
As determined using the Targetscan software.
Figure 1.
Percentages of transcripts belonging to the six classes of transcripts encoding proteins involved in IB and gut immunity (listed in Table 2) that are potentially targeted by microRNAs whose expression changes in IBD. ‘All microRNAs’ refers to microRNAs in Table 2. The name of five microRNAs representative of microRNAs that show class-preference are given under the pie charts.
MicroRNAs in inflammation-related cancers
Chronic UC has been associated with increased risk of colonic neoplasia, and microRNAs are critical players in molecular circuitries linking inflammation and cancer [37]. Changes in expression or glycosylation of proteins of tight junction, adherens junction or desmosomes have the potential to turn them from tumor suppressors to tumor promoters [38–40]. The expression of several microRNAs implicated in IBD is deregulated in colon cancer. For example, the levels of microRNAs of the miR-200 family increased in UC dysplastic lesions [11], and miR-200-3p showed higher expression in colorectal cancer complications [11]. In IBD, downregulation of miR-200 microRNAs correlated with increased Snail and Slug, a hallmark of epithelium-to-mesenchyme transition [41]. Archetypal oncogenic miR-21 was found upregulated in patients with colitis-associated colorectal cancer as well as in a mouse model of this pathology [42,43]. Inflammation can trigger a miR-34a-Numb feed-forward loop that enhances asymmetric stem cell division in intestine and colon cancer [44•]. Inhibiting miR-214, a microRNA highly expressed in colon samples of patients with UC or UC-associated colorectal cancer, reduced the number and size of tumors of mice given azoxymethane in a DSS-induced colitis model [45]. MiR-301a targets BTG anti-proliferation factor 1 (BTG1), thus favoring intestinal inflammation and colitis-associated cancer development [46••]. Neuropeptide Y from enteric neurons promotes inflammation-induced cancer, both by activating cell proliferation through the PI3-K/AKT pathway and by down-regulating miR-375, a microRNA pro-apoptotic in IECs [47]. By contrast, microRNAs such as miR-193a-3p and 4728-3p were down-regulated in patients with UC-associated colorectal cancer, and behaved as tumor suppressor in colon cancer cells, by respectively targeting interleukin 17 receptor D (ILR17D) and focal adhesion signaling [48,49]. Nevertheless, miR-31, not express in normal IECs but highly deregulated in mouse and human colorectal cancers, can function both as an oncogenic or a tumor-suppressor microRNA depending on the context, and its deletion has been shown to promote UC-associated cancer in mouse [50]. Interestingly, two different E-cadherin- p120 complexes have been identified at the junctions of polarized epithelial cells, the apical one containing the factor PLEKHA7 that is implicated in stabilization of adherens junctions. Remarkably, apical PLEKHA7 complex, through its association with the microprocessor components Drosha and DGCR8, was able to regulated the levels of a subset of microRNAs such as miR-24, miR-30a, miR-30b and let-7g. Owing to the presence of microprocessor, apical PLEKHA7 complex was able to control the processing of at least the precursor (pri-miR-30b) of miR-30b [51••], a microRNA, known to inhibit epithelium-to-mesenchyme transitions. It is now clear that microRNAs targeting transcripts encoding components of cell–cell junctions are instrumental in IB dysfunction associated with both IBD and IBD-associated tumors (Figure 2).
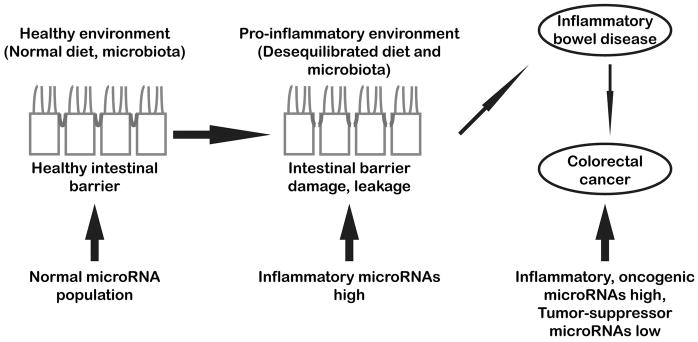
Figure 2.
Schematic diagram summarizing the main results and concepts discussed in this review. MicroRNAs are powerful regulators of cell homeostasis and function. Following IB damage, persistent inflammatory conditions can alter microRNA expression, initiating or accelerating the development of IBD. On the long run, chronic deregulation of microRNAs targeting transcripts encoding key components of IB as well as of oncogenic and tumor-suppressors microRNAs may lead to the development of colorectal cancer.
MicroRNAs as biomarkers and therapeutic targets
In the light of the above considerations, one can expect microRNAs to represent valuable biomarkers. Indeed, miR-26b, up-regulated in colon biopsies of patients with UC and UC-associated cancer, was conversely down-regulated in sporadic colon cancers [52]. The levels of miR-320a, a microRNA that inhibits Escherichia coli-induced damage to IB, were increased in blood samples of CD patients and UC mice [53•]. The expression of miR-221-5p, that reduces IL6-receptor expression and decreases inflammation, was higher in biopsies of UC but not of CD patients [54]. Polymorphisms in miR-122, miR-196a2 and miR-124a genes have been associated with different IBD clinical phenotypes [55]. MicroRNA mimics and antisense inhibitory RNAs (antagomiRs) should allow to compensate for endogenous microRNA deregulation, and several pre-clinical and clinical assays are already under way. For example, microvesicles-delivered miR-200b attenuated experimental colitis-associated intestinal fibrosis by inhibiting TGF-β1 mediated epithelium-to-mesenchyme transition in IECs and in a rat model of 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced intestinal fibrosis [56]. Nevertheless, caution should be exerted for the treatment of long-lasting pathologies. In particular, in long-lasting treatments, the risk of off-target effects should not be minimized, especially given the potential of microRNAs to be secondly transported toward non-target cells. Also caution should be exerted considering that microRNAs have repeatedly been shown to reduce the levels of their target transcripts in a dose-dependent manner. For example, only low levels of pro-inflammatory miR-155 translated into targeting Quaking transcripts in lipopolysaccharide (LPS)-challenged macrophages [57]. Thus, except when mutations incapacitate endogenous microRNA(s) of interest, it remains of great importance to develop new drugs capable to modulate ‘on demand’ the expression of particular microRNA(s) while causing minimal harmful consequences.
Conclusion
During the last 15 years, many publications have unraveled the enormous potential of microRNAs to provide new, powerful biomarkers that can be used to define ever more subtle sub-categories within patients with major pathologies such as cancer or autoimmune, metabolic, neuro-degenerative and behavioral diseases. This versatility brings hopes to develop new drugs capable, through the manipulation of endogenous microRNA populations, to deliver maximal beneficial effects with minimal iatrogenic consequences, or, in specific contexts, to directly deliver microRNA mimics or antagomiRs to pathological cells, ultimately leading to truly ‘personalized’ medicine. On the other hand, a number of phytochemicals that can be provided by dietary intake or food supplementation have been identified as being beneficial for the body. Although few epidemiological studies and intervention trials are available, experiments conducted on cell cultures or in animals have established that some of these phytochemicals deliver at least some of their antioxidant, anti-inflammatory and/or anti-proliferative effects through the modification of the composition of endogenous microRNA populations. For example, catalpol, an iridoid glucoside found in plants of genus Rehmannia, has been shown to reduce endoplasmic reticulum stress in UC through the down-regulation of miR-132 [58], while salvianolic acid B, extracted from Salvia miltiorrhiza, restored impaired IB in a rat model of TNBS-induced colitis likely through inducing downregulation of MLCK by miR-1 [59]. Also, flavonoid and non-flavonoid poly-phenols found in different fruits and berries present antioxidant, anti-inflammatory and/or anti-proliferative effects on cell cultures or in murine models, including cases of gut inflammation or colon cancer. Many of these polyphenols appear to modify the expression of endogenous microRNAs [60]. Considering that polyphenols do not behave as classical, purified drugs and seem to act in many different ways through many different mechanisms, they are often considered as non-effective and not relevant. Nevertheless, polyphenols such as resveratrol that do not display harmful properties might possibly be used as diet supplement to regulate the function of micro-RNAs, especially those present in EICs. Altogether, maintaining or restoring normal microRNA expression in IECs should prove beneficial in health as well as in disease.
Acknowledgments
This paper was founded by grants: AM Cancer Soc IRG-67-003-50 and NIH R03 NS102861 to ET, and NIH R35 CAI97706 to CMC.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 2.Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res. 2017;10:63–73. doi: 10.2147/JIR.S116088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol. 2017;11:821–834. doi: 10.1080/17474124.2017.1343143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi W, Yeruva S, Turner JR. Contributions of intestinal epithelial barriers to health and disease. Exp Cell Res. 2017;358:71–77. doi: 10.1016/j.yexcr.2017.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.France MM, Turner JR. The mucosal barrier at a glance. J Cell Sci. 2017;130:307–314. doi: 10.1242/jcs.193482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka H, Yamamoto Y, Kashihara H, Yamazaki Y, Tani K, Fujiyoshi Y, Mineta K, Takeuchi K, Tamura A, Tsukita S. Claudin-21 has a paracellular channel role at tight junctions. Mol Cell Biol. 2016;36:954–964. doi: 10.1128/MCB.00758-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Lin J, Zhang X, Zhao Z, Welker NC, Li Y, Liu Y, Bronner MP. Novel microRNA signature to differentiate ulcerative colitis from Crohn disease: a genome-wide study using next generation sequencing. MicroRNA. 2016;5:222–229. doi: 10.2174/2211536605666161117113031. Important paper presenting data based on next generation sequencing. Analysis was done on frozen nondysplastic colonic mucosa originating from distal-most colectomy of 10 UC patients, 9 CD patients and 18 patients with diverticular disease used as controls. These results suggest that the altered expression of different microRNAs, including miR-147b, miR-194-2, miR-383, miR-615 and miR-1826, may be at play in pathophysiologic mechanisms underlying UC and CD. [DOI] [PubMed] [Google Scholar]
- 10••.Wu LY, Ma XP, Shi Y, Bao CH, Jin XM, Lu Y, Zhao JM, Zhou CL, Chen D, Liu HR. Alterations in microRNA expression profiles in inflamed and non-inflamed ascending colon mucosae of patients with active Crohn’s disease. J Gastroenterol Hepatol. 2017;32:1706–1715. doi: 10.1111/jgh.13778. Another important paper analyzing the effects of inflammation on micro-RNAs expressed in ascending colon of adult patients with CD. Authors identified 43 miRNAs that were significantly up-regulated and 35 that were down-regulated in the mucosa of inflammatory CD as compared with non-inflammatory CD group of patients. Experimental and computer analyzes show these microRNAs and mRNAs differentially expressed between the two groups to be implicated in inflammatory response and interactions with intestinal flora. [DOI] [PubMed] [Google Scholar]
- 11.Lewis A, Felice C, Kumagai T, Lai C, Singh K, Jeffery RR, Feakins R, Giannoulatou E, Armuzzi A, Jawad N, Lindsay JO, Silver A. The miR-200 family is increased in dysplastic lesions in ulcerative colitis patients. PLoS One. 2017;12:e0173664. doi: 10.1371/journal.pone.0173664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Shachar S, Yanai H, Sherman Horev H, Elad H, Baram L, Issakov O, Tulchinsky H, Pasmanik-Chor M, Shomron N, Dotan I. MicroRNAs expression in the ileal pouch of patients with ulcerative colitis is robustly up-regulated and correlates with disease phenotypes. PLoS One. 2016;11:e0159956. doi: 10.1371/journal.pone.0159956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szűcs D, Béres NJ, Rokonay R, Boros K, Borka K, Kiss Z, Arató A, Szabó AJ, Vannay Á, Sziksz E, Bereczki C, Veres G. Increased duodenal expression of miR-146a and -155 in pediatric Crohn’s disease. World J Gastroenterol. 2016;22:6027–6035. doi: 10.3748/wjg.v22.i26.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Béres NJ, Szabó D, Kocsis D, Szűcs D, Kiss Z, Müller KE, Lendvai G, Kiss A, Arató A, Sziksz E, Vannay Á, Szabó AJ, Veres G. Role of altered expression of miR-146a, miR-155, and miR-122 in pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2016;22:327–335. doi: 10.1097/MIB.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 15.Mohammadnia-Afrouzi M, Hosseini AZ, Khalili A, Abediankenari S, Amari A, Aghili B, Nataj HH. Altered microRNA expression and immunosuppressive cytokine production by regulatory T cells of ulcerative colitis patients. Immunol Invest. 2016;45:63–74. doi: 10.3109/08820139.2015.1103749. [DOI] [PubMed] [Google Scholar]
- 16.Xu M, Zuo D, Liu X, Fan H, Chen Q, Deng S, Shou Z, Tang Q, Yang J, Nan Z, et al. MiR-155 contributes to Th17 cells differentiation in dextran sulfate sodium (DSS)-induced colitis mice via Jarid2. Biochem Biophys Res Commun. 2017;488:6–14. doi: 10.1016/j.bbrc.2017.04.143. [DOI] [PubMed] [Google Scholar]
- 17•.Heinsbroek SE, Squadrito ML, Schilderink R, Hilbers FW, Verseijden C, Hofmann M, Helmke A, Boon L, Wildenberg ME, Roelofs JJ, et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. 2016;9:960–973. doi: 10.1038/mi.2015.113. In this paper, authors analyze the role of CD206/MRC1 (mannose receptor C-type 1) in experimental colitis, using a knockout mouse model. Their result establish that the reduction of inflammation in the gut of knockout mice does not arise from the deletion of CD206 itself, but rather from the deletion of miR-511-3p, a microRNA located within intron 5 of the CD206/MRC1. Namely, overexpressing miR-511-3p increased by 50% the expression of TLR4, a receptor implicated in the innate immune response. [DOI] [PubMed] [Google Scholar]
- 18.Ando Y, Mazzurana L, Forkel M, Okazaki K, Aoi M, Schmidt PT, Mjösberg J, Bresso F. Downregulation of microRNA-21 in colonic CD3+ T cells in UC remission. Inflamm Bowel Dis. 2016;22:2788–2793. doi: 10.1097/MIB.0000000000000969. [DOI] [PubMed] [Google Scholar]
- 19.Kim HY, Kwon HY, Ha Thi HT, Lee HJ, Kim GI, Hahm KB, Hong S. MicroRNA-132 and microRNA-223 control positive feedback circuit by regulating FOXO3a in inflammatory bowel disease. J Gastroenterol Hepatol. 2016;31:1727–1735. doi: 10.1111/jgh.13321. [DOI] [PubMed] [Google Scholar]
- 20•.Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS, Gerich ME, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214:1737–1752. doi: 10.1084/jem.20160462. Mice bearing a X chromosome (−/y) where miR-223 has been deleted, show enhanced myeloid-driven experimental colitis and the production of higher IL-1β levels. Authors establish that miR-223 targets transcripts encoding NLRP3 (NLR family pyrin domain containing 3), a component of inflammasome. Mutating miR-223 target site in the 3′-UTR of NLRP3 transcripts generates a phenocopy of (−/y) mice, indicating that this microRNA is implicated in controlling gut inflammatory response, which was further demonstrated using a miR-223 mimic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21• •.Pierdomenico M, Cesi V, Cucchiara S, Vitali R, Prete E, Costanzo M, Aloi M, Oliva S, Stronati L. NOD2 is regulated by Mir-320 in physiological conditions but this control is altered in inflamed tissues of patients with inflammatory bowel disease. Inflamm Bowel Dis. 2016;22:315–326. doi: 10.1097/MIB.0000000000000659. Authors show that inflammation reduces the expression of miR-320, and increases the expression of NOD2, a gene encoding a molecular pattern recognition receptor implicated in bacterial sensing and induction of autophagy. Inhibition of miR-320 causes translocation of NF-κB into the nucleus and production of downstream pro-inflammatory cytokines. Authors unravel an inverse correlation between the levels of miR-320 and NOD2 in pediatric patients with IBD. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Zhang S, Qiu Y, He Y, Chen B, Mao R, Cui Y, Zeng Z, Chen M. Upregulation of miR-665 promotes apoptosis and colitis in inflammatory bowel disease by repressing the endoplasmic reticulum stress components XBP1 and ORMDL3. Cell Death Dis. 2017;8:e2699. doi: 10.1038/cddis.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi T, Xie Y, Fu Y, Zhou Q, Ma Z, Ma J, Huang Z, Zhang J, Chen J. The signaling axis of microRNA-31/interleukin-25 regulates Th1/Th17-mediated inflammation response in colitis. Mucosal Immunol. 2017;10:983–995. doi: 10.1038/mi.2016.102. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Ma T, Chen W, Chen Y, Li M, Ren L, Chen J, Cao R, Feng Y, Zhang H, Shi R. MicroRNA-124 promotes intestinal inflammation by targeting aryl hydrocarbon receptor in Crohn’s disease. J Crohns Colitis. 2016;10:703–712. doi: 10.1093/ecco-jcc/jjw010. [DOI] [PubMed] [Google Scholar]
- 25.Nijhuis A, Curciarello R, Mehta S, Feakins R, Bishop CL, Lindsay JO, Silver A. MCL-1 is modulated in Crohn’s disease fibrosis by miR-29b via IL-6 and IL-8. Cell Tissue Res. 2017;368:325–335. doi: 10.1007/s00441-017-2576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian T, Zhou Y, Feng X, Ye S, Wang H, Wu W, Tan W, Yu C, Hu J, Zheng R, et al. MicroRNA-16 is putatively involved in the NF-κB pathway regulation in ulcerative colitis through adenosine A2a receptor (A2aAR) mRNA targeting. Sci Rep. 2016;6:30824. doi: 10.1038/srep30824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu W, He Y, Feng X, Ye S, Wang H, Tan W, Yu C, Hu J, Zheng R, Zhou Y. MicroRNA-206 is involved in the pathogenesis of ulcerative colitis via regulation of adenosine A3 receptor. Oncotarget. 2017;8:705–721. doi: 10.18632/oncotarget.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo JS, Alam KJ, Kim HS, Lee YM, Yun KJ, Chae SC. MicroRNA 429 regulates mucin gene expression and secretion in murine model of colitis. J Crohns Colitis. 2016;10:837–849. doi: 10.1093/ecco-jcc/jjw033. [DOI] [PubMed] [Google Scholar]
- 29.Min M, Peng L, Yang Y, Guo M, Wang W, Sun G. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm Bowel Dis. 2014;20:652–659. doi: 10.1097/MIB.0000000000000009. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Tian Y, Zhu W, Gong J, Guo Z, Guo F, Gu L, Li J. IL-10/microRNA-155/SHIP-1 signaling pathway is crucial for commensal bacteria induced spontaneous colitis. Biochem Pharmacol. 2016;116:100–106. doi: 10.1016/j.bcp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Dai X, Chen X, Chen Q, Shi L, Liang H, Zhou Z, Liu Q, Pang W, Hou D, Wang C, et al. MicroRNA-193a-3p reduces intestinal inflammation in response to microbiota via down-regulation of colonic PepT1. J Biol Chem. 2015;290:16099–16115. doi: 10.1074/jbc.M115.659318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J, Li M, Cui Y, Chen M, Hu JF, Zhang S. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016;17:58. doi: 10.1186/s13059-016-0901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antoni L, Nuding S, Wehkamp J, Stange EF. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20:1165–1179. doi: 10.3748/wjg.v20.i5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odenwald MA, Turner JR. Intestinal permeability defects: is it time to treat? Clin Gastroenterol Hepatol. 2013;11:1075–1083. doi: 10.1016/j.cgh.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buckley A, Turner JR. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a029314. http://dx.doi.org/10.1101/cshperspect.a029314. [DOI] [PMC free article] [PubMed]
- 36.Lee J, Park EJ, Yuki Y, Ahmad S, Mizuguchi K, Ishii KJ, Shimaoka M, Kiyono H. Profiles of microRNA networks in intestinal epithelial cells in a mouse model of colitis. Sci Rep. 2015;5:18174. doi: 10.1038/srep18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253:167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- 38.Salvador E, Burek M, Förster CY. Tight junctions and the tumor microenvironment. Curr Pathobiol Rep. 2016;4:135–145. doi: 10.1007/s40139-016-0106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho S, Reis CA, Pinho SS. Cadherins glycans in cancer: sweet players in a bitter process. Trends Cancer. 2016;2:519–531. doi: 10.1016/j.trecan.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Zhou G, Yang L, Gray A, Srivastava AK, Li C, Zhang G, Cui T. The role of desmosomes in carcinogenesis. Onco Targets Ther. 2017;10:4059–4063. doi: 10.2147/OTT.S136367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zidar N, Botjančič E, Jerala M, Kojc N, Drobne D, tabuc B, Glavač D. Down-regulation of microRNAs of the miR-200 family and up-regulation of Snail and Slug in inflammatory bowel diseases — hallmark of epithelial–mesenchymal transition. J Cell Mol Med. 2016;20:1813–1820. doi: 10.1111/jcmm.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, Chen H, Zhang P, Wang F, Han H, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470–1481. doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 43.Ludwig K, Fassan M, Mescoli C, Pizzi M, Balistreri M, Albertoni L, Pucciarelli S, Scarpa M, Sturniolo GC, Angriman I, Rugge M. PDCD4/miR-21 dysregulation in inflammatory bowel disease-associated carcinogenesis. Virchows Arch. 2013;462:57–63. doi: 10.1007/s00428-012-1345-5. [DOI] [PubMed] [Google Scholar]
- 44•.Bu P, Wang L, Chen KY, Srinivasan T, Murthy PK, Tung KL, Varanko AK, Chen HJ, Ai Y, King S, Lipkin SM, Shen X. A miR-34a-Numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18:189–202. doi: 10.1016/j.stem.2016.01.006. This manuscript establishes that direct suppression of transcripts encoding endocytic adaptor protein Numb by miR-34a in early-stage colon cancer stem cells induces a feed-forward regulatory loop targeting Notch, causing a clear bifurcation between stem cell and non-stem cell fate. Especially, they show that this cell fate transition is caused by miR-34a-dependent Numb down-regulation, that induces Notch degradation. This cell fate determination mechanism is critical for maintenance of mouse intestinal stem cell homeostasis, and its disruption contribute to stem cell proliferation and tumor development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, van der Meulen-de Jong AE, Oikonomopoulos A, van Deen WK, Vorvis C, et al. MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology. 2015;149:981–992. doi: 10.1053/j.gastro.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.He C, Yu T, Shi Y, Ma C, Yang W, Fang L, Sun M, Wu W, Xiao F, Guo F, et al. MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhibiting BTG1. Gastroenterology. 2017;152:1434–1448. doi: 10.1053/j.gastro.2017.01.049. This paper shows that miR-301a knockout mice are resistant to DSS-induced colitis. In particular, miR-301a targets BTG anti-proliferation factor 1 (BTG1), known to inhibiting NF-κB activation. MiR-301a knockout mice showed better preservation of IB integrity following colitis induction, and developed fewer colitis-associated tumors. This phenotype was associated with a reduction of CDH1/E-cadherin expression, suggesting that miR-301a is implicated in inflammation-associated destabilization of IB and induction of colorectal cancer. [DOI] [PubMed] [Google Scholar]
- 47.Jeppsson S, Srinivasan S, Chandrasekharan B. Neuropeptide Y (NPY) promotes inflammation-induced tumorigenesis by enhancing epithelial cell proliferation. Am J Physiol Gastrointest Liver Physiol. 2017;312:G103–G111. doi: 10.1152/ajpgi.00410.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pekow J, Meckel K, Dougherty U, Huang Y, Chen X, Almoghrabi A, Mustafi R, Ayaloglu-Butun F, Deng Z, Haider HI, et al. miR-193a-3p is a key tumor suppressor in ulcerative colitis-associated colon cancer and promotes carcinogenesis through upregulation of IL17RD. Clin Cancer Res. 2017;23:5281–5291. doi: 10.1158/1078-0432.CCR-17-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pekow J, Hutchison AL, Meckel K, Harrington K, Deng Z, Talasila N, Rubin DT, Hanauer SB, Hurst R, Umanskiy K, et al. miR-4728-3p functions as a tumor suppressor in ulcerative colitis-associated colorectal neoplasia through regulation of focal adhesion signaling. Inflamm Bowel Dis. 2017;23:1328–1337. doi: 10.1097/MIB.0000000000001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Bai J, Zhang L, Lou F, Ke F, Cai W, Wang H. Conditional knockout of microRNA-31 promotes the development of colitis associated cancer. Biochem Biophys Res Commun. 2017;490:62–68. doi: 10.1016/j.bbrc.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 51••.Kourtidis A, Ngok SP, Pulimeno P, Feathers RW, Carpio LR, Baker TR, Carr JM, Yan IK, Borges S, Perez EA, et al. Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat Cell Biol. 2015;17:1145–1157. doi: 10.1038/ncb3227. This paper is the first to show the presence of an active microprocessor complex with a cytoplasmic localization. Authors identify two different E-cadherin–p120 complexes, among whose the apical complex, but not the basolateral one, contains the factor PLEKHA7 implicated in stabilization of adherens junctions. Authors further show that PLEKHA7-anchored microprocessor contains miR-24, miR-30a, 30b and let-7 microRNAs, and at the minimum allows the processing of miR-30a from its precursor RNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benderska N, Dittrich AL, Knaup S, Rau TT, Neufert C, Wach S, Fahlbusch FB, Rauh M, Wirtz RM, Agaimy A, et al. miRNA-26b overexpression in ulcerative colitis-associated carcinogenesis. Inflamm Bowel Dis. 2015;21:2039–2051. doi: 10.1097/MIB.0000000000000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Cordes F, Brückner M, Lenz P, Veltman K, Glauben R, Siegmund B, Hengst K, Schmidt MA, Cichon C, Bettenworth D. MicroRNA-320a strengthens intestinal barrier function and follows the course of experimental colitis. Inflamm Bowel Dis. 2016;22:2341–2355. doi: 10.1097/MIB.0000000000000917. In this paper, authors compare the effects of a probiotic and of an enteropathogenic E. coli strain on the structure of epithelial tight-junctions as measured by trans-epithelial resistance. They show that the expression of miR-320 in colon cells is increased by co-incubation with probiotic E. coli but decreased by co-incubation with enteropathogenic E. coli. Authors establish that miR-320 targets PP2A/PP2R5B, a factor that reduces the amount of JAM-A, a phosphatase regulating tight junction stability and IB impermeability. [DOI] [PubMed] [Google Scholar]
- 54.Fang K, Sideri A, Law IK, Bakirtzi K, Polytarchou C, Iliopoulos D, Pothoulakis C. Identification of a novel substance P (SP)-neurokinin-1 receptor (NK-1R) microRNA-221-5p inflammatory network in human colonic epithelial cells. Cell Mol Gastroenterol Hepatol. 2015;1:503–515. doi: 10.1016/j.jcmgh.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciccacci C, Politi C, Biancone L, Latini A, Novelli G, Calabrese E, Borgiani P. Polymorphisms in MIR122, MIR196A2, and MIR124A genes are associated with clinical phenotypes in inflammatory bowel diseases. Mol Diagn Ther. 2017;21:107–114. doi: 10.1007/s40291-016-0240-1. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Zhou CZ, Zhu R, Fan H, Liu XX, Duan XY, Tang Q, Shou ZX, Zuo DM. Microvesicles shuttled miR-200b attenuate experimental colitis associated intestinal fibrosis by inhibiting the development of EMT. J Gastroenterol Hepatol. 2017 doi: 10.1111/jgh.13797. http://dx.doi.org/10.1111/jgh.13797. [DOI] [PubMed]
- 57.Tili E, Chiabai M, Palmieri D, Brown M, Cui R, Fernandes C, Richmond T, Kim T, Sheetz T, Sun HL, et al. Quaking and miR-155 interactions in inflammation and leukemogenesis. Oncotarget. 2015;6:24599–24610. doi: 10.18632/oncotarget.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong Y, Shi L, Wang L, Zhou Z, Wang C, Lin Y, Luo D, Qiu J, Chen D. Activation of sirtuin 1 by catalpol-induced down-regulation of microRNA-132 attenuates endoplasmic reticulum stress in colitis. Pharmacol Res. 2017;123:73–82. doi: 10.1016/j.phrs.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 59.Xiong Y, Wang J, Chu H, Chen D, Guo H. Salvianolic acid B restored impaired barrier function via downregulation of MLCK by microRNA-1 in rat colitis model. Front Pharmacol. 2016;7:134. doi: 10.3389/fphar.2016.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tili E, Michaille JJ. Promiscuous effects of some phenolic natural products on inflammation at least in part arise from their ability to modulate the expression of global regulators, namely microRNAs. Molecules. 2016;21 doi: 10.3390/molecules21091263. pii:E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]