Abstract
The oral fissure is immensely inhabited with a number of polymicrobial colonies similar to the intestinal system. Periodontitis is a dysbiotic disease resulting from deviation in subgingival Gram-positive bacteria to Gram-negative bacteria shift from Gram-positive bacteria. The development of periodontal dysbiosis occurs over a broadened timeframe, which slowly turns the symbiotic association of host and microbe to pathogenic. This review highlights a recent paradigm of periodontitis progression has been postulated which challenges the traditional concept of periodontitis being induced by few particular periopathogens such as belonging to red complex, but by a more comprehensive dysbiotic-synergistic community.
Key words: Dysbiosis, keystone pathogens, polymicrobial synergy, polymicrobial synergy and dysbiosis model
INTRODUCTION
The infant, from birth, is exposed to extensive microbiota acquired principally from the mother, which get colonized though only a few are able to manifest affluently. The colonization, growth, and predominance of the microorganisms and among them which will be crucial or trivial parts of the occupant microbiota of a site is controlled by the organic properties of every natural surrounding which brings about various surfaces having particular yet trademark microbiotas.[1,2,3,4,5] The human microbes and the host have co-advanced to have a harmonious or reciprocal relationship.[6] The resident microorganisms contribute to nutrition, food digestion, differentiation of the host mucosa, regulation of human metabolism, development of immunity and its role, and aversion of colonization by different exogenic, infectious microorganisms, and consequently pick up warm, secure, and nutritious natural surroundings from the host.[7] The association among the resident microbiota and the host is constantly changing or dynamic.
THE BENEFICIAL MICROBIOTA
The oral fissure is immensely inhabited with a number of polymicrobial colonies because of the skilled ability of microbes to acclimatize to various niches through increased rates of recombination of genes.
Robust commensal bacterial colonization prevents the pathogenic bacterial colonization by a process called as colonization resistance. Commensal microbiota not just solely shields the host by colonization of the niches, but its interaction with the host also boosts the appropriate tissue structure development and function.[8] Colonization resistance is an essential objective of commensal microbiota, and broad-spectrum antibiotics usage have documented to have detrimental effects on commensal depletion.[9,10] Most of the significant studies with respect to the intestine have shown the ability of polymicrobial commensal communities to actively mature proper tissue structure and function.[11,12,13] The oral fissure of humans is analogous to the enteral system and contains highly evolved polymicrobial communities, and also has a state of “controlled” inflammation.[14,15] One of the crucial defensive characteristics of periodontium tissue is neutrophil migration into the surrounding tissue which is highly regulated such that both extreme and scarce migration can both result in disease.[16] Studies have additionally demonstrated the impact of bacterial colonization of the periodontium is more intricate than that ascertained in enteral tissue.[17] Two autonomous examinations have discovered that commensal colonization altogether significantly expands the quantity of neutrophils found in the gingival tissue of sans germ mice.[18,19] Hence, a vital defensive mechanism established in periodontium tissue, the passage of polymorphoneutrophils into the gingival tissue, is coordinated by both host formative and bacterial prompted programs. Moreover, it has been discovered that the chemokine ligands, drawing in and coordinating neutrophil movement in periodontal tissue of the mouse, too are customized by both host and microbially incited programs. Nonetheless, in such circumstances, the host developing affair is accorded with chemokine ligand 1 (C-X-C motif) (CXCL1) while commensal microbial colonization, particularly initiates the dissemination of the intently associated neutrophil chemokine ligand CXCL2 through the differentiation of toll-like receptor and myeloid primary response gene 88 activation pathway. Hence, it evidently indicates that in healthy periodontium the commensal community of the oral cavity contributes remarkably to its innate immunity; however, their bequest is stratified on the developmental programs of the host.[19,20]
DYSBIOSIS
Dysbiosis means symbiosis gone astray. According to this concept, few diseases are caused because of diminishing quantity of advantageous symbionts as well as an expansion in the quantity of pathogenic life forms.[21] Rather than causative to healthy functioning of the tissues, the polymicrobial oral community meddles with the functioning of normal periodontal tissue, and hence normal tissue function is disturbed. Periodontitis is a dysbiotic disease resulting from deviation in subgingival Gram-positive bacteria to Gram-negative bacteria.[21,22]
In humans, other than dental plaque periodontitis, risk factors such as environmental or genetic can produce dysbiosis. Ecological factors such as obesity or smoking have been reported to be cohorted with subgingival dysbiotic colonies. Smoking, de facto, creates the healthy subgingival microbiome to more intimately simulate pathogenic sites, advocating that it creates a risky environment for periodontitis. The relationship between diabetes and periodontitis is well documented and has been hypothesized to be confederated with dysbiosis in the periodontal communities. Regarding human genetics, studies still have to assess whether polymorphisms in the nucleotide of pathogenic bacteria of the host and various other genetic risk facets are associated with dysbiosis in the periodontal community.[23,24]
The development of periodontal dysbiosis occurs over a broadened timeframe, which slowly turns the symbiotic association of host and microbe to pathogenic. Among microbial complexes,first such complex that has been related to disease is the orange complex consisting of anaerobic Gram-negative species such as Prevotella intermedia, Prevotella nigrescens, Prevotella micros, and Fusobacterium nucleatum, which on disease progression shifts toward red complex consisting of Tannerella forsythia, Tannerella Denticola, and Porphyromonas gingivalis.[25]
Nonetheless, this paradigm has been impugned. For example, in 2009, it was discovered by Riep et al. that known periodontal pathogenic microbes such as T. forsythia and P. gingivalis could often be retrieved from healthy gingival tissues.[26] Kumar et al. in 2006 directly contradicted the current pattern as they could contemplate contrasting association between Veillonella, a Gram-negative microbe, and healthy periodontium; and Filifactor alocis, an anaerobic Gram-positive organism with periodontal disease.[27] Synergism between pathogenic bacteria and viruses such as Epstein–Barr virus, human cytomegalovirus, and species of herpetic viruses has also been hypothesized. This suggests that, other than bacterial causation, aspects such as bacterial–viral coinfection, genetic, and immunological factors too contribute to periodontitis.
RE-VISITING KOCH'S POSTULATES
Koch's postulates though serving excellently to medical microbiologists have posed certain limitations considering chronic infectious diseases. Nonetheless, this concern may be resolved by two notions.
Pathogenic microbial community
In 2009, Siqueira and Rocas explained the first concept of pathogenic microbial community.[28] The authors suggested “community as pathogen” paradigm rather than customary “single pathogen” paradigm to approach the polymicrobial etiology of periodontitis. Hence, bacterial communities as a whole derived from diseased and periodontally healthy sites could be assessed for “pathogenic genes,” which could be then correlated with periodontitis.[25]
Hill's criteria of casualty
This is the second notion or concept to tackle the limitations of Koch's postulates. The relation between Helicobacter pylori and peptic ulcer is accepted worldwide because of its fulfillment of Hill's causality criteria and not Koch's postulates.[29] The establishment of causation of periodontal disease would be more facile if there is amalgamation of prevailing research and pathogenic microbial dysbiotic community concept with criteria of causality by Hill as given in Table 1.[25]
Table 1.
Causality criteria for periodontitis by Hill (Source: Nath and Raveendran[25])

KEYSTONE PATHOGENS
Indeed during the dawn of the 21st century, the liaison of microorganisms inhabiting the gingival sulcus was profoundly studied and the “red complex” hypothesis was staunchly followed until the hypothesis of “keystone pathogen” was proposed by Hajishengallis et al., which further excogitated to the theory of polymicrobial synergy and dysbiosis.[30] This theory proposes that the homeostasis of the periodontal tissue can be disrupted by little or abundant keystone pathogenic species through both characteristic and assessable variations to the commensal microflora. The keystone or keystone-like pathogens include microbes such as P. gingivalis and T. forsythia, of which the effect of P. gingivalis on downregulation of host responses and elevating the virulence of the entire microbial community is most extensively studied as it is a prime organism associated with chronic severity of the periodontal disease.[31] Keystone pathogens communicate with the accessory pathogens facilitating synergism and progression toward pathogenesis.
Moore et al.,1982, in early bacteriological studies noted the occurrence of P. gingivalis in low abundance in plaque associated with periodontitis. Along with this the authors also reported an apparent paradox with respect to P. gingivalis and its role in an inflammatory disease. P. gingivalis, independently, could not strongly promote inflammation.[32] This paradox was recently explained by a study performed in mice suggesting a disease model accordant with the alternate perspective of shift in the microbiota associated with periodontitis. The study reported that innate immunity can be impaired by P. gingivalis in manners that can influence the alteration of the microbial composition and its growth in periodontal tissues. Specifically, it was found that P. gingivalis, even at lower concentration, could transform a symbiotic community to a pathogenic dysbiotic one triggering loss of bone by inflammation. P. gingivalis though could colonize the host (germfree mice) was unable to produce periodontitis in the absenteeism of commensal microbes. This capacity of P. gingivalis, although being characteristically a smaller component of bacterial community, to produce a community-wide impact tipping the balance toward a more pathogenic dysbiosis has earned it the title of a keystone pathogen.[30,33]
P. gingivalis exerts its keystone effects through bacterial synergy as well as modulation of the host. In terms of host modulation, P. gingivalis facilitates the colonization and growth of other organisms, for example, F. nucleatum, by delaying neutrophil recruitment by transiently inhibiting the initiation of chemokines like gingival IL8 and T-cell chemokine-like IP 10 [Figure 1]. It has also shown to affect the function of neutrophils by coactivating toll-like receptor (TLR) 2 and C5aR [Figure 2]. However, the persistence of P. gingivalis in the periodontium is dependent on the instigation of incendiary crosstalk seen between receptor of complement C5a and TLR 2 and also the ability of its gingipains to produce C5 convertase activity, which has shown to retard the annihilating ability of leukocytes. This was substantiated by a study in which dysbiosis and periodontitis could not be caused by P. gingivalis in C5aR sans host (mice).[30,31,33]
Figure 1.
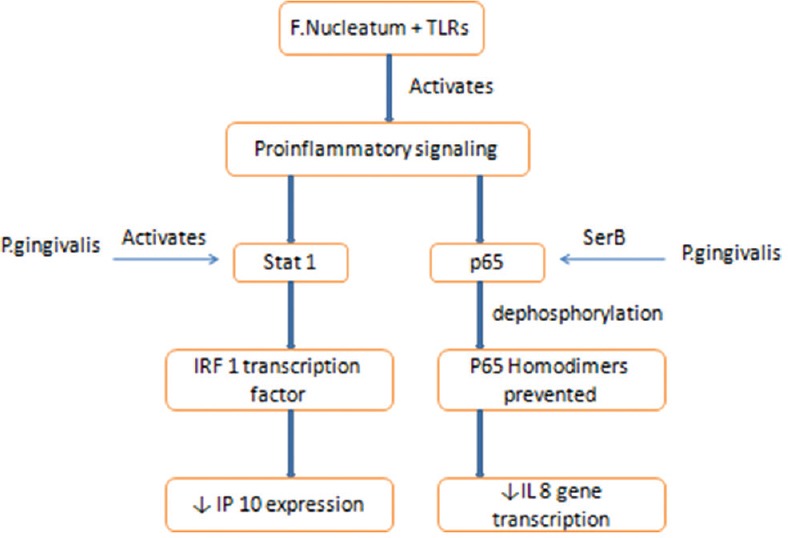
Downregulation of chemokines by Porphyromonas gingivalis. TLR – Toll like receptors; F. Nucleatum – Fusobacterium nucleatum; IRF – Interferon-Regulatory Factor; IP – Interferon gamma-induced protein; IL – Interleukin
Figure 2.
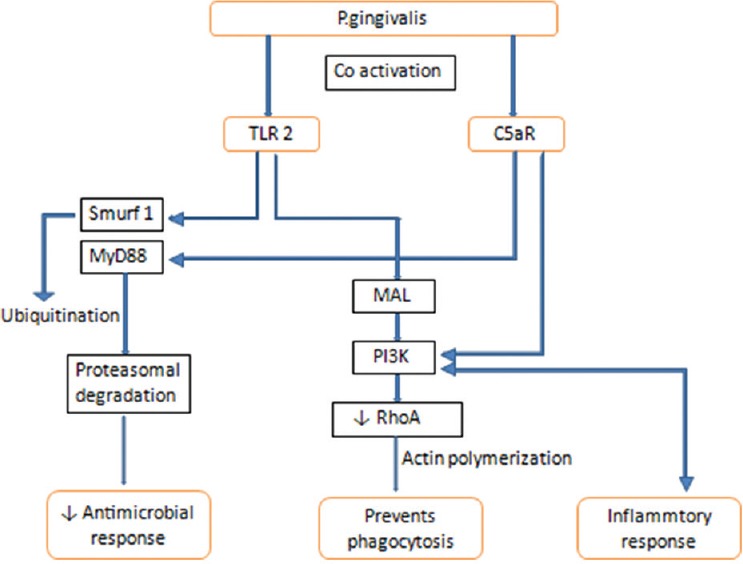
Disruption of neutrophil function and dysbiosis by Porphyromonas gingivalis. TLR – Toll like receptors; MAL – MyD88-Adaptor Like; C5aR – Complement 5a Receptor; Smurf – Smad ubiquitination regulatory factor; P13K – Phosphoinositide 3-kinase; RhoA – Ras homolog gene family, member A; MyD88 – Myeloid differentiation primary response 88
Pathobionts, the preceding underacknowledged species, have unveiled good or even better relationship with periodontitis that the traditional red complex species.[31]
One such pathobiont is F. alocis. It is identified to be a fastidious, asaccharolytic, Gram-positive, obligate anaerobe inhabiting the gingival sulcus. This organism is difficult to culture because of it slow-growing ability and hence cannot be detected by traditional culture-based methodologies. Nonetheless, through new culture-independent techniques, it has been discovered abundantly in the periodontal pockets. Kumar et al., in 2005, detected F. alocis in increased quantities than other gram-negative bacteria.[34] F. alocis might play a significant part in periodontitis pathogenesis as it can resist oxidative stress in the periodontal pocket, stimulating its increased growth in such conditions and also allowing it to exist with other periodontal species through genes coding for a highly evolved metabolic pathway of amino acid. Likewise, it also has the ability to form a synergistic polymicrobial community with various other microorganisms which enhances its capacity to invade the host tissues and cause chronic periodontal inflammation by triggering the production of pro-inflammatory cytokines such as tumor necrosis factor α, interleukin (IL) 1 β, and IL 6.[34,35] A co-culture of F. alocis and P. gingivalis showed increased adherence and invasion of gingival epithelial cells. Furthermore, F. nucleatum appeared to positively affect the colonization of F. alocis, whereas the presence of Streptococcus gordonii was found to be antagonistic. The relation between F. alocis and Aggregatibacter actinomycetemcomitans was found to be strain specific.[35,36]
COMMUNITY FORMED BY BACTERIAL SYNERGISM: THE REAL CULPRIT
From various studies, it has become apparent that pathogenic periodontal microbes exert their pathogenicity only in environmental conditions favoring synergism. Likewise, contribution is seen within various Gram-positive as well as Gram-negative bacteria in the progression of periodontal disease where they either incite or endure inflammation or allocate any other beneficial support to this microbial community. These interbacterial species through signaling among them modulate opportunistic actions such as expression of genes, exchange of DNA, and also attainment of nourishment. In this manner, polymicrobial synergism can be displayed by the bacterial communities.[33]
As proved earlier P. gingivalis in incapable of pathogenicity alone and hence requires a community for exerting its detrimental effects P. intermedia by producing Interpain A and P. gingivalis using its HmuY hameophore work in synchrony to obtain haem from hemoglobin. Furthermore, co-culture of P. gingivalis and T. denticola has shown increased pathogenicity in animal models exhibiting increase alveolar bone loss. P. gingivalis also shows adherence to an accessory pathogen such as S. gordonii causing an increased virulence and bone loss. P. gingivalis, T. forsythia, and P. intermedia have reported to synergistically downregulate the classical, alternate as well as lectin pathway by releasing proteases that deteriorates C3-C4 mannose-binding protein or ficolins.[31,37]
Comparatively, a few organisms have found to be antagonistic to P. gingivalis. For instance, the correlation between S. cristatus and P. gingivalis extenuates the bone loss caused by P. gingivalis in mice as S. cristatus downregulates the expression of fimbriae gene in P. gingivalis.[31]
THE POLYMICROBIAL SYNERGY AND DYSBIOSIS MODEL
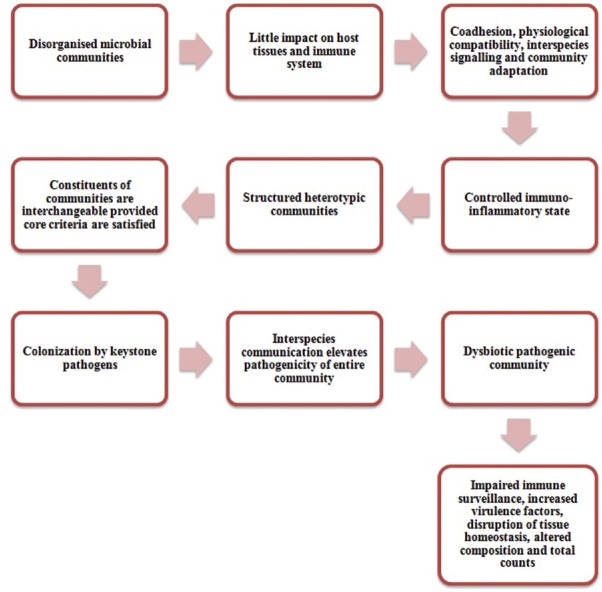
A recent paradigm of periodontitis progression has been postulated which challenges the traditional concept of periodontitis being induced by few particular periopathogens such as belonging to RED COMPLEX, but by a more comprehensive dysbiotic-synergistic community. These heterogenous microbiota inhabiting the periodontium or distinct gene integrations execute discrete functions that shape and stabilitate the infection eliciting pathogens. Consequently, there will be several fundamental prerequisites for a conceivably pathogenic assemblage to emerge (i) To form a diverse community, microbial components will exhibit the appropriate receptors and adhesions, (ii) each representative of the assemblage will be co-operative and harmonious, and (iii) the assemblage as a whole will withstand the inherent as well as attained host immunity and add to the inflammation of the periodontal tissues. It is quite pertinent that subgingival plaque procured from the healthy periodontal sites possess the same potential as that obtained from the diseased sites to produce robust inflammation through the activation of toll-like receptors. However, it does not eventuate always as it requires certain keystone pathogens such as P. gingivalis that will impede the host's immune response and boost the pathogenic potential of the whole community. Hence, the complete comprehension of the pathogenesis of periodontitis based on the polymicrobial synergy and dysbiosis model may provide newer and better therapeutic interventions.[33]
The causation of periodontitis by the diverse bacteria in gingival sulcus as explained by the polymicrobial synergy and dysbiosis model [Figure 3].[33]
Figure 3.
The polymicrobial synergy and dysbiosis model of periodontal disease etiology
CONCLUSION
As it appears from the current literature, periodontitis to a certain extent is caused by a deflection from the harmonious symbiotic bacterial community to a dysbiotic one. Rising attainments have demonstrated that a single organism is no good in causing pathogenesis, what instigates the disease process is the activity of community as a whole. However, other factors related to hereditary, immune system, and ecological conditions must also be researched. New strategies for bacterial counteraction have also been suggested by the polymicrobial synergy and dysbiosis model which mainly target P. gingivalis. Salivary diagnostics can be used to identify these organisms which can then be eliminated by target antibiotics. However, the complete eradication of organisms such as P. gingivalis is not possible as this organism is deep-seated in biofilms as well as epithelial cells. Hence, management of the entire community is a more logical choice and therefore scaling and root planing still remains the gold standard. In addition, influencing the growth of beneficial microbes using probiotics and also antagonist organisms to harbor the periodontal tissues will disrupt the growth of P. gingivalis and other accessory microbes. Boosting the host immune response by modulating the immunity such as neutralizing the complement pathways in the periodontal tissues will prevent the propagation of the disease and restore the tissues to health. Altogether, both clinicians as well as researchers aim at finding the optimum therapeutics for the patients. An amalgamation of research and clinical work to bring forth efficacious treatment modalities that can penetrate the complex biofilm as well as modulate the host response and at the same time be simple and cost-effective to the patients is currently the need of hour.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
We would like to thank Dr. Vinayak Joshi for his valuable insight and assistance that greatly improved the manuscript.
REFERENCES
- 1.Sampaio-Maia B, Monteiro-Silva F. Acquisition and maturation of oral microbiome throughout childhood: An update. Dent Res J (Isfahan) 2014;11:291–301. [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen GJ, Brüggemann H. Bacterial skin commensals and their role as host guardians. Benef Microbes. 2014;5:201–15. doi: 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 3.Cogen AL, Nizet V, Gallo RL. Skin microbiota: A source of disease or defence? Br J Dermatol. 2008;158:442–55. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: Health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ, et al. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–74. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Relman DA. The human microbiome: Ecosystem resilience and health. Nutr Rev. 2012;70(Suppl 1):S2–9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brook I. Bacterial interference. Crit Rev Microbiol. 1999;25:155–72. doi: 10.1080/10408419991299211. [DOI] [PubMed] [Google Scholar]
- 10.He X, McLean JS, Guo L, Lux R, Shi W. The social structure of microbial community involved in colonization resistance. ISME J. 2014;8:564–74. doi: 10.1038/ismej.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 13.Umesaki Y, Setoyama H. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2000;2:1343–51. doi: 10.1016/s1286-4579(00)01288-0. [DOI] [PubMed] [Google Scholar]
- 14.Kornman KS, Page RC, Tonetti MS. The host response to the microbial challenge in periodontitis: Assembling the players. Periodontol 2000. 1997;14:33–53. doi: 10.1111/j.1600-0757.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 15.Page RC, Schroeder HE. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab Invest. 1976;34:235–49. [PubMed] [Google Scholar]
- 16.Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, et al. The leukocyte integrin antagonist del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol. 2012;13:465–73. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10:302–6. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukamoto Y, Usui M, Yamamoto G, Takagi Y, Tachikawa T, Yamamoto M, et al. Role of the junctional epithelium in periodontal innate defense and homeostasis. J Periodontal Res. 2012;47:750–7. doi: 10.1111/j.1600-0765.2012.01490.x. [DOI] [PubMed] [Google Scholar]
- 19.Zenobia C, Luo XL, Hashim A, Abe T, Jin L, Chang Y, et al. Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cell Microbiol. 2013;15:1419–26. doi: 10.1111/cmi.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart TC, Shapira L, Van Dyke TE. Neutrophil defects as risk factors for periodontal diseases. J Periodontol. 1994;65:521–9. doi: 10.1902/jop.1994.65.5s.521. [DOI] [PubMed] [Google Scholar]
- 21.Berezow AB, Darveau RP. Microbial shift and periodontitis. Periodontol 2000. 2011;55:36–47. doi: 10.1111/j.1600-0757.2010.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–71. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 23.Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000. 2013;62:59–94. doi: 10.1111/j.1600-0757.2012.00457.x. [DOI] [PubMed] [Google Scholar]
- 24.Stabholz A, Soskolne WA, Shapira L. Genetic and environmental risk factors for chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:138–53. doi: 10.1111/j.1600-0757.2010.00340.x. [DOI] [PubMed] [Google Scholar]
- 25.Nath SG, Raveendran R. Microbial dysbiosis in periodontitis. J Indian Soc Periodontol. 2013;17:543–5. doi: 10.4103/0972-124X.118334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riep B, Edesi-Neuss L, Claessen F, Skarabis H, Ehmke B, Flemmig TF, et al. Are putative periodontal pathogens reliable diagnostic markers? J Clin Microbiol. 2009;47:1705–11. doi: 10.1128/JCM.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL, et al. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–73. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siqueira JF, Jr, Rôças IN. Community as the unit of pathogenicity: An emerging concept as to the microbial pathogenesis of apical periodontitis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:870–8. doi: 10.1016/j.tripleo.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 29.Lowe AM, Yansouni CP, Behr MA. Causality and gastrointestinal infections: Koch, Hill, and Crohn's. Lancet Infect Dis. 2008;8:720–6. doi: 10.1016/S1473-3099(08)70257-3. [DOI] [PubMed] [Google Scholar]
- 30.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–83. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR, et al. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1982;38:1137–48. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–19. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–55. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aruni AW, Roy F, Fletcher HM. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun. 2011;79:3872–86. doi: 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. Oral community interactions of Filifactor alocis in vitro. PLoS One. 2013;8:e76271. doi: 10.1371/journal.pone.0076271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendrickson EL, Beck DA, Miller DP, Wang Q, Whiteley M, Lamont RJ, et al. Insights into dynamic polymicrobial synergy revealed by time-coursed RNA-Seq. Front Microbiol. 2017;8:261. doi: 10.3389/fmicb.2017.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]