Abstract
The major objective of this study was to investigate the association of genetic and nongenetic factors with variability in protein abundance and in vitro activity of the androgen-metabolizing enzyme UGT2B17 in human liver microsomes (n = 455). UGT2B17 abundance was quantified by liquid chromatography-tandem mass spectrometry proteomics, and enzyme activity was determined by using testosterone and dihydrotestosterone as in vitro probe substrates. Genotyping or gene resequencing and mRNA expression were also evaluated. Multivariate analysis was used to test the association of UGT2B17 copy number variation, single nucleotide polymorphisms (SNPs), age, and sex with its mRNA expression, abundance, and activity. UGT2B17 gene copy number and SNPs (rs7436962, rs9996186, rs28374627, and rs4860305) were associated with gene expression, protein levels, and androgen glucuronidation rates in a gene dose-dependent manner. UGT2B17 protein (mean ± S.D. picomoles per milligram of microsomal protein) is sparsely expressed in children younger than 9 years (0.12 ± 0.24 years) but profoundly increases from age 9 years to adults (∼10-fold) with ∼2.6-fold greater abundance in males than in females (1.2 vs. 0.47). Association of androgen glucuronidation with UGT2B15 abundance was observed only in the low UGT2B17 expressers. These data can be used to predict variability in the metabolism of UGT2B17 substrates. Drug companies should include UGT2B17 in early phenotyping assays during drug discovery to avoid late clinical failures.
Introduction
Uridine 5′-diphospho-glucuronosyltransferases (UGTs; EC 2.4.1.17) facilitate excretion of a wide variety of lipophilic drugs, environmental chemicals, and endogenous substrates containing hydroxyl, carboxyl, amino, and sulfur-containing functional groups by catalyzing conjugation of these substrates with glucuronic acid to increase hydrophilicity. UGT enzymes belong to distinct subfamilies of more than 26 genes with 19 well characterized functional proteins. The functional isoforms belong to the UGT1 and UGT2 superfamilies, which are further divided into three subfamilies, based on their sequence similarities, into UGT1As (UGT1A1, UGT1A3–UGT1A10), UGT2As (UGT2A1–UGT2A3), and UGT2Bs (UGT2B4, UGT2B7, UGT2B10, UGT2B11, UGT2B15, UGT2B17, and UGT2B28) (Guillemette, 2003; Oda et al., 2015; Yuan et al., 2016).
Although all hepatic UGT isoforms are generally variable, UGT2B17 shows extensively greater interindividual variability in its protein abundance and activity (Fallon et al., 2013; Neumann et al., 2016), and it is expressed in a variety of tissues, such as liver, intestine, kidney, testis, uterus, placenta, mammary gland, adrenal gland, skin, and prostate (Beaulieu et al., 1996; Ekstrom et al., 2013). Numerous endogenous steroids, including testosterone (T), dihydrotestosterone (DHT), androstane-3-α, 17-β-diol (3-α-diol), androsterone and estradiol, and xenobiotics (e.g., 17-dihydroexemestane, vorinostat, lorcaserin) have been identified as substrates of UGT2B17 (Beaulieu et al., 1996; Wong et al., 2011; Sadeque et al., 2012; Chen et al., 2016b; Neumann et al., 2016). The expression of UGT2B17 in sex hormone–sensitive organs also indicates its role in sex hormone homeostasis. For example, although controversial, gene deletion in UGT2B17 is associated with greater risk of developing androgen-sensitive prostate diseases (Barbier and Belanger, 2008; Paquet et al., 2012; Kpoghomou et al., 2013; Gauthier-Landry et al., 2015). The UGT2B17 gene-deletion allele has been shown to be associated with several other pathophysiological conditions, such as obesity (Zhu et al., 2015), chronic lymphocytic leukemia (Gruber et al., 2013), and endometrial cancer (Hirata et al., 2010). UGT2B17 also appears to play a critical role in the metabolism of tobacco-specific carcinogens and the risk of lung cancer (Lazarus et al., 2005; Chen et al., 2016a). In addition, high intratumoral UGT2B17 expression levels correlate with better survival outcomes in patients with breast cancer (Hu et al., 2016). Besides its role in disease pathophysiology, the UGT2B17 gene deletion is associated with false-negative doping test results, which in turn is linked to variable testosterone metabolism (Schulze et al., 2008). An investigational drug developed by Merck, MK-7246, a selective CRTH2 (prostaglandin D2 receptor 2) antagonist, was discontinued from development after unpredicted variability observed in its pharmacokinetics (PK). MK-7246 was later characterized as a selective UGT2B17 substrate (Wang et al., 2012). Similarly, the PK and anticancer effectiveness of UGT2B17 substrates 17-dihydroexemestane and vorinostat are highly variable (Wong et al., 2011; Chen et al., 2016b). Particularly, the normalized 17-dihydroexemestane and vorinostat levels were 28% and 26% higher, respectively, in subjects carrying the UGT2B17 gene deletion compared with those carrying the reference allele (Wong et al., 2011; Luo et al., 2017). The in vitro glucuronidation rate of 17-dihydroexemestane is significantly decreased (14-fold) in human liver microsomes (HLMs) exhibiting the UGT2B17 deletion genotype versus wild-type UGT2B17 HLMs (Sun et al., 2010). Taken together, high variability in UGT2B17 abundance significantly contributes to an unpredictable fate of its substrates that may lead to adverse pathophysiologic consequences and drug toxicity or lack of efficacy.
To understand more completely the underlying causes of UGT2B17 variability, we investigated the association of genetic and nongenetic factors with variability in protein abundance and in vitro activity of UGT2B17 in HLMs. The knowledge of individual contribution of population factors in UGT2B17 variability can be applied to the prediction of the metabolism of androgens and other UGT2B17 substrates. Further, because androgens are also metabolized by UGT2B15 (minor pathway), we studied the effect of its genetic polymorphism (UGT2B15*2) on the metabolism of testosterone and DHT, particularly in the poor expressers of UGT2B17.
Materials and Methods
Chemicals and Reagents.
Iodoacetamide, dithiothreitol, and Pierce trypsin protease (MS grade) were purchased from Thermo Fisher Scientific (Rockford, IL). Ammonium bicarbonate buffer (98% purity) was purchased from Acros Organics (Geel, Belgium). Chloroform, MS-grade acetonitrile, methanol, and formic acid were purchased from Fischer Scientific (Fair Lawn, NJ). Human serum albumin (HSA) and bovine serum albumin (BSA) were obtained from Calbiochem (Billerica, MA) and Thermo Fisher Scientific, respectively. The purified light peptides were purchased from New England Peptides (Cambridge, MA). Synthetic isotopically pure heavy stable isotope-labeled peptides were produced by Thermo Fisher Scientific. UDPGA and MgCl2 were purchased from Sigma-Aldrich (St. Louis, MO). Tesosterone (1 mg/ml in 100% acetonitrile) and testosterone-glucuronide were purchased from Cerilliant (Round Rock, TX). Testosterone-glucuronide was dissolved in 100% methanol (1 mg/ml). DHT (1 mg/ml in methanol) and DHT-glucuronide were purchased from Sigma-Aldrich and Cerilliant, respectively. Testosterone-glucuronide-d3 and DHT-glucuronide were procured from Cerilliant.
Human Liver Tissue and Preparation of Microsomes.
Previously prepared HLM samples (Pearce et al., 2016; Shirasaka et al., 2016) were used in this study. The liver tissue samples for HLM preparation were received from three liver tissue banks: 1) the University of Washington Human Liver Bank (Seattle, WA) (n = 56), 2) Children’s Mercy Kansas City (Kansas City, MO) (n = 128), and 3) the Liver Bank at the St. Jude Children’s Research Hospital (Memphis, TN) (n = 271). The Children’s Mercy Kansas City samples were originally obtained from the University of Maryland Brain and Tissue Bank for Developmental Disorders and the Liver Tissue Cell Distribution System. Additional details on the selection, procurement, and storage of the livers and investigator blinding for sample analyses have been described previously (Prasad et al., 2014; Shirasaka et al., 2016; Tanner et al., 2017). Age, sex, and ethnicity were known for 96%, 98%, and 88% of the liver donors, respectively (Supplemental Table S1). The age range for donors was from 0 to 87 years (median age, 24 years). Of the 455 samples analyzed for UGT2B abundance, demographic association analyses were conducted on 423 samples for which UGT2B17 copy number variation (CNV) information was available. Sex distribution of these 423 samples was 252 males, 163 females, and eight unknown. The ethnicity distribution was 333 Caucasian, 26 African American, four Hispanic, one Native American, one Asian, one Pacific Islander, and 56 unknown. Cause of death, medications used, and liver pathology were known for less than 50% of the donors. The collection and use of these tissues for research purposes were approved by the human subjects Institutional Review Boards of the University of Washington (Seattle, WA), the St. Jude Children’s Research Hospital (Memphis, TN), and the Pediatric Institutional Review Board of Children’s Mercy Kansas City (Kansas City, MO).
UGT2B17 and UGT2B15 Protein Quantification in HLM Samples.
Total protein quantification in HLM samples was performed using a bicinchonic acid assay kit (BCA Protein Assay Kit; Pierce Biotechnology, Waltham, MA). HLMs (80 μl, 2 mg/ml total protein) were digested as described in the Supplemental Materials. The surrogate peptides of UGT2B17 (FSVGYTVEK and SVINDPIYK) and UGT2B15 (SVINDPVYK) were quantified in the digested samples using a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method (Vrana et al., 2017), described in the Supplemental Materials.
UGT2B17 mRNA Quantification.
A subset of liver tissue samples (n = 230) were available for UGT2B17 mRNA expression analysis (Supplemental Table S1). Details of the RNA-seq procedures, including RNA isolation, TruSeq-stranded mRNA preparation, and read processing and analysis pipeline have been described previously (Tanner et al., 2017). mRNA transcript levels are presented in fragments per kilobase per million reads (FPKM) values.
UGT2B17 Sequencing, Genotyping, Haplotype and Copy Number Variation Analysis.
Because liver samples were obtained from different sources, two approaches, gene sequencing and genotyping, were used for genetic characterization of the liver tissue samples, as discussed in the Supplemental Materials. The University of Washington and St. Jude Liver Bank samples were sequenced using the Pharmacogenomics Research Network (PGRN)-Seq platform, a targeted sequencing approach, as described elsewhere (Gordon et al., 2016), whereas the samples provided by Children’s Mercy were genotyped on DMET or PharmacoScan arrays (Affymetrix, Santa Clara, CA). Linkage disequilibrium (LD) analysis of UGT2B17 variants and inferred haplotypes were determined using Haploview 4.2 (Cambridge, MA).
UGT2B17 and UGT2B15 Enzyme Activity Assay.
For activity assays, 346 HLM samples (donor age ranges from 0 to 87 years; median age = 18 years, Supplemental Table S1) were available. Glucuronidation activity was determined by quantifying the rates of testosterone- and DHT-glucuronide formation (picomoles per minute per milligram of microsomal protein) in triplicate. The assay reactions contained 0.1 mg/ml HLM protein, 0.1 M phosphate buffer (pH 7.4), a mix of 1 μM testosterone and 1 μM DHT, BSA (0.01%), and alamethicin (0.1 mg/ml) (final volume of 95 μl). Final vehicle (methanol or ethanol) concentration was less than 1%. Reactions were preincubated for 15 minutes on ice. UDPGA (5 μl; final concentration, 2.5 mM) was added to initiate reactions, and mixtures were gently agitated for 30 minutes at 37°C before being quenched with ice-cold acetonitrile containing 50 ng/ml progesterone (200 μl, internal standard) and subjected to centrifugation for 5 minutes at ∼1300g. Supernatants were analyzed by an optimized LC-MS/MS method provided in the Supplemental Materials.
Data Analysis.
We used a robust strategy to ensure optimum reproducibility of UGT2B17 and UGT2B15 protein quantification (Bhatt and Prasad, 2018). For example, ion suppression was addressed by using heavy peptide internal standards. BSA or HSA was used as an exogenous protein internal standard, which was added to each sample in a fixed quantity before desalting by methanol-chloroform-water extraction and trypsin digestion. The addition of BSA or HSA addresses the variability introduced during predigestion processing, such as 1) protein loss during methanol-chloroform-water extraction and 2) sample-to-sample trypsin digestion artifacts. To address interbatch variability, two to three sets of pooled representative HLM samples were processed each day, which served as quality controls across the entire study. In total, a three-step data normalization approach was used; first, average light peak areas for specific peptide daughter fragments were divided by corresponding average heavy peak areas. Next, this ratio was further divided by the BSA or HSA light/heavy area ratio. Finally, for each day, these data were further normalized by mean values of the quality control values run with each individual batch to adjust for any interday variability.
Ontogeny was measured by categorical and continuous analyses of age versus UGT2B17 protein abundance data. For categorical analysis, the samples were grouped based on the following age categories: neonatal (0–27 days), infancy (28–364 days), early childhood (1 to <6 years), middle childhood (6 to <12 years), adolescence (12–18 years), and adulthood (>18 years).
Statistical analyses were performed using GraphPad Prism 5 (La Jolla, CA) and Microsoft Excel (365 ProPlus; Redmond, WA). Nonparametric tests were used to test age, sex, and genotype dependence. To compare two groups (e.g., male vs. female), the Mann-Whitney test was used. The Kruskal-Wallis test followed by Dunn’s multiple comparison tests were used to perform the age-dependent data analyses and determine associations between the genotype and mRNA expression, protein abundance, and enzyme activity. For correlation analysis, the nonparametric Spearman regression test was used. Additionally, the Jonckheere-Terpstra test and multivariate analysis were performed by using RStudio (version 1.0.136, R version 3.3.2, Boston, MA).
A nonlinear regression equation (eq. 1) was used to fit the ontogeny data, as described previously (Bhatt et al., 2017), where Adultmax is the maximum average relative protein abundance, Age is the age in years of the subject at the time of sample collection, Age50 is the age in years at which half-maximum adult protein abundance is obtained, E is protein abundance at any given age, Ebirth is protein abundance at birth, and n is the exponential factor.
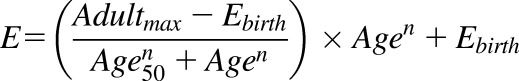 |
(1) |
For haplotype analysis, the number of variants was directly counted. Hardy-Weinberg equilibrium (HWE) was determined by comparing the variant frequencies with the expected values using a contingency table χ2 statistic with Yates’ correction. The numbers of haplotype and statistics D, D’, and LD were estimated by Haploview 4.2 software. Relationships were considered significant at P < 0.05.
Results
Hierarchical Clustering of Major Hepatic Drug-Metabolizing Enzymes and Correlation Analysis of UGT2B17 Protein, mRNA, and Activity.
Hierarchical clustering analysis of quantitative proteomics results of major hepatic drug-metabolizing enzymes from a preliminary study conducted in first 165 samples (out of a total 455 samples) suggested unique and highly variable protein abundance for UGT2B17 (Supplemental Fig. S1). The correlation between UGT2B17 mRNA and protein abundance or activity was moderate (r2 = 0.17 and 0.19, respectively) but statistically significant (P < 0.0001) (Supplemental Fig. S2A). Consistent with the literature (Ohtsuki et al., 2012), the correlation between mRNA and protein expression in tissues was weak; in contrast, a strong correlation between protein abundance and activity was observed (Supplemental Fig. S2, B and C). Although our quantitative proteomics method was very sensitive for UGT2B17 detection [lower limit of quantification (LLOQ) = 0.17 pmol/mg of microsomal protein], the protein was not detected in 48% of the samples, indicating a rather large range of variability in protein expression. For statistical analysis, samples below the limit of detection (<LOD) were assigned a value of 0.06 pmol/mg of microsomal protein (i.e., one third of LLOQ) instead of zero. In low UGT2B17 expressers, rates of testosterone- and DHT-glucuronide formation were also consistently low. The average human liver UGT2B17 protein abundance in liver microsomal samples (n = 370, excluding zero copy number samples) was 0.92 ± 1.6 pmol/mg microsomal protein with 162-fold interindividual variability (0.06–9.7 pmol/mg microsomal protein). Mean rates of testosterone- and DHT-glucuronide formation (range; fold difference) were 15.4 (0.3–184; 558) and 41.8 (1.0–233; 233) pmol/min per milligram microsomal protein, respectively (Table 1).
TABLE 1.
Effect of genetic variations, age, and gender on UGT2B17 protein abundance and activity
| Glucuronide Formation Activity (pmol/min per mg of Microsomal Protein) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Abundance (pmol/mg of Microsomal Protein) |
T-Glucuronide |
DHT-Glucuronide |
|||||||||||
| n | Mean | Median | Range (Min–Max, Fold Difference) | No. of <LOD Samples | n | Mean | Median | Range (Min-Max, Fold Difference) | n | Mean | Median | Range (Min–Max, Fold Difference) | |
| All samplesa | 370 | 0.92 | 0.06 | 0.06–9.7, 162 | 202 | 325 | 15.4 | 6.4 | 0.3–184, 558 | 325 | 41.8 | 26.2 | 1.0–233, 233 |
| CN = 0 | 53 | 0 | 0 | — | — | 46 | 5.8 | 3.7 | 0.6–20.5, 34 | 46 | 27.8 | 21.2 | 2.6–90.2, 35 |
| CN = 1 | 172 | 0.67 | 0.06 | 0.06–6.5, 108 | 107 | 133 | 13.8 | 6.2 | 0.3–73.6, 223 | 133 | 40.8 | 27.4 | 1.0–165, 165 |
| CN = 2 | 198 | 1.14 | 0.19 | 0.06–9.7, 162 | 95 | 146 | 19.9 | 7.8 | 0.5–184, 368 | 146 | 47 | 29.5 | 1.5–233, 155 |
| Diplotypes | |||||||||||||
| Reference CAGA/CAGA | 131 | 0.57 | 0.06 | 0.06–7.9, 132 | 94 | 91 | 13.8 | 5.7 | 0.5–123, 246 | 91 | 38.7 | 25.3 | 2.1–181.4, 86.4 |
| Heterozygous TCAA/CAGG | 62 | 1.59 | 1.04 | 0.06–6.02, 100 | 15 | 35 | 27.5 | 21.4 | 0.5–98.2, 196 | 35 | 63.4 | 58.8 | 1.5–160, 107 |
| Homozygous TCAG/TCAG | 58 | 1.63 | 1.03 | 0.06–9.7, 162 | 11 | 31 | 30.3 | 23.2 | 1.9–184, 96.8 | 31 | 66 | 57 | 8.3–233, 28 |
| Age categories | |||||||||||||
| Neonatal | 3 | 0.06 | 0.06 | 0.06–0.06 | 3 | 3 | 1.1 | 0.94 | 0.4–2.0, 5 | 3 | 4.8 | 3.8 | 1.6–9.0, 5.6 |
| Infancy | 23 | 0.11 | 0.06 | 0.06–0.86, 14.3 | 20 | 19 | 3.3 | 1.3 | 0.4–11.5, 29 | 19 | 14.1 | 6.2 | 1.0–55.1, 55.1 |
| Early childhood | 38 | 0.13 | 0.06 | 0.06–1.19, 21 | 34 | 29 | 6.6 | 3.7 | 0.3–33.1, 110 | 29 | 24.8 | 17.0 | 1.0–101, 101 |
| Middle childhood | 44 | 0.15 | 0.06 | 0.06–1.49, 24.8 | 34 | 35 | 6.0 | 3.7 | 0.7–38.9, 56 | 35 | 25.0 | 16.9 | 2.6–116, 44.6 |
| Adolescence | 61 | 0.81 | 0.06 | 0.06–6.9, 115 | 33 | 54 | 18.8 | 7.9 | 0.5–131.3, 263 | 54 | 48.5 | 32.1 | 2.1–194, 92 |
| Adulthood | 185 | 1.33 | 0.46 | 0.06–9.7, 162 | 76 | 127 | 23.2 | 12.6 | 0.8–185.8, 232 | 127 | 56.3 | 47.4 | 3.0–235, 78.3 |
| Gender (age 12 yr) | |||||||||||||
| Male | 149 | 1.60 | 0.91 | 0.06–9.7, 162 | 60 | 108 | 26.9 | 18.1 | 0.5–184, 368 | 108 | 60.3 | 47.6 | 2.1–232, 110 |
| Female | 96 | 0.57 | 0.06 | 0.06–5.5, 92 | 49 | 73 | 13.9 | 9.2 | 0.79–98, 124 | 73 | 43.3 | 36.2 | 2.9–160, 55 |
The samples with zero copy number were excluded from the analysis in all categories except the second row. Thirty-two samples (of 455) were excluded from these analyses because copy number (CN) variation data were not available on these samples.
Association of UGT2B17 Protein Abundance, Testosterone/DHT Glucuronide Formation, and Genetic Variation.
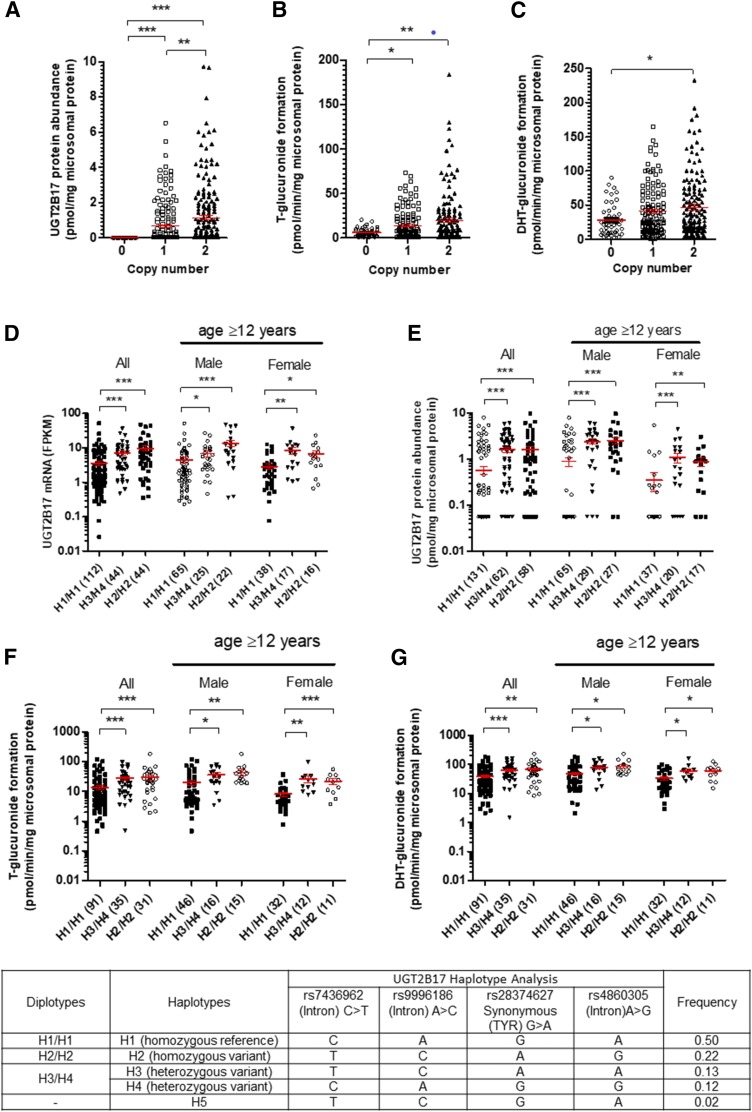
UGT2B17 protein was detected in 38% and 52% of the samples carrying one and two gene copies, respectively. UGT2B17 protein was undetectable in samples homozygous for the UGT2B17 gene deletion (CNV = 0). This variability was also reflected by mRNA data demonstrating that FPKM values were significantly higher (P < 0.0001) in samples with one or two UGT2B17 gene copies; however, FPKM values did not differ among the samples with one and two gene copies owing to the high variability (>205-fold) within each group. Samples carrying two UGT2B17 gene copies showed a 1.7-fold higher mean UGT2B17 protein abundance compared with single gene copy samples (Fig. 1; Table 1). The UGT2B17 gene-dose-dependent effects on rates of testosterone- and DHT-glucuronide formation rates were consistent with the abundance data (Fig. 1; Table 1). Of the 11 UGT2B17 SNPs detected in our samples, only four variants (rs7436962, rs9996186, rs28374627, and rs7668258) were associated with mRNA expression, protein abundance, and activity (Supplemental Table S2). Haplotype analyses suggested significant LD between three intronic SNPs (rs7436962, rs9996186, and rs4860305) and a missense SNP rs28374627 (Fig. 1). We identified four haplotypes (H1–H4; Fig. 1) representing combinations of these SNPs with frequency >10% in our sample set. When the diplotypes were compared, samples harboring H3/H4 and H2/H2 haplotypes showed higher UGT2B17 mRNA expression levels, protein abundance, and activity compared with the reference H1/H1 diplotype (Fig. 1; Table 1). The gene-dose effect was also verified by using multivariate analysis (Table 2) and Jonckheere-Terpstra test (Supplemental Table S5)
Fig. 1.
The UGT2B17 gene deletion is associated with its protein abundance (A), rates of testosterone- and DHT-glucuronide formation (B and C, respectively). UGT2B17 diplotypes (haplotype pairs on homologous chromosomes) are associated with UGT2B17 mRNA expression (D), protein abundance (E), testosterone-glucuronide formation (F), and DHT-glucuronide formation (G). Confounding factor, that is, samples from subjects aged younger than 12 years, were excluded from the subanalysis. *P < 0.05; **P < 0.01; ***P < 0.0001. Sample number in each group is shown in parentheses in the x-axis.
TABLE 2.
Multivariate linear regression analysis of predictors associated with interindividual variability of UGT2B17 protein abundance and UGT2B17-mediated testosterone and DHT-glucuronide formation
Statistically significant correlations are shown in bold.
| Dependent Variable | Independent Variable | Effect Size β (Coefficient) | S.E. | t-Stat | P Value |
|---|---|---|---|---|---|
| Protein abundance (pmol/mg of microsomal protein) | Intercept | −1.0 | 0.28 | −3.5 | 5.8 × 10−4 |
| Male | 0.84 | 0.18 | 4.5 | 9.7 × 10−6 | |
| Adolescence | 0.48 | 0.38 | 1.3 | 0.21 | |
| Adulthood | 1.67 | 0.25 | 4.7 | 4.2 × 10−6 | |
| Diplotype H3/H4 | 0.95 | 0.25 | 3.8 | 1.8 × 10−4 | |
| Diplotype H2/H2 | 1.1 | 0.23 | 4.7 | 4.2 × 10−6 | |
| Copy no.: 2 | 0.22 | 0.22 | 1.1 | 0.31 | |
| Testosterone -glucuronide formation (pmol/min per mg of microsomal protein) | Intercept | −13.9 | 7.0 | −2.0 | <0.05 |
| Male | 11.7 | 4.0 | 2.9 | 0.004 | |
| Adolescence | 15.2 | 8.8 | 1.7 | 0.09 | |
| Adulthood | 21.0 | 6.2 | 3.4 | 9.8 × 10−4 | |
| Diplotype H3/H4 | 12.7 | 5.5 | 2.3 | 0.02 | |
| Diplotype H2/H2 | 20.1 | 5.3 | 3.8 | 2.2 × 10−4 | |
| Copy no.: 2 | 5.5 | 4.8 | 1.1 | 0.25 | |
| DHT-glucuronide formation (pmol/min per mg of microsomal protein) | Intercept | −8.5 | 11 | −0.8 | 0.44 |
| Male | 15.6 | 6.4 | 2.45 | 0.015 | |
| Adolescence | 27.5 | 14 | 2.0 | <0.05 | |
| Adulthood | 42.8 | 9.9 | 4.3 | 2.7 × 10−5 | |
| Diplotype H3/H4 | 26.5 | 8.7 | 3.1 | 0.003 | |
| Diplotype H2/H2 | 31.5 | 8.4 | 3.8 | 0.0002 | |
| Copy no.: 2 | 2.8 | 7.6 | 0.4 | 0.71 |
Association of UGT2B17 Abundance and Testosterone/DHT Glucuronide Formation with Age and Sex.
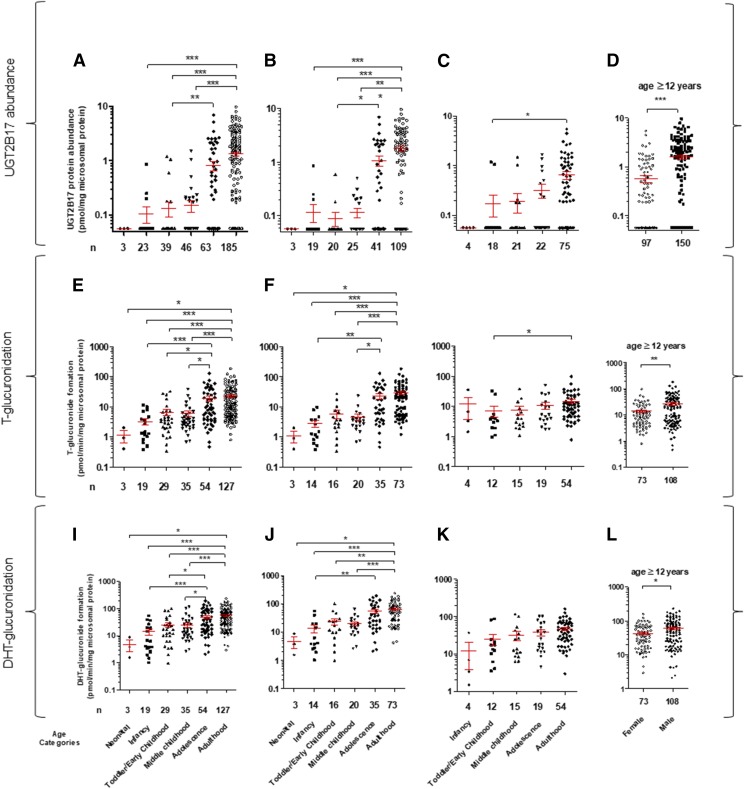
UGT2B17 protein abundance was significantly higher in adulthood compared with infancy and early or middle childhood (Fig. 2A; Supplemental Fig. S3; P values are marked in the figures). Noticeably, the age at which protein expression reaches 50% of that observed in adults (Age50) was >10 years in both male and female samples (Supplemental Fig. S3). Trend analysis (Jonckheere-Terpstra test) showed that there was a statistically significant higher median UGT2B17 abundance with increasing age category (neonatal-infant-early childhood-middle childhood-adolescence- adulthood) (Supplemental Table S5). Male liver donors have a 2.8-fold higher mean UGT2B17 protein level (P < 0.0001) compared with females in samples from donors ≥12 years of age (Fig. 2D; Table 1). Consistent with the protein abundance data, rates of testosterone- and DHT-glucuronide formation were significantly higher in adulthood compared with neonatal, infancy, early childhood, and middle childhood age groups (Fig. 2, E and I). Age-dependent increase in UGT2B17 abundance and activity was greater in male versus female livers (Fig. 2, B, F, and J vs. Fig. 2, C, G, and K). Overall, male liver showed 2- and 1.4-fold higher rates of testosterone- and DHT-glucuronide formation compared with females in samples ≥12 years of age (Fig. 2, H and L).
Fig. 2.
Categorical age-dependent UGT2B17 protein abundance (A–D), testosterone (T)-glucuronide formation (E–H), and DHT-glucuronide formation (I–L) data in all (A, E, and I), male (B, F and J) and female (C, G, and K) livers. The x-axis labels identifying data categories in the bottom panel (I–L) are also applicable to the corresponding top two panels (A–H). Number of samples is presented either as main label in the x-axis (A–H) or the x-axis parentheses (I–L). Donors with zero UGT2B17 gene copy were excluded from this analysis. Of 375 samples (male plus female), 205 were lower than the LOD of UGT2B17 protein measurement. For statistical analysis, samples <LOD (excluding zero copy number) were assigned a value of 0.06 pmol/mg of microsomal protein, which was one-third the LLOQ (0.17 pmol/mg of microsomal protein). UGT2B17 was sparsely (12 of 92 samples) detected in children younger than the age of 9 years. An association of age with UGT2B17 abundance or testosterone and DHT-glucuronide formation was more prominent in male versus. female. Mean UGT2B17 protein abundance and testosterone- and DHT-glucuronide formation in these samples was 2.8-, 1.9-, and 1.4-fold greater in male versus female donors aged ≥12 years, respectively (D). *P < 0.05; **P < 0.01; ***P < 0.0001.
Association of UGT2B15 Abundance and Testosterone/DHT-Glucuronide Formation with UGT2B15*2 in Poor UGT2B17 Expressers.
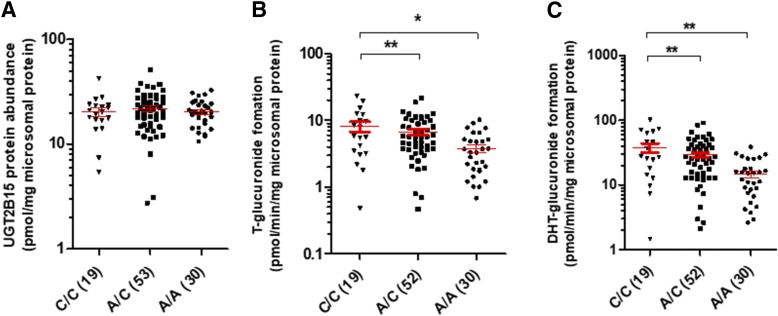
An association of the rates of testosterone- or DHT-glucuronide formation with UGT2B15 protein abundance was observed only in samples with low UGT2B17 levels (i.e., <LOD) (Supplemental Fig. S4). Importantly, in these low UGT2B17 expressers, rs1902023, a nonsynonymous UGT2B15 SNP (Court et al., 2004), was not associated with UGT2B15 protein abundance; however, a significant gene-dose dependent association was found between this SNP and the rates of testosterone- and DHT-glucuronide formation (Fig. 3), respectively. These data suggest that this nonsynonymous UGT2B15 genotype likely affects substrate affinity (km) or catalytic activity (kcat) due to a change in amino acid residue in the active or cofactor binding site. Furthermore, the correlation between protein level and activity improved in samples that did not carry heterozygous or homozygous variants of rs1902023 (Supplemental Fig. S4).
Fig. 3.
The UGT2B15 SNP (rs1902023) is not associated with UGT2B15 protein abundance in low UGT2B17 expressers (A), but it is significantly associated with increased rates of testosterone-glucuronide (B) and DHT-glucuronide (C) formation (picomoles per milligram per minute microsomal protein). Only samples with UGT2B17 abundance <LOD (0.06 pmol/mg of microsomal protein) samples were included in this analysis.
Multivariate Linear Regression Analyses.
The results were further verified using multivariate linear regression analyses to evaluate associations with UGT2B17 protein abundance and rates of testosterone- or DHT-glucuronide formation as the outcomes and CNV, diplotype, age category, and sex as the predictors. Baseline parameters were set as females, children, reference diplotype (H1/H1), and a copy number of 1 (0 copy number samples were excluded from the analyses). The results of the multivariate linear regression analyses, with missing values imputed, are presented in Table 2. For UGT protein abundance, the coefficient of determination (R2 value) for the multivariate linear regression was 0.26, indicating that 26% of the variability in UGT2B17 abundance is explained by the predictors in the model. Sex, age categories, and diplotypes were significant predictors of UGT2B17 protein abundance. For testosterone- and DHT-glucuronide formation, the R2 values for the multivariate linear regression were 0.21 and 0.24, indicating that 21% and 24% of the variability in testosterone- and DHT-glucuronide formation is explained by the predictors in the model, respectively. All other multivariate analysis output parameters are presented in Table 2.
Discussion
Distinct from other major drug-metabolizing enzymes, unusually high interindividual variability was observed for UGT2B17 in our large cohort of human liver tissue samples. The major factors impacting the observed variability include CNVs, SNPs, age, and sex. Developmental UGT2B17 gene expression and association of SNPs located in the UGT2B17 gene with its mRNA expression have been previously reported (Burgess et al., 2015; Neumann et al., 2016). Similarly, highly variable protein abundance of UGTs in adult liver is known (Fallon et al., 2013); however, our data are novel in respect to measuring protein abundance by selective LC-MS/MS proteomics, enzyme activity using two probe substrates (testosterone and DHT), and comprehensive CNV and diplotype analyses in the same set of samples. The large cohort of samples allowed comprehensive multivariate analyses, which revealed the individual contributions of many factors impacting UGT2B17 protein abundance and androgen glucuronidation activities. These protein abundance and activity data are important to predict variability in the metabolism of UGT2B17 xenobiotic substrates and sex steroids.
With respect to drug metabolism, UGT2B17 is a less studied enzyme, and no regulatory guidance for industry currently exists for this enzyme. The US Food and Drug Administration and European Medicines Agency recommend in vitro testing for the likelihood of a new chemical entity to be a substrate or inhibitor of other UGT isoforms, such as UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, UGT2B7 and UGT2B15. Our data predict that overlooking UGT2B17 could lead to clinical failure of a UGT2B17 substrate drug owing to high PK variability. Indeed, the UGT2B17 substrate MK-7246 was discontinued from clinical trials for high PK variability (Wang et al., 2012). Consistent with the literature (Gallagher et al., 2010), females have lower UGT2B17 expression levels compared with males. Likewise, the distinct ontogeny of UGT2B17 compared with the common drug-metabolizing enzymes, P450s, and other UGTs is an important finding of this study. These data predict that the use of UGT2B17 substrate drugs (e.g., vorinostat and lorcaserin) in women and children younger than 12 years could lead to supratherapeutic drug levels. We therefore recommend that UGT2B17 should be included in the in vitro UGT screening panel during early drug discovery, and caution should be taken when designing clinical studies of UGT2B17 substrate drugs in females and children along with consideration of genetic polymorphisms. UGT2B17 is expressed in other organs, such as intestine, appendix, bone marrow, and prostate; however, liver is considered an effective elimination organ for UGT2B17 substrates because of its larger size (resulting high total abundance) and high blood flow. The liver microsomes used in this study were isolated in two different laboratories, and other factors, such as medication use and storage conditions, could affect protein abundance in the microsomes; however, the lack of correlation between UGT2B17 and other proteins (e.g., UGT2B15, Supplemental Fig. S1) in the same samples indicates that the observed UGT2B17 data primarily reflect biologic or interindividual variability.
By regulating testosterone metabolism, UGT2B17 is linked to multiple pathophysiological conditions, such as obesity (Zhu et al., 2015) and prostate cancer (Barbier and Belanger, 2008; Paquet et al., 2012; Kpoghomou et al., 2013; Gauthier-Landry et al., 2015). For example, the UGT2B17 gene deletion (homozygous) is associated with decreases in fat mass (P < 0.01) and insulin sensitivity (P < 0.05) (Swanson et al., 2007), and the males with lower testosterone levels are 2.4 times more likely to be obese than males with higher testosterone levels (Mulligan et al., 2006). On the other hand, high UGT2B17 protein levels have been identified as the strongest independent molecular prognostic marker of overall survival in mutated chronic lymphocytic leukemia patients (Bhoi et al., 2016). The UGT2B17 deletion (homozygous) genotype is associated with a decreased risk of colorectal cancer in men, but it was nonpredictive in women (Angstadt et al., 2013). It is also noteworthy that association studies on UGT2B17 gene deletion and disease risks (e.g., prostate cancer) are controversial. These contradictions in literature could be explained as the published association studies do not acknowledge the effect of confounding factors other than gene deletion (e.g., SNPs and nongenetic factors) on UGT2B17 variability. Further, UGT2B15, which affects testosterone glucuronidation in UGT2B17 poor expressers, has not been considered in these association studies. Therefore, the data presented here will help in designing better clinical studies to investigate association of individual factors affecting UGT2B17 with disease risk.
Although testosterone and its glucuronides are believed to be transported by organic anion polypeptide transporters (OATPs) and multidrug resistance-associated protein (MRPs) (Hamada et al., 2008), the UGT2B17 interindividual variability is significantly greater compared with the variability in transporter abundance previously reported by us in a subset of these samples (Prasad et al., 2014, 2016). Nevertheless, genetic polymorphism in transporters should also be considered when designing clinical studies to investigate association of UGT2B17 variability with testosterone-related clinical outcomes.
UGT2B17 interindividual variability data could also be used to develop a better doping test approach to avoid false-negative or -positive test results. The urinary testosterone (T) to epitestosterone (E) ratio (T/E) has a cutoff limit of 4 and is used to detect T doping in all cases. T is metabolized by UGT2B17, and E is metabolized by UGT2B7. Individuals homozygous for the UGT2B17 deletion allele excrete negligible amounts of T in urine compared with subjects with one or two gene copies (Bao et al., 2008) and rarely reach the T/E cutoff value of 4 after T doping, indicating that genetic testing for the UGT2B17 deletion allele may increase the chances of identifying atypical findings, especially in dissecting false-negative test results. Moreover, based on our study, there should be different cutoff values for males versus females, adolescents versus adults, and different haplotypes.
The sex- or age-dependent expression of UGT2B17 may be explained by its regulation by androgens and estradiol. For example, Bao et al. demonstrated that UGT2B15 and 2B17 are androgen-regulated genes and that androgen receptor (AR) is required for both their basal and androgen-regulated expression (Bao et al., 2008). Similarly, UGT2B17 is 5-fold more abundant in metastatic versus benign prostate cancer samples (Paquet et al., 2012). UGT2B17 and myeloid cell leukemia-1 (Mcl-1) expression is upregulated in endometrial cancer (EC) tissues, and UGT2B17 depletion induces inhibition of cell growth and apoptosis in EC cells through Mcl-1 downregulation (Hirata et al., 2010); however, UGT2B17 variability within a single group (e.g., adult males) indicates the involvement of multiple other epigenetic and transcriptional mechanisms. For example, UGT2B15 and UGT2B17 are both negatively regulated by the miR-376c microRNA that binds to the 3′-UTRs of UGT2B15 and UGT2B17 mRNA in prostate cancer cells (Wijayakumara et al., 2015). It has also been shown that Forkhead Box Protein A1 (FOXA1) regulates UGT2B17 gene transcription in LNCaP prostate cancer cells (Hu et al., 2010). Similarly, polymorphic PXR and CAR are associated with altered expression of UGT2Bs, respectively (Verreault et al., 2010). Therefore, SNPs in these polymorphic transcriptional factors [e.g., -298G/G and 11193C/C in PXR (Du et al., 2013) and IVS2-99C>T in CAR (Urano et al., 2009)] can also indirectly influence UGT2B17 expression. Clearly, further research characterizing the underlying mechanisms contributing to interindividual variability in UGT2B17 abundance and activity is warranted.
Despite the significant physiologic role of testosterone, testosterone replacement therapy (TRT) is controversial. For example, a meta-analysis suggested an association of TRT with prostate cancer; however, many independent studies failed to reproduce this finding (Barbier and Belanger, 2008; Paquet et al., 2012; Kpoghomou et al., 2013; Gauthier-Landry et al., 2015). The Food and Drug Administration recently reissued a black-box warning on TRT because of its association with cardiovascular side effects. UGT2B17 variability should be considered during TRT to ensure safe and effective testosterone use. A similar strategy may also be considered to improve high-testosterone therapy (also referred to as bipolar androgen therapy) in prostate cancer patients (Schweizer et al., 2015).
Taken together, the findings of this study are of clinical importance and can be directly translated to individualize drug therapy of UGT2B17 substrates by stratifying patients based on UGT2B17 genotype and predicted phenotype. Moreover, physiologically based PK models can be developed based on these data to predict more accurately the UGT2B17-mediated glucuronidation of endobiotics and xenobiotics and translate such data to predicting in vivo disposition of these substrates.
Acknowledgments
We thank Prachi Choudhari and Mikael Boberg for assistance with LC-MS/MS sample analysis; Dr. Kenneth Thummel and Dr. Allan Rettie, UW School of Pharmacy for their valuable suggestions throughout this project; and Amy Turner, Praful Aggarwal, and Rachel Lorier from Dr. Ulrich Broeckel’s laboratory to generate a part of the genotyping data.
Abbreviations
- BSA
bovine serum albumin
- CNV
copy number variation
- DHT
dihydrotestosterone
- FPKM
fragments per kilobase per million reads
- HLM
human liver microsome
- HSA
human serum albumin
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LD
linkage disequilibrium
- LLOQ
lower limit of quantification
- LOD
limit of detection
- PK
pharmacokinetics
- P450
cytochrome P450 enzyme
- PGRN
Pharmacogenomics Research Network
- SNPs
single nucleotide polymorphisms
- T
testosterone
- UGT
uridine 5′-diphospho-glucuronosyltransferases
Authorship Contributions
Participated in research design: Bhatt, Gaedigk, Leeder, Prasad.
Conducted experiments: Bhatt, Basit, Zhang, Claw, Lee, Mehrotra, Prasad, Gaedigk, Pearce, Gaedigk, Broeckel.
Contributed new reagents or analytic tools: Chaudhry, Schuetz, Leeder, Prasad.
Performed data analysis: Bhatt, Basit, Zhang, Claw, Lee, Mehrotra, Gaedigk, Broeckel, Thornton, Prasad, Nickerson.
Wrote or contributed to the writing of the manuscript: Bhatt, Basit, Zhang, Claw, Mehrotra, Gaedigk, Lee, Pearce, Gaedigk, Broeckel, Nickerson, Thornton, Amory, Leeder, Prasad.
Footnotes
Most of this work was supported by the National Institute of Child Health and Human Development (NICHD), National Institutes of Health (NIH) grant [R01 HD081299]. The data collection was also supported by NIH grants, [U01 GM092676] and [P01 GM116691]. The NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland is funded by the NIH contract, [N01-HD-9-0011/HHSN275200900011C] and the Liver Tissue Cell Distribution System is funded by NIH contract number, [N01-DK-7-0004/HHSN267200700004C].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Angstadt AY, Berg A, Zhu J, Miller P, Hartman TJ, Lesko SM, Muscat JE, Lazarus P, Gallagher CJ. (2013) The effect of copy number variation in the phase II detoxification genes UGT2B17 and UGT2B28 on colorectal cancer risk. Cancer 119:2477–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao BY, Chuang BF, Wang Q, Sartor O, Balk SP, Brown M, Kantoff PW, Lee GSM. (2008) Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate 68:839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier O, Bélanger A. (2008) Inactivation of androgens by UDP-glucuronosyltransferases in the human prostate. Best Pract Res Clin Endocrinol Metab 22:259–270. [DOI] [PubMed] [Google Scholar]
- Beaulieu M, Lévesque E, Hum DW, Bélanger A. (1996) Isolation and characterization of a novel cDNA encoding a human UDP-glucuronosyltransferase active on C19 steroids. J Biol Chem 271:22855–22862. [DOI] [PubMed] [Google Scholar]
- Bhatt DK, Gaedigk A, Pearce RE, Leeder JS, Prasad B. (2017) Age-dependent protein abundance of cytosolic alcohol and aldehyde dehydrogenases in human liver. Drug Metab Dispos 45:1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DK, Prasad B. (2018) Critical issues and optimized practices in quantification of protein abundance level to determine inter-individual variability in DMET proteins by LC-MS/MS proteomics. Clin Pharmacol Ther 103:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoi S, Baliakas P, Cortese D, Mattsson M, Engvall M, Smedby KE, Juliusson G, Sutton LA, Mansouri L. (2016) UGT2B17 expression: a novel prognostic marker within IGHV-mutated chronic lymphocytic leukemia? Haematologica 101:e63–e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess KS, Philips S, Benson EA, Desta Z, Gaedigk A, Gaedigk R, Segar MW, Liu Y, Skaar TC. (2015) Age-related changes in microRNA expression and pharmacogenes in human liver. Clin Pharmacol Ther 98:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Luo S, Kozlovich S, Lazarus P. (2016a) Association between glucuronidation genotypes and urinary NNAL metabolic phenotypes in smokers. Cancer Epidemiol Biomarkers Prev 25:1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SM, Atchley DH, Murphy MA, Gurley BJ, Kamdem LK. (2016b) Impact of UGT2B17 gene deletion on the pharmacokinetics of 17-hydroexemestane in healthy volunteers. J Clin Pharmacol 56:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, von Moltke LL, Greenblatt DJ. (2004) UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther 310:656–665. [DOI] [PubMed] [Google Scholar]
- Du QQ, Wang ZJ, He L, Jiang XH, Wang L. (2013) PXR polymorphisms and their impact on pharmacokinetics/pharmacodynamics of repaglinide in healthy Chinese volunteers. Eur J Clin Pharmacol 69:1917–1925. [DOI] [PubMed] [Google Scholar]
- Ekström L, Johansson M, Rane A. (2013) Tissue distribution and relative gene expression of UDP-glucuronosyltransferases (2B7, 2B15, 2B17) in the human fetus. Drug Metab Dispos 41:291–295. [DOI] [PubMed] [Google Scholar]
- Fallon JK, Neubert H, Hyland R, Goosen TC, Smith PC. (2013) Targeted quantitative proteomics for the analysis of 14 UGT1As and -2Bs in human liver using NanoUPLC-MS/MS with selected reaction monitoring. J Proteome Res 12:4402–4413. [DOI] [PubMed] [Google Scholar]
- Gallagher CJ, Balliet RM, Sun D, Chen G, Lazarus P. (2010) Sex differences in UDP-glucuronosyltransferase 2B17 expression and activity. Drug Metab Dispos 38:2204–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Landry L, Bélanger A, Barbier O. (2015) Multiple roles for UDP-glucuronosyltransferase (UGT)2B15 and UGT2B17 enzymes in androgen metabolism and prostate cancer evolution. J Steroid Biochem Mol Biol 145:187–192. [DOI] [PubMed] [Google Scholar]
- Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. (2016) PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics 26:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M, Bellemare J, Hoermann G, Gleiss A, Porpaczy E, Bilban M, Le T, Zehetmayer S, Mannhalter C, Gaiger A, et al. (2013) Overexpression of uridine diphospho glucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood 121:1175–1183. [DOI] [PubMed] [Google Scholar]
- Guillemette C. (2003) Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J 3:136–158. [DOI] [PubMed] [Google Scholar]
- Hamada A, Sissung T, Price DK, Danesi R, Chau CH, Sharifi N, Venzon D, Maeda K, Nagao K, Sparreboom A, et al. (2008) Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res 14:3312–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Zaman MS, Chen Y, Ueno K, Majid S, Tripsas C, Rubin M, Chen LM, Dahiya R. (2010) Function of UDP-glucuronosyltransferase 2B17 (UGT2B17) is involved in endometrial cancer. Carcinogenesis 31:1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DG, Gardner-Stephen D, Severi G, Gregory PA, Treloar J, Giles GG, English DR, Hopper JL, Tilley WD, Mackenzie PI. (2010) A novel polymorphism in a forkhead box A1 (FOXA1) binding site of the human UDP glucuronosyltransferase 2B17 gene modulates promoter activity and is associated with altered levels of circulating androstane-3α,17β-diol glucuronide. Mol Pharmacol 78:714–722. [DOI] [PubMed] [Google Scholar]
- Hu DG, Selth LA, Tarulli GA, Meech R, Wijayakumara D, Chanawong A, Russell R, Caldas C, Robinson JL, Carroll JS, et al. (2016) Androgen and estrogen receptors in breast cancer coregulate human UDP-glucuronosyltransferases 2B15 and 2B17. Cancer Res 76:5881–5893. [DOI] [PubMed] [Google Scholar]
- Kpoghomou MA, Soatiana JE, Kalembo FW, Bishwajit G, Sheng W. (2013) UGT2B17 polymorphism and risk of prostate cancer: a meta-analysis. ISRN Oncol 2013:465916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus P, Zheng Y, Runkle EA, Muscat JE, Wiener D. (2005) Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics 15:769–778. [DOI] [PubMed] [Google Scholar]
- Luo S, Chen G, Truica C, Baird CC, Leitzel K, Lazarus P. (2017) Role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenomics J DOI: 10.1038/tpj.2017.18 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan T, Frick MF, Zuraw QC, Stemhagen A, McWhirter C. (2006) Prevalence of hypogonadism in males aged at least 45 years: the HIM study. Int J Clin Pract 60:762–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann E, Mehboob H, Ramírez J, Mirkov S, Zhang M, Liu W. (2016) Age-dependent hepatic UDP-glucuronosyltransferase gene expression and activity in children. Front Pharmacol 7:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S, Fukami T, Yokoi T, Nakajima M. (2015) A comprehensive review of UDP-glucuronosyltransferase and esterases for drug development. Drug Metab Pharmacokinet 30:30–51. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, et al. (2012) Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 40:83–92. [DOI] [PubMed] [Google Scholar]
- Pâquet S, Fazli L, Grosse L, Verreault M, Têtu B, Rennie PS, Bélanger A, Barbier O. (2012) Differential expression of the androgen-conjugating UGT2B15 and UGT2B17 enzymes in prostate tumor cells during cancer progression. J Clin Endocrinol Metab 97:E428–E432. [DOI] [PubMed] [Google Scholar]
- Pearce RE, Gaedigk R, Twist GP, Dai H, Riffel AK, Leeder JS, Gaedigk A. (2016) Developmental expression of CYP2B6: a comprehensive analysis of mRNA expression, protein content and bupropion hydroxylase activity and the impact of genetic variation. Drug Metab Dispos 44:948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, Ambudkar SV, Unadkat JD. (2014) Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos 42:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Gaedigk A, Vrana M, Gaedigk R, Leeder JS, Salphati L, Chu X, Xiao G, Hop C, Evers R, et al. (2016) Ontogeny of hepatic drug transporters as quantified by LC-MS/MS proteomics. Clin Pharmacol Ther 100:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeque AJ, Usmani KA, Palamar S, Cerny MA, Chen WG. (2012) Identification of human UDP-glucuronosyltransferases involved in N-carbamoyl glucuronidation of lorcaserin. Drug Metab Dispos 40:772–778. [DOI] [PubMed] [Google Scholar]
- Schulze JJ, Lundmark J, Garle M, Skilving I, Ekström L, Rane A. (2008) Doping test results dependent on genotype of uridine diphospho-glucuronosyl transferase 2B17, the major enzyme for testosterone glucuronidation. J Clin Endocrinol Metab 93:2500–2506. [DOI] [PubMed] [Google Scholar]
- Schweizer MT, Antonarakis ES, Wang H, Ajiboye AS, Spitz A, Cao H, Luo J, Haffner MC, Yegnasubramanian S, Carducci MA, et al. (2015) Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med 7:269ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2016) Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J 16:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Chen G, Dellinger RW, Sharma AK, Lazarus P. (2010) Characterization of 17-dihydroexemestane glucuronidation: potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet Genomics 20:575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson C, Mellström D, Lorentzon M, Vandenput L, Jakobsson J, Rane A, Karlsson M, Ljunggren O, Smith U, Eriksson AL, et al. (2007) The uridine diphosphate glucuronosyltransferase 2B15 D85Y and 2B17 deletion polymorphisms predict the glucuronidation pattern of androgens and fat mass in men. J Clin Endocrinol Metab 92:4878–4882. [DOI] [PubMed] [Google Scholar]
- Tanner JA, Prasad B, Claw KG, Stapleton P, Chaudhry A, Schuetz EG, Thummel KE, Tyndale RF. (2017) Predictors of variation in CYP2A6 mRNA, protein, and enzyme activity in a human liver bank: influence of genetic and nongenetic factors. J Pharmacol Exp Ther 360:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano T, Usui T, Shiraki M, Ouchi Y, Inoue S. (2009) Association of a single nucleotide polymorphism in the constitutive androstane receptor gene with bone mineral density. Geriatr Gerontol Int 9:235–241. [DOI] [PubMed] [Google Scholar]
- Verreault M, Kaeding J, Caron P, Trottier J, Grosse L, Houssin E, Pâquet S, Perreault M, Barbier O. (2010) Regulation of endobiotics glucuronidation by ligand-activated transcription factors: physiological function and therapeutic potential. Drug Metab Rev 42:110–122. [DOI] [PubMed] [Google Scholar]
- Vrana M, Whittington D, Nautiyal V, Prasad B. (2017) Database of optimized proteomic quantitative methods for human drug disposition-related proteins for applications in physiologically based pharmacokinetic modeling. CPT Pharmacometrics Syst Pharmacol 6:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Trucksis M, McElwee JJ, Wong PH, Maciolek C, Thompson CD, Prueksaritanont T, Garrett GC, Declercq R, Vets E, et al. (2012) UGT2B17 genetic polymorphisms dramatically affect the pharmacokinetics of MK-7246 in healthy subjects in a first-in-human study. Clin Pharmacol Ther 92:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayakumara DD, Hu DG, Meech R, McKinnon RA, Mackenzie PI. (2015) Regulation of human UGT2B15 and UGT2B17 by miR-376c in prostate cancer cell lines. J Pharmacol Exp Ther 354:417–425. [DOI] [PubMed] [Google Scholar]
- Wong NS, Seah EZh, Wang LZ, Yeo WL, Yap HL, Chuah B, Lim YW, Ang PC, Tai BC, Lim R, et al. (2011) Impact of UDP-gluconoryltransferase 2B17 genotype on vorinostat metabolism and clinical outcomes in Asian women with breast cancer. Pharmacogenet Genomics 21:760–768. [DOI] [PubMed] [Google Scholar]
- Yuan LM, Gao ZZ, Sun HY, Qian SN, Xiao YS, Sun LL, Zeng S. (2016) Inter-isoform hetero-dimerization of human UDP-glucuronosyltransferases (UGTs) 1A1, 1A9, and 2B7 and impacts on glucuronidation activity. Sci Rep 6:34450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AZ, Cox LS, Ahluwalia JS, Renner CC, Hatsukami DK, Benowitz NL, Tyndale RF. (2015) Genetic and phenotypic variation in UGT2B17, a testosterone-metabolizing enzyme, is associated with BMI in males. Pharmacogenet Genomics 25:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]