Abstract
The histone H3 lysine 36 methyltransferase SETD2 is frequently mutated in various cancers, including leukemia. However, there has not been any functional model to show the contribution of SETD2 in hematopoiesis or the causal role of SETD2 mutation in tumorigenesis. In this study, using a conditional Setd2 knockout mouse model, we show that Setd2 deficiency skews hematopoietic differentiation and reduces the number of multipotent progenitors; although the number of phenotypic hematopoietic stem cells (HSCs) in Setd2-deleted mice is unchanged, functional assays, including serial BM transplantation, reveal that the self-renewal and competitiveness of HSCs are impaired. Intriguingly, Setd2-deleted HSCs, through a latency period, can acquire abilities to overcome the growth disadvantage and eventually give rise to hematopoietic malignancy characteristic of myelodysplastic syndrome. Gene expression profile of Setd2-deleted hematopoietic stem/progenitor cells (HSPCs) partially resembles that of Dnmt3a/Tet2 double knockout HSPCs, showing activation of the erythroid transcription factor Klf1-related pathway, which plays an important role in hematopoietic malignant transformation. Setd2 deficiency also induces DNA replication stress in HSCs, as reflected by an activated E2F gene regulatory network and repressed expression of the ribonucleotide reductase subunit Rrm2b, which results in proliferation and cell cycle abnormalities and genomic instability, allowing accumulation of secondary mutation(s) that synergistically contributes to tumorigenesis. Thus, our results demonstrate that Setd2 is required for HSC self-renewal, and provide evidence supporting the causal role of Setd2 deficiency in tumorigenesis. The underlying mechanism shall advance our understanding of epigenetic regulation of cancer and provide potential new therapeutic targets.
Introduction
Hematopoietic homeostasis relies on a balance between hematopoietic stem cell (HSC) self-renewal and differentiation. This process is regulated cooperatively by both genetic and epigenetic mechanisms.1, 2 Accumulating cancer genomic data suggest that the mutations in some key regulatory genes may perturb this homeostasis and eventually cause hematopoietic malignancies. The most frequently mutated genes roughly fell into several categories: transcription factors (e.g., RUNX1, CEBPα), cellular signal transducers (e.g., JAK2, RAS), chromatin/epigenetic regulators (e.g., DNMT3A, TET2), and mRNA splicing/translation-related proteins (e.g., SF3B1, RPS14).3–5 Consistent with the fact that the hematopoietic malignancies are highly heterogeneous groups of diseases, the mechanisms by which the mutated genes cause these diseases are diverse. Many mutated genes contribute to tumorigenesis by conferring on the cells enhanced self-renewal and proliferation capacities,6 which represent a growth advantage over normal cells. However, an intriguing observation is that some mutated genes apparently reduce the self-renewal and proliferation capacities, but still lead to malignancies eventually.7–10 One support of this notion is the “Dameshek’s Riddle”, a paradox first described by Dameshek in 1967, in which some patients who initially develop aplastic anemia have a high risk of leukemia later in their life.11 This paradox currently has been more broadly applicable in clinical representations of hematopoietic malignancies, such as myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN) and acute leukemia. While the underlying mechanism remains elusive, it is conceivable that some of the original abnormal cell clones may intrinsically be able to gain an uncontrolled advantage of proliferation through time, thereby developing into cancer.
The epigenetic regulator SETD2 has been found being frequently mutated in several types of leukemia,12–15 as well as various solid tumors.16, 17 SETD2 is the major enzyme responsible for histone H3 lysine 36 trimethylation (H3K36me3),18–20 which is implicated in many aspects of genome regulation, including DNA methylation,21, 22 nucleosome dynamics,23 DNA repair,24 suppression of intragenic transcriptional initiation,25, 26 transcriptional elongation,27–29 and alternative mRNA splicing.30 Furthermore, SETD2 catalyzes microtubulin methylation during mitosis, suggesting another layer of the mechanism regulating genome stability.31 Setd2 constitutive knockout (KO) mice are embryonic lethal due to defects in blood vessel development.18 In mouse embryonic stem cells, Setd2 is required for endoderm differentiation.32 Recently, intratumor heterogeneity studies of kidney cancer revealed that SETD2 is one of the earliest mutated tumor suppressors and that its loss-of-function promotes cancer evolution through DNA replication stress (RS) and impaired genome integrity.33, 34 Importantly, in agreement with the role of SETD2 in DNA replication, inhibition of WEE1, a coordinator of the transition between DNA replication and mitosis, exerts a synthetic lethal effect with SETD2 deficiency in cancer cells, implying a great potential for targeted therapy.35 However, there has not been any functional model to show the contribution of SETD2 in hematopoiesis or the causal role of SETD2 mutation in tumorigenesis. In this study, we generated hematopoietic-specific Setd2 KO mice and found that Setd2 plays an important role in maintaining the balance between HSC self-renewal and differentiation and that the Setd2-deficient HSPCs can consistently evolve into hematopoietic malignancies. This study thus provides the first animal model that demonstrates the causal role of SETD2 deficiency in tumorigenesis.
Results
Generation of the conditional Setd2 KO mouse
To construct inducible, hematopoietic tissue-specific Setd2 KO mice, we inserted a LoxP-Lox2272 flanked, reversed IRES-EGFP-polyA cassette into the first intron of Setd2. When expressing a Cre enzyme, this cassette can be inverted in the genome, leading to Setd2 promoter-driven EGFP expression and termination of the Setd2 transcription. The EGFP expression thus not only indicates the deletion of Setd2 but also serves as a reporter for the endogenous Setd2 expression (Supplementary information, Figure S1A). This mouse (Setd2FL/FL) was then crossed with our previously reported Setd2+/− conventional KO mice18 and Mx1-Cre mice to generate Setd2FL/−Cre+ (KO) and Setd2FL/+Cre−(wild type; WT) mice. Upon injection of Polyinosinic–polycytidylic acid (pI–pC), the Mx1 promoter-driven Cre was induced and EGFP was detected in different hematopoietic compartments of Setd2 KO mice (Supplementary information, Figure S1B). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis showed that mRNA level of Setd2 was significantly downregulated in KO LSK (Lineage− c-Kit+ Sca1+) cells (Supplementary information, Figure S1C). Meanwhile, a dramatic reduction of H3K36me3 was detected in KO LSK and HSC (slam-LSK: CD150+ CD48−Lin−c-Kit+ Sca1+), compared with that of WT mice (Supplementary information, Figure S1D and E). These results indicate an efficient deletion of Setd2 in KO mice.
Setd2 deletion leads to a differentiation bias and a decrease of MPPs
RT-qPCR analysis of different bone marrow (BM) hematopoietic cells revealed that Setd2 is broadly expressed, though relatively lower in granulo-monocytes (GM, Gr1+Mac1+) (Fig. 1a), implying that Setd2 deletion may affect multiple-hematopoietic compartments. We first performed a routine examination of the peripheral blood (PB) at 4–5 weeks after pI–pC induction. The results showed that KO mice had a decreased white blood cell (WBC) count (Fig. 1b), but normal red blood cell (RBC), hemoglobin (HGB) and platelet (PLT) counts (Supplementary information, Figure S2A-C). The reduced WBC was likely due to a reduction of B cells, as indicated by flow cytometry analysis (FACS) (Supplementary information, Figure S2D and E). Furthermore, B cell reduction was also observed in KO spleen (Supplementary information, Figure S2F). In contrast, the T cell number of KO mice was not significantly changed (although the percentage of T cells was relative higher in KO PB), whereas the number of myeloid cells was slightly increased (Fig. 1c). Although their T cell number in PB was unchanged, KO mice had fewer cells in thymus and the developmental stages of thymus T cells were altered, as indicated by FACS analysis, suggesting a minor defect in T cell development (Supplementary information, Figure S2G-J). In contrast to the reduced WBC number in PB, total BM cell count was normal in KO mice (Supplementary information, Figure S2K). However, the proportional changes of B and GM cells were observed (i.e., decreased B and increased GM cells) (Fig. 1d; Supplementary information, Figure S2L). We also performed colony-forming unit (CFU) assays to evaluate the function of BM hematopoietic cells. Comparing with the WT control, KO BM cells produced less GEMM (granulocyte, erythroid, macrophage, megakaryocyte mixed colonies), GM (granulocyte and macrophage mixed colonies), and G (granulocyte) colonies during serial passaging (Fig. 1e), indicating that Setd2 deletion impaired the proliferation and/or differentiation capacities of HSPCs. Collectively, these data suggest that Setd2 regulates multiple aspects of different hematopoietic cell compartments.
Fig. 1.
Setd2 deficiency impairs multiple-hematopoietic compartment development. a qPCR detection of Setd2 expression level in different BM cell subsets, including HSCs (LT-HSC, CD150+CD48-Lin−c-Kit+Sca1+), MPPs (CD48+Lin−c-Kit+Sca1+), B cells (B220+), T cells (CD3ε+), Ery cells (Ter119+), GM cells (Gr1+Mac1+). b, c The absolute number of PB leukocyte and lymphocyte subsets. WT, n = 18; KO, n = 19. d Statistical comparison of the proportions of different lineages between WT and KO BM. n = 4. e Colony-forming unit (CFU) assay of total BM cells. n = 3. f, g Representative FACS analysis (f) and absolute cell count (g) of BM HSPCs from bilateral femurs and tibias. WT, n = 8; KO, n = 9. h CFU assay of BM FLT3−-LSKs. n = 3. i In vitro growth curve of BM FLT3−-LSK cells. n = 3. All the samples are prepared using mice at 4−5 weeks after pI–pC induction. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
To further define the role of Setd2 in HSPCs, we used FACS to analyze the numbers and frequencies of different HSPC subpopulations in WT and KO BM. The results showed that KO mice had decreased LSK cells. However, the numbers of phenotypic HSCs showed no difference between WT and KO mice (Fig. 1f, g; Supplementary information, Figure S2M).The decrease of LSK cells thus was attributed to a decline in the multipotent progenitor (MPP) subpopulations, including MPP1 and MPP2.36 The unchanged number and frequency of the cell surface marker-defined HSCs (phenotypic HSCs) in KO mice does not necessarily mean that the function of Setd2 deleted HSCs are normal. To clarify this issue, we employed antibody against FLT3 to mark FLT3− and FLT3+ LSKs, which are rich in HSCs and HPCs, respectively. We then performed CFU assays with sorted FLT3− and FLT3+ LSK cells separately and found that FLT3− LSK cells displayed a similar change as total BM cells during serial passaging compared with WT control (Fig. 1h, Supplementary information, Figure S2N). Subsequently, we conducted a short-term culture with sorted FLT3− LSK cells in liquid media, cell count results showing that Setd2 disruption dramatically reduced cell number of FLT3− LSK cells (Fig. 1i), which should be attributed to the defects of cell proliferation rather than the increment of cell apoptosis (Supplementary information, Figure S2O and P). These results suggest that Setd2 regulates HSC proliferation in this in vitro setting.
Setd2 deletion impairs HSC self-renewal
BM transplantation provides a powerful tool to evaluate the capacities of HSC self-renewal and differentiation, while excluding any cell non-autonomous effect. To this end, we first performed a competitive BM transplantation (cBMT) assay. Sorted Lin−cells from WT and KO BMs (CD45.2 genetic background) were mixed with total BM cells (CD45.1) and then transplanted into CD45.1 recipient mice (Supplementary information, Figure S3A). The donor-derived (CD45.2) cells in PB were monitored every 4 weeks, and the results showed that, in the total PB cells, as well as in the B, T, and GM subpopulations, the percentages of CD45.2 cells in KO transplants were much lower than that in WT transplants. This trend was consistently observed from week 4 through week 16 (Fig. 2a; Supplementary information, Figure S3B-E). Interestingly, at 16 weeks post transplantation, although all the other cell types derived from KO BM were significantly lower than those from WT BM, the HSCs exhibited a comparable frequency between KO and WT transplants (Fig. 2b). This is consistent with the previously observed phenotype of unchanged count of KO HSCs in the steady state (Fig. 1g). Meanwhile, the hematopoietic differentiation bias in KO mice under steady-state condition (i.e., decreased B and increased GM cells; Fig. 1c) was also observed under the stress of BM transplantation (Supplementary information, Figure S3F). In order to eliminate the possibility that the decline of KO transplantation was attributed to the impaired homing capacity, we performed homing assay with total BM cells and found no significant distinction between WT and KO group at 6 h and 18 h after transplantation (Supplementary information, Figure S3G and H).
Fig. 2.
Setd2 deficiency abolishes HSC self-renewal capacity. a Long-term follow-up of donor-derived cell (CD45.2) proportion in PB of competitive BM transplanted (cBMT) mice. n = 16. b Proportion analysis of donor-derived cells in different BM subsets of cBMT mice. n = 7. c Long-term follow-up of donor-derived cell proportion in PB of primary (WT, n = 19; KO, n = 18) and secondary (n = 14) noncompetitive BM transplanted (ncBMT) mice. d Proportion analysis of donor-derived cells in different BM subsets of primary ncBMT mice. WT, n = 5, KO, n = 4. e Absolute cell count of donor-derived HSPC subsets in BM of primary ncBMT mice. WT, n = 5; KO, n = 4. f Proportion analysis of donor-derived cells in different BM subsets of secondary ncBMT mice. n = 4. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
As a complement to the cBMT assay, a noncompetitive BM transplantation (ncBMT) assay was also performed. WT and KO BM cells, without competitor cells, were transplanted into recipient mice, and pI–pC was injected to induce Setd2 deletion 5 weeks later (Supplementary information, Figure S3I). This strategy excluded the possibility that the defects in HSC reconstitution were attributed to the impaired homing capacity or the cell dysfunction occurred before transplantation. As a result, monitoring the frequency of donor-derived CD45.2 cells in PB revealed limited hematopoietic reconstitution ability of KO cells in recipient mice (Fig. 2c; Supplementary information, Figure S3J). However, the frequency and the absolute number of donor-derived HSCs showed no significant difference between WT and KO mice (Fig. 2d, e), which was in accord with the results from cBMT assay described above. Since serial HSC transplantation is a golden standard for evaluating HSC self-renewal ability, we next transplanted the BM cells of primary ncBMT mice into secondary recipient mice and analyzed the PB and BM cells. The results indicated that all the KO-derived CD45.2+ cell populations, including the HSCs, were barely detected, whereas the WT-derived cells showed a normal frequency in the reconstituted hematopoietic systems (Fig. 2f; Supplementary information, Figure S3K). Furthermore, the hematopoietic differentiation bias was also observed in serial ncBMT assay (Supplementary information, Figure S3L). Taken together, these results suggest that Setd2 may regulate both self-renewal and differentiation of HSCs probably through a cell type-specific regulation, and that Setd2 deletion therefore induces a stepwise impairment of HSC functions.
Setd2-deficient HSCs evolve into MDS-like disease
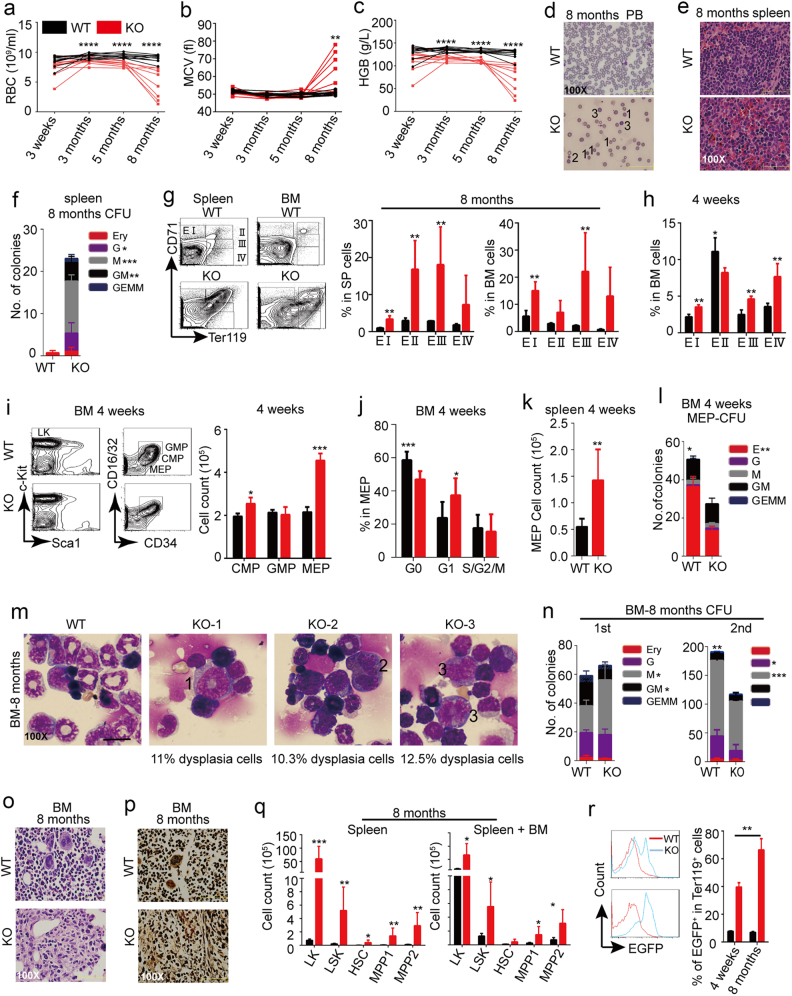
As previously reported, long-term self-renewal defects of HSCs can lead to HSC aging or neoplastic transformation. Both KO and WT mice were then followed up with blood routine examinations. Indeed, some abnormal cytological features were emerged in KO mice at about 8 months after pI–pC induction. First, KO mice presented symptoms of macrocytic anemia (Fig. 3a–c). Abnormal morphological features of RBC dysplasia, such as Howell–Jolly bodies, anisopoikilocytosis, and teardrop cells appeared in KO blood smears (Fig. 3d; Supplementary information, Figure S4A). Five of eight KO mice also showed leukocytopenia and thrombopenia at month 8, and the other three had thrombocythemia and neutrophilia probably due to enhanced myeloid cell differentiation (Supplementary information, Figure S4B-D). This disease was fatal to some mice (Supplementary information, Figure S4E). Second, splenomegaly and hepatomegaly were observed in 8-month-old KO mice (Supplementary information, Figure S4F). Third, extramedullary hematopoiesis occurred in spleens of KO mice. Histopathology revealed that expansions of erythroid and myeloid cells resulted in splenomegaly (Fig. 3e; Supplementary information, Figure S4G-I). Splenocytes derived from KO mice formed more myeloid colonies when plated in methylcellulose (Fig. 3f). FACS analyses also revealed expansion of erythroid and myeloid cells in KO spleen (Fig. 3g left; Supplementary information, Figure S4H). Abnormal erythropoiesis and decrement of GM cells were observed in KO BM (Fig. 3g right; Supplementary information, Figure S4J). To reason the abnormal erythroid phenotype, we detected the development process of young mice at 4 weeks after Setd2 excision, and found that Setd2 deletion perturbed erythroid differentiation at all stages of erythroid development (Fig. 3h). erythroid–megakaryocytic progenitors (MEPs), which give rise to RBC and PLT, were significantly increased in BM (Fig. 3i). The expansion of MEP numbers in BM might be partially ascribed to the increment of cycling MEP cells, but not the abnormal erythropoiesis in spleen, because the growth of MEP number in spleen was insufficient to compensate for that in BM (Fig. 3j, k). However, the capacity of CFU-E formation of MEPs sorted from BM at 4 weeks after Setd2 excision was dramatically decreased compared with WT control, which may account for the abnormal erythropoiesis in spleen and PB (Fig. 3l). These results indicated that an early inefficient compensatory increase in erythropoiesis caused a development of erythroid dysplasia. Furthermore, histopathology and CD45 immunohistochemistry analyses also confirmed that extramedullary hematopoiesis existed in liver (Supplementary information, Figure S4K and L). Fourth, bones of KO mice were much paler than WT mice, and the number of total BM cells was decreased (Supplementary information, Figure S4M). Giemsa staining of BM smears revealed the obvious decrement of the myeloid/erythroid ratio in KO BM (Supplementary information, Figure S4N), which is consistent with the FACS analysis (Fig. 3g right; Supplementary information, Figure S4J). Meanwhile, KO BM presented myeloid and erythroid dysplasia such as megaloblastic change and bilobulated cell (Fig. 3m). We also performed serial CFU assays to evaluate the function of BM cells collected from 8 months-old mice. The experiment results showed that the colony-forming ability of CFU-GM was decreased and that of CFU-M was increased, and no distinction was observed in total colony number between KO and WT group in the first plating (Fig. 3n, left), which is different from that in young mice. The replating potential of KO cells was decreased (Fig. 3n, right). Furthermore, at least four of five analyzed KO mice showed myelofibrosis (MF) features by histopathology and silver staining (Fig. 3o, p; Supplementary information, Figure S4O and P), and fibrosis also occurred in KO spleens (Supplementary information, Figure S4Q). In order to verify whether the fibrosis was induced by hematopoietic changes or BM micro-environmental defects, we checked the EGFP expression in CD45 and Ter119 double negative cells and found only 0.72% cells expressing EGFP (Supplementary information, Figure S4R), which thus may not be able to contribute to the profound hematopoietic defects. Fifth, we estimated the proportion of HSPCs in three pairs of 8-month-old WT and KO mice. Compared with the WT control, HSPCs in KO BM presented a decreased tendency due to the reduction of total BM cells, though there was no statistical difference between two groups (Supplementary information, Figure S4S). Of note, the frequency and number of HSPCs were increased observably in spleen, especially the LK (Lin− c-Kit+) subset, and the total number of HSPCs in the whole mouse was also increased (Fig. 3q; Supplementary information, Figure S4T). As mentioned above, in this KO model, the efficiency of Setd2 deletion can be measured by monitoring the percentage of EGFP+ cells in different hematopoietic subsets. Our results showed that EGFP signals could be detected in 40% spleen erythroid cells of young mice at 4 weeks after pI–pC induction, whereas 80% spleen erythroid cells in 8-month-old KO mice displayed EGFP-positive signal (Fig. 3r), indicating that Setd2-deficient erythroid cells acquired a growth advantage. Taken together, based on the criteria of Bethesda proposals and some other reports for classification of nonlymphoid hematopoietic neoplasms in mice,37–39 these data indicate that the hematopoietic disease induced by Setd2 deficiency resembles human MDS in many features.
Fig. 3.
Long-term Setd2 deficiency is sufficient to induce MDS-like disease. a–c Routine blood test follow-up. n = 10. d Representative displays of Giemsa staining of blood smears. 1: Howell–Jolly body; 2: Microspherocyte; 3: large spherocyte. e Representative displays of hematoxylin–eosin (HE) staining of spleen tissue sections. f CFU assay of total spleen cells at 8 months after pI–pC induction. n = 3. g Proportion analysis of spleen and BM erythrocytes in different development stages at 8 months after pI–pC induction. n = 3. h Proportion analysis of BM erythrocytes in different development stages at 4 weeks after pI–pC induction. n = 4. i FACS and absolute cell count analyses of BM myeloid progenitor compartments at 4 weeks after pI–pC induction. GMP: CD34+CD16/32+LK, CMP: CD34+CD16/32lowLK, MEP: CD34−CD16/32−LK. n = 3. j Ki67 staining for the detection of MEP cell cycle at 4 weeks after pI–pC induction. n = 9. k Absolute cell count analyses of spleen MEP cells at 4 weeks after pI–pC induction. n = 6. l CFU assay of BM MEPs at 4 weeks after pI–pC induction. n = 3. m Representative display of BM smears. 1: the myeloid cell with megaloblastic change; 2: bilobulated cell; 3: the erythroid cell with megaloblastic change. n CFU assay of total BM cells at 8 months after pI–pC induction. n = 3. o, p Representative displays of HE (o) and silver staining (p) of BM tissue sections. q Absolute cell count of HSPCs in spleen and BM. n = 3. r Proportion analysis of EGFP-positive cells in spleen erythroid cells of WT and KO mice at different ages. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
The MDS-like phenotype is recapitulated in the transplanted mice
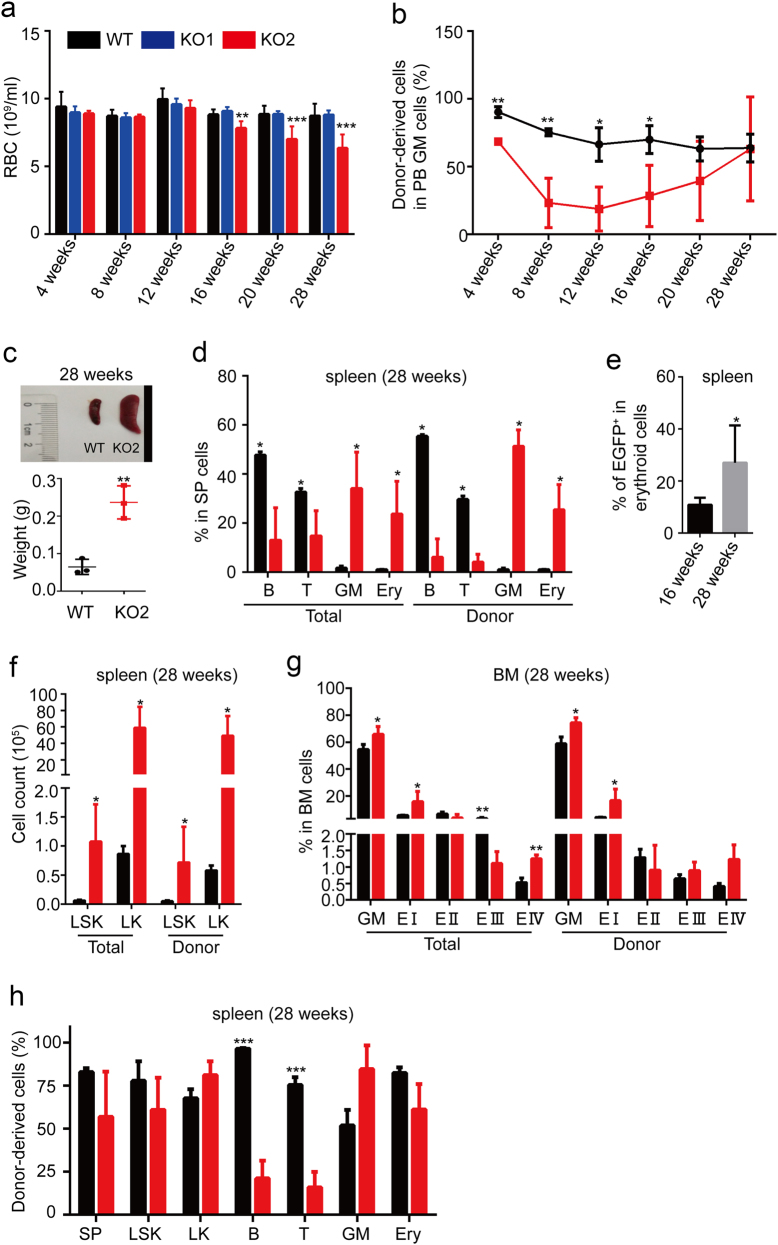
To further clarify whether the MDS-like disease in our Setd2 KO mouse model is intrinsic to the Setd2-deficient hematopoietic compartments, we performed long-term follow-up of ten pairs of ncBMT mice. At 16 weeks after pI–pC induction, four of ten KO ncBMT mice displayed progressive anemia and gradual WBC elevation in PB (Fig. 4a; Supplementary information, Figure S5A-C). We then carefully analyzed these four KO ncBMT mice and found that the donor-derived KO myeloid cells, but not B and T lineages, started to exhibit a competitive advantage from 12 weeks (Fig. 4b; Supplementary information, Figure S5D-F). These mice also displayed splenomegaly (Fig. 4c) and increment of erythroid and myeloid cells at week 28 (Fig. 4d). And their EGFP+ erythroid cells in spleen also displayed a significant expansion at week 28 relative to week 16 (Fig. 4e). In addition, the number of HSPCs dramatically increased in their spleens (Fig. 4f). The increase of erythroid and myeloid cells was also observed in BM (Fig. 4g). Although the proportion of LS and LSK in KO ncBMT Lin− BM cells was dramatically increased, the total number of HSPCs did not show a significant difference between two groups, which may be due to the decrement of the total number of BM cells in KO ncBMT mice (Supplementary information, Figure S5G-J). Notably, the majority of KO HSPCs, myeloid and erythroid cells in spleen and BM were derived from donor cells (Fig. 4h; Supplementary information, Figure S5K and L). All these data indicate that Setd2 deficiency is a cell-autonomous requirement for the development of the MDS-like disease.
Fig. 4.
MDS-like disease of Setd2 deficiency is reproducible in primary ncBMT mice. a Routine blood test follow-up. Setd2 KO ncBMT mice are divided to two groups: mice in group1 (KO1) did not develop MDS-like disease (n = 6), whereas mice in group2 (KO2) did (n = 4). WT = 10. b Proportion follow-up of donor-derived myeloid cells. WT, n = 10; KO2, n = 4. c Spleen size and morphology analysis. Representative morphology is displayed. n = 3. d Proportion analysis of different mature lineages in total or donor-derived spleen cells. n = 3. e Proportion analysis of EGFP-positive cells in spleen erythroid cells. n = 3. f Absolute cell count of LSK and LK cells in total or donor-derived spleen cells. n = 3. g Proportion analysis of different erythroid subsets in total and donor-derived BM cells. n = 3. h Proportion analysis of donor-derived cells in different spleen cell subsets. n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Setd2 deletion alters HSC transcriptional signature and partially resembles Dnmt3a/Tet2 double KO
To gain insight into the molecular mechanism underlying the impaired self-renewal and the subsequent neoplastic transformation caused by Setd2 loss, we performed RNA-seq analysis of BM LSK cells at 4 weeks after pI–pC induction. As a result, 472 genes, including 246 genes downregulated and 226 upregulated, showed significant differential expression levels (fold change, FC ≤ 0.5 or FC ≥ 2; P < 0.05) between two groups (Supplementary information, Figure S6A). Gene Ontology (GO) analysis revealed that the downregulated genes are enriched in cytokines, Wnt, interferon γ, and retinoic acid signaling pathways (Supplementary information, Figure S6B), implying insufficient activation of these signaling pathways which may contribute to the impaired HSC function.6 On the other hand, the upregulated genes are enriched in the pathways related to hematopoietic lineage differentiation and functions, as well as the cellular response to ROS (reactive oxygen species) (Supplementary information, Figure S6C). Intracellular ROS levels dictate different states of hematopoietic cells—e.g., a low ROS level is required for maintaining LT-HSC self-renewal, whereas a high ROS level represents a stress condition and drives HSCs to proliferate and further on to biased differentiation.40–43 Therefore, the increased expression of the ROS-responsive genes reflects an accumulation of ROS in Setd2 KO cells and subsequent abnormal differentiation. The herein identified Setd2-regulated genes involved in these signaling pathways were validated by RT-qPCR (Supplementary information, Figure S6D). Besides the analysis of this relatively small number of upregulated and downregulated genes, we also performed a gene set enrichment analysis (GSEA) to gain a global view of the transcriptome profile regulated by Setd2. Consistent with our finding that Setd2 deletion impaired HSC self-renewal, a gene set associated with HSCs was significantly depleted, whereas a hematopoietic mature cell gene set was markedly enriched in the gene expression profile of Setd2 KO LSKs (Fig. 5a). Thus, the altered gene expression denotes the functional defects of the phenotypical HSPCs (i.e., they retain identical cellular surface markers).
Fig. 5.
Setd2 KO HSPCs share some common transcriptional signatures with Dnmt3a/Tet2DKO HSPCs. a GSEA analysis of the transcriptional signature of HSCs (left) and mature cells (right) in KO BM LSKs relative to WT group. NES normalized enrichment score. b Plot representing the comparison of upregulated and downregulated genes between Setd2 KO LSKs and other epigenetic regulator KO HSPCs cells. *Nom P < 0.05, **Nom P < 0.01, ***Nom P < 0.001. Nom P, Nominal P-Value. c Number of hypo-methylated and hyper-methylated DMRs in Setd2 KO LSKs. d The comparison of average methylation level of all DMRs between WT and KO LSKs. e qPCR validation of deregulated genes associated with erythroid cells. n = 3. f GSEA analysis of Klf1 target genes (top) and heme metabolism-related genes (bottom) in Setd2 KO BM LSKs relative to WT control. g qPCR detection of Klf1, Hbb-b1, and Hba-a1 expression in Setd2 KO Lin− BM cells transfected with Klf1-shRNA. *P < 0.05, **P < 0.01, ***P < 0.001
To understand the mechanism by which Setd2-deleted HSPCs evolved into MDS, we compared the Setd2 KO with several previously reported models, in which deletion of the key epigenetic regulator(s), including Dnmt3a, Tet2, Asxl1, and Bap1, led to hematopoietic malignancies.8, 10, 44 GSEA analysis revealed that Setd2 KO resembled the DKO of Dnmt3a and Tet2; the upregulated and downregulated genes by Dnmt3a/Tet2 DKO were significantly enriched and depleted, respectively, in Setd2 KO LSKs (Fig. 5b). In striking contrast, Dnmt3a or Tet2 single KOs did not show such a correlation with Setd2 KO, neither did the Asxl1 nor Bap1 KOs (Fig. 5b). Given the functional connection between Setd2 and the DNA methylation that are co-regulated by Dnmt3a and Tet2 (via a direct recognition of Setd2-mediated H3K36me3 by Dnmt3a), the Setd2 KO and Dnmt3a/Tet2 DKO may share a common DNA methylation-dependent mechanism underlying the development of hematopoietic malignancy. To investigate the changes of DNA methylation associated with Setd2 deficiency, we performed a whole-genome bisulfate sequencing analysis of WT and KO LSK cells (Supplementary information, Figure S6E). We identified all the differentially methylated regions (DMRs), and found that the Setd2 KO genome carries a much larger number of hypo-methylated DMRs relative to hyper-methylated ones (5839 versus 1294; Fig. 5c), and that the methylation level of DMRs in the KO genome is significantly lower than WT (Fig. 5d). Some genes dysregulated in the KO LSK cells were found to be associated with these DMRs (e.g., Ltbp3 and Gata3, as shown in Supplementary information, Figure S6F). Furthermore, a profiling of genome-wide distribution of DNA methylation signals in different genomic regions revealed that the KO genome has decreased CpG methylation levels in many regions, including CG island shore, promoter, gene body and repeat regions (Supplementary information, Figure S6G). These results suggest that DNA methylation may play an important role in Setd2-mediated regulation of gene expression and HSPC function.
In Dnmt3a/Tet2 DKO mouse model, Dnmt3a-depletions and Tet2-depletions synergistically derepressed lineage-specific transcription factors such as Klf1 and Ikzf1. The activation of Klf1 in the DKO HSCs, accompanied with an upregulation of erythroid transcriptional signature, resulted in enhanced HSCs self-renewal and deficient erythropoiesis and marked anemia. Accordingly, we analyzed our RNA-seq data and detected a significant upregulation of Klf1 (FC = 2.14; P < 0.001), although it does not fall into the category of genes whose expression is associated with the DMRs (Supplementary information, Figure S6H). The Klf1 upregulation and altered expression of several related genes were subsequently confirmed in KO LSKs by RT-qPCR (Fig. 5e). The Klf1 target genes, as well as other erythroid-specific gene sets, were also significantly enriched in Setd2 KO transcriptome (Fig. 5f), which may account for the abnormal increment of early erythrocytes in young KO mice and the subsequent anemia in old ones. To understand the role of Klf1 in KO HSPCs, we used short hairpin RNA (shRNA) to knockdown Klf1 in KO Lin− BM cells. After culturing the cells for 3 days with the StemSpan medium containing stem cell factor (SCF) and erythropoietin (EPO), we determined the expression of markers for HSC maintenance and erythroid differentiation. As a result, while the expression of c-Kit did not show significant change (Supplementary information, Figure S6I), the HGB beta adult major chain (Hbb-b1), an activated gene by Setd2 KO (FC = 4.32; P < 0.0001) and a known Klf1 target gene, was downregulated by Klf1 knockdown in the Setd2 KO cells. In contrast, hemoglobin alpha adult chain 1 (Hba-a1) was not affected by either Setd2 KO or Klf1 knockdown (Fig. 5g). The successfully rescued expression of erythroid genes by Klf1 knockdown in the Setd2 KO cells suggests that the Klf1 upregulation contributes to the apparently increased, but ineffective, erythropoiesis in the Setd2 KO mice.
Setd2 loss triggers replication stress and genomic instability
The HSPCs from both Setd2 KO and Dnmt3a/Tet2 DKO mice, as shown in transplantation assays, could evolve into hematopoietic malignancies in the recipient mice. However, a notable difference between these two types of HSPC is that the competitiveness of HSPCs was reduced by Setd2 KO but increased by Dnmt3a/Tet2 DKO, as reflected by comparing the percentages of donor-derived hematopoietic cells (Fig. 2). We reasoned that Setd2 KO HSPCs should be able to acquire abilities through the months of latency to overcome their growth disadvantage and to gain uncontrolled competitiveness, allowing them to evolve into malignant cells. Previous studies have established that the Setd2 deficiency in cancer cells results in replication stress, leading to subsequent DNA damage and impaired genome integrity.34 RRM2, a ribonucleotide reductase (RNR) subunit that is directly regulated by SETD2-mediated H3K36me3, plays an important role in the induction of DNA RS, as well as in the synthetic lethality between SETD2 deficiency and WEE1 inhibition.35 We therefore speculated that Setd2 loss could also cause DNA RS in HSPCs. Indeed, we found a set of E2F3 target genes were significantly enriched in KO LSKs (Fig. 6a), which indicates a transcriptional response to DNA replication and denotes a blockage of cells from quiescence to cell cycling.44
Fig. 6.
Setd2 deficiency leads to HSPC proliferation defects. a, b GSEA analysis of E2F3 target genes (a) and deoxyribonucleotide (dNTP) metabolic process (b) in Setd2 KO BM LSKs relative to WT control. c qPCR detection of dNTP metabolism-related genes in KO and WT BM LSKs at 4 weeks after pI–pC induction. d CFU assay of Setd2 KO Lin− BM cells transfected with empty vector or Rrm2b. e Cell cycle analysis of BM FLT3−-LSKs (left) and FLT3+-LSKs (right) from WT and KO mice at 4 weeks after pI–pC induction by using Ki67 staining. n = 4. f Statistical comparison of absolute cell count of BM HSPCs between WT or KO group at day 20 after single-time 5-FU treatment. n = 5. g Proliferation potential analysis of BM FLT3−-LSKs (left) and FLT3+-LSKs (right) by in vivo BrdU administration at day 20 after single-time 5-FU treatment. n = 4. h Genome distribution of mutations (SNVs and indels) in KO BM cells at 8 months after pI–pC induction. i Percentages of distinct mutation type of SNVs in KO BM cells. j Measurement of the ROS level in WT and KO LSKs at 4 weeks after pI–pC induction by using CellROX Deep Red Reagent staining. k Genome distribution of CNVs in KO BM cells at 8 months after pI–pC induction. l, m GSEA analysis of NUP98-HOXA9 targets (l) and interferon gamma pathway (m) in 8 months-old KO BM LSK cells relative to WT group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
We next investigated whether the Setd2 deletion in HSPCs affected RNRs, which play important roles in providing deoxyribonucleoside triphosphates (dNTPs) for DNA replication and repair. The RNR holoenzyme consists of a large subunit RRM1 and two small subunits RRM2 and RRM2B, in which the RRM2-containing and RRM2B-containing holoenzymes preferentially regulate dNTP synthesis in S and G0/G1 phases, respectively.45–47 Notably, we found that Rrm2b, but not Rrm2, was downregulated (FC = 0.5; P = 0.001) in Setd2 KO LSKs, suggesting an insufficient supply of dNTPs in at least one subset of these cells. Accordingly, the GO collection of genes involved in the dNTP metabolic process were modestly depleted in Setd2 KO gene expression data (Fig. 6b), qPCR also confirmed the downregulation of some dNTP metabolism-related genes, including Rrm2b (Fig. 6c). Further investigation revealed that enforced overexpression of Rrm2b could significantly increase colony-forming capacity of KO lin− cells (Fig. 6d). Thus, in parallel with the direct transcriptional regulation of RRM2 by SETD2 in carcinoma cells, our data suggest that, in HSPCs, the Setd2 deletion inactivated Rrmb2b, leading to DNA replication defects and genome instability. The upregulation of E2F3 target genes and repression of the RNRs suggest a possibility that Setd2-deleted HSPCs (or a subset of which) were confronted with a problem with their proliferation and cell cycle. To examine this possibility, we set out to analyze these properties of the Setd2 KO and WT HSPCs. As it is known, HSCs largely reside in a quiescent state, whereas HPCs are more actively dividing. Using Ki67 and Hoechst staining, we detected that the proportion of FLT3+-LSKs, but not the FLT3−-LSKs of the Setd2 KO mice, increased in G0 phase and decreased in G1 and S phases (Fig. 6e; Supplementary information, Figure S6J). In contrast, Annexin V staining of these cells showed that the Setd2 deficiency had no effect on apoptosis of either FLT3−-LSK or FLT3+-LSK cells (Supplementary information, Figure S6K). Thus, these results support the notion that Setd2 deficiency-related DNA RS preferentially impairs the proliferation and cell cycle of HPCs, but not quiescent HSCs. To further validate the replication defects of Setd2-deleted HSCs in vivo, we used 5-fluorouracil (5-FU), an inhibitor for thymidylate synthase, to treat KO and WT mice and to induce RS in HSCs. 20 days after 5-FU treatment, the frequency and number of HSCs and MPPs of KO mice were much lower than that of WT mice (Fig. 6f; Supplementary information, Figure S6L). 5-bromo-2-deoxyuridine (BrdU) incorporation assays indicated that Setd2-deleted HSCs, under this stress state, exhibit defects in proliferation and cell cycle (Fig. 6g; Supplementary information, Figure S6M). These results suggest that, even in quiescent HSCs, Setd2 deletion could cause DNA replication defects as well.
In order to validate the genome instability caused by Setd2 deletion and find some clues about the secondary mutation(s) that cause the malignant transformation, we performed a whole-genome sequence of BM cells from a Setd2 KO mouse with MDS-like symptom, paired tail tissue was used as a normal reference. Proximately 3494 single-nucleotide variants (SNVs) and 496 small indels (small insertions or deletions) were detected, and the distribution of these mutations on genome did not show obvious bias (Fig. 6h). It is noted that the most common mutation type is C∙G → T∙A transition, followed by A∙T → G∙C transition and C∙G → A∙T transversion (Fig. 6i), which mirrored the mutation pattern seen in CD34+ cell population from MDS patients.48 The C∙G → T∙A transition may be ascribed to DNA methylation change caused by Setd2 deletion. Smoking-exposure as well as 8-oxo-2’-deoxyguanosine, one of the major products of DNA oxidation that favored hydrogen bonding with adenine, were inclined to cause C∙G → A∙T transversion.49 As described above, GO analysis of upregulated genes had revealed that ROS was activated in Setd2 KO cells (Supplementary information, Figure S6C), and consistent with this, a high level of ROS was detected in KO LSKs by staining with CellROX dye (Fig. 6j). In view of the possibility of smoking exposure was ruled out in this study, 8-oxo-2’-deoxyguanosine is more likely to be involved in this transversion. Similar to the data generated from MDS patients,48 copy number variations (CNVs) also existed in KO BM cells, which were analyzed with regard to their chromosomal positions (Fig. 6k). Finally, to identify the malignant transformation of HSPCs at the whole transcriptome level, we also performed RNA-seq analysis of BM LSKs from Setd2 KO mouse with MDS-like symptom. GSEA analysis revealed that KO HSPCs displayed a distinct transcriptional signature of malignant transformation, target genes of NUP98-HOXA9 were significantly enriched in KO HSPCs50 (Fig. 6l), and interferon pathway was also dramatically activated in KO HSPCs51 (Fig. 6m), indicating these pathways may be involved in MDS transformation. Taken together, Setd2 deficiency-induced DNA RS and genome instability might contribute to the functional defects of HSPCs and subsequent development of hematopoietic malignancies.
Discussion
In this study, we found that Setd2 plays an important role in hematopoietic homeostasis, and that its loss-of-function causes malignant transformation. Notably, while Setd2 deletion broadly affects multiple-hematopoietic compartments in steady state, its impact on HSC self-renewal is only seen when the cells are under certain stress such as serial transplantation, induction of DNA RS, and aging. We found that the aged Setd2 KO mice develop characteristic symptoms of MDS; this is the first animal model that interprets the function of SETD2 as a tumor suppressor and is reminiscent of the frequent SETD2 inactive mutations discovered in cancer patients. Our close analyses of the Setd2-deficient HSPCs at molecular and cellular levels provide insights into the mechanism of how the Setd2 loss causes hematopoietic defects and tumorigenesis.
Regarding the role of Setd2 in normal hematopoiesis, an interesting finding in this study is that the Setd2 loss showed distinct effects on HSCs relative to progenitors and differentiated cells. As shown by our PCR and reporter gene assays, Setd2 is broadly expressed in multiple-hematopoietic subsets, with a relatively higher level in HSPCs and lower in mature myeloid and erythroid cells. It is usually difficult to clarify the definite role of such a broadly expressed gene in HSC self-renewal, because the deficiency of the gene could affect the activities of many hematopoietic compartments and thereby result in a severe defect in hematopoietic repopulation in HSC transplantation.6 However, we found that, in steady state, while remarkably decreased progenitors and skewed lineage differentiation were evident in the Setd2 KO mice, their LT-HSCs remained intact. This result suggests that Setd2 plays a cell type-specific role in different hematopoietic stages. To unearth the role of Setd2 in HSC function, we performed serial BM transplantation, which represents a golden standard for measuring HSC self-renewal and differentiation potentials. Interestingly, in both cBMT and ncBMT assays, unchanged number of Setd2 KO LT-HSCs, again, was similarly observed in the first-round transplanted mice, although the donor-derived progenitors and differentiated cells decreased. Nevertheless, in the secondarily transplanted mice, Setd2 KO donor-derived HSCs were barely detected, while the WT donor-derived HSCs still showed a normal number and percentage, indicating impaired HSC self-renewal caused by the Setd2 loss in this serial transplantation settings. Collectively, these results suggest that Setd2 is a bona fide regulator of HSC self-renewal, in which Setd2 may fulfill its function through a mechanism that is distinct from its regulation of the downstream hematopoietic differentiation.
While the Setd2-deficient HSCs initially exhibited impaired self-renewal and reduced competitiveness, these cells were observed to be able to overcome these disadvantages through time, and eventually developed into hematopoietic malignancy in mice. This phenomenon strikingly fits the foresighted “Dameshek’s Riddle”, which illustrates the transition from aplastic anemia to leukemia in patients.11, 52 Apart from the high relevance of this paradox in clinical representations of many types of hematopoietic malignancies in patients, several mouse models with deletions of certain cancer-causing genes have shown a similar transition process during cancer progression. Among these models, loss of the epigenetic regulators Bap1 or Asxl1, both of which physically and functionally interact with polycomb repressive complex 2 (PRC2), results in impaired HSC self-renewal and subsequent MDS symptoms; the underlying mechanisms are shown to involve alterations of PRC2-mediated histone H3 lysine 27 trimethylation (H3K27me3).8, 10, 53 Another interesting model is the deficiency of Xist, a noncoding RNA that initiates X chromosome inactivation in females, which causes female-specific hematopoietic malignancy with mixed MDS and MPN features. The Xist-deficient HSCs also show intrinsic defects in self-renewal and competitiveness, likely due to X chromosome reactivation and consequent genome-wide changes.54 Our study of the Setd2 KO mouse thus provides another excellent animal model for the “Dameshek’s Riddle” and, in combination with the above-mentioned examples, suggests that the epigenetic regulation of the fate of HSCs could be one of the principles behind this clinically relevant paradox.
Human MDS is a heterogeneous group of HSC-based hematopoietic malignancies with poor prognosis and are characterized by PB cytopenias of one or more lineages, cytologic dysplasia, and BM hypercellularity.39 Compared with acute and chronic leukemia, our understanding of the pathogenesis of MDS has lagged behind, which hampers the development of efficacious therapeutics.55 As the attempts to xenograft human MDS cells in mice have not been successful, generation of genetically engineered mouse models is of great importance, even though it is still difficult to model all the clinical features of human MDS in a single model.39 In this study, we found that the phenotype of Setd2 deficiency recapitulates many necessary and supportive diagnostic criteria of human MDS.37, 39 All the Setd2-deficient mice showed anemia, and most of them also showed leukocytopenia and thrombopenia. Ineffective erythropoiesis was shown in BM and spleen, and, importantly, myeloid and erythroid dysplasia were clearly observed in BM. Outgrowth of the diseased clones was reflected by the increase of EGFP+ (Setd2-deleted) erythroid cells in the old relative to the young mice. Although Setd2-deficient mice did not show increased HSPCs and hypercellularity in BM, all of them had increased total HSPCs in BM plus spleen. Notably, MF was observed in some Setd2-deficient mice, which could be an indication of concomitant MPN. However, accumulation of megakaryocytes in BM, which is another marker for human MPN,56, 57 was not observed, thus we speculate that the MF may be secondary to the MDS.
Based on this study and previously published studies of ours and others, we propose three different, albeit potentially overlapping, mechanisms by which Setd2 regulates cell fates in hematopoiesis and malignant transformation. (1) Setd2 facilitates gene transcription by catalyzing H3K36me3 in the promoters of target genes. This mechanism has been shown in our previous study to regulate the Fgfr3 gene, which is required for endodermal differentiation of mouse ES cells. In hematopoiesis, Setd2 may similarly regulate some genes that are important for hematopoietic differentiation and thereby play a role in specific differentiation steps (e.g., B cell differentiation). (2) Setd2 regulation of genes may depend, at least partially, on the DNA methylation regulatory system, which is supported by the herein observed correlation between Setd2 KO and Dnmt3a/Tet2 DKO transcriptional profiles. It has been shown that, in mouse ES cells, Dnmt3b directly recognizes Setd2-catalyzed H3K36me3 on the gene body and that the gene-body DNA methylation prevents intragenic transcriptional initiation.26 Given the high similarity between Dnmt3a and Dnmt3b in their sequences and domain architectures, Dnmt3a shall be able to recognize H3K36me3 as well, though probably in different cell types and/or genome sites. Indeed, our results show that the Setd2 deficiency leads to a dramatic change of genome-wide DNA methylation, especially in the genomic regions known to be rich in Setd2-mediated H3K36 trimethylation. Many of these DNA methylation features were similarly observed in the Dnmt3a/Tet2-deficient mouse model. Given that the DNA methylation regulators (including TET2, DNMT3A, IDH1, and IDH2) are among the most frequently mutated genes in human MDS,5 the DNA methylation-dependent function of Setd2 could be highly relevant to the MDS phenotypes of Setd2 KO mice and carry important clinical implications. (3) We show that Setd2 loss induces DNA replication stress in HSCs, which is possibly responsible for the proliferation and cell cycle defects of the cells, as well as the impaired genome instability observed in the malignancies. This is consistent with previous studies performed in carcinoma cells. One proposed mechanism is that the Setd2 loss results in aberrant nucleosome compaction and thereby affects the DNA replication factories.58 Some of our results suggest that this mechanism may also occur in the Setd2-deletion HSCs, although in this study we have not yet focused on verifying the details of it. However, our study suggests another possible mechanism underlying the Setd2 loss-induced DNA RS. We have found that the Setd2 loss represses the RNR subunit Rrmb2, accompanied with a repression of a set of dNTP metabolism genes; the insufficient dNTP synthesis in the cells thus may also contribute to the DNA RS. Taken together, Setd2 plays an important role in maintaining hematopoietic homeostasis and preventing malignant transformation, likely through cell type-specific and stage-specific mechanisms. Further clarify the details of the mechanisms shall advance our understanding of epigenetic regulation of development and cancer and provide potential new therapeutic targets.
Materials and methods
Mouse model construction
Setd2 conditional KO mouse model (Setd2FL/FL) was constructed by Shanghai Research Center for Model Organisms. Briefly, an element which contains lox2272-loxp-pA-eGFP-IRES-En2SA-lox2272-loxp was inserted into intron 1 of the Setd2 locus. The heterozygous ES cells were microinjected into C57BL/6 blastocysts followed by implantation into pseudopregnant B6CBF1 foster mothers to acquire chimeras. Chimeras were mated with C57BL/6 to generate F1 mice, and F1 offsprings were backcrossed to C57BL/6 strain for several generations. Setd2 conventional heterozygous mouse model (HE, Setd2+/−) was constructed by our group previously. Mx1-cre transgenic mice were purchased from Model Animal Resource Information Platform (China). To obtain hematopoiesis-specific conditional Setd2 KO mice, Setd2 conventional HE mice were mated with Mx1-cre mice first, Setd2+/− Mx1-cre+ mice were then intercrossed with Setd2FL/FL mice to obtain Setd2FL/−Mx1-cre+ mice (KO), Setd2FL/+Mx1-cre− mice (WT) were used as a control. Both WT and KO mice were intraperitoneally (i.p.) injected with polyinosinic–polycytidylic acid (pI–pC, Invivogen, tlrl-pic-5, 70 mg/kg) every other day for three times. Mice were used according to animal care standards and all protocols were approved by the Committee of Animal Use for Research at Shanghai Jiao Tong University School of Medicine (China).
Flow cytometry
BM, spleen (SP), thymus (Thy), and PB cells were harvested from KO mice. Cells were stained in PBS with 1% FCS at 4 °C for 30 min to detect Hematopoietic stem/progenitor cells (HSPCs) and mature lineage cells with following antibodies: anti-CD117/C-Kit, anti-Sca1, anti-CD48, anti-CD150, anti-CD135/FLT3, anti-CD34, anti-CD41, anti-CD16/32, anti-CD105, anti-CD71, anti-Ter119, anti-CD45R/B220, anti-CD19, anti-CD11b/Mac1, anti-Ly6C/Gr1, anti-CD3ε, anti-CD4, anti-CD8, anti-CD44, anti-CD25,anti-CD45, anti-CD45.1, anti-CD45.2, anti-F4/80, and biotin lineage cocktail. All antibodies were purchased from BD Biosciences, Ebioscience, and Biolegnd. Flow cytometry experiments were performed on BD LSRFortessa™ X-20 (BD Biosciences) and analyzed with FlowJo Software.
Hematopoietic stem/progenitor cell culture
1.5 × 104 sorted FLT3−LSK and FLT3+LSK cells were cultured in StemSpan™ SFEM (CAT#09600, Stem Cell Technologies) with SCF (50 ng/mL), TPO (10 ng/mL), IL3 (10 ng/mL), IL6 (10 ng/mL), and Flt3L (50 ng/mL) for 4 days. Cell number was counted at day 2 and day 4. All cytokines were purchased from R&D Company.
For Rrm2b overexpression assay, MigRI-mcherry-vector and MigRI-mcherry-Rrm2b were transfected into BOSC23 cells, virus particles in the medium were harvested 48 h later. Meanwhile, KO Lin− BM cells were harvested and cultured in StemSpan™ SFEM with SCF (50 ng/mL), TPO (10 ng/mL), IL3 (10 ng/mL), IL6 (10 ng/mL), and Flt3L (50 ng/mL) for 24 h, followed by virus infection. Medium was changed using StemSpan™ SFEM with above cytokines at 8 h after infection, and cells were cultured for another 3 days.
For Klf1 shRNA knockdown assay, two shRNA sequences of Klf1 were acquired from Broad institute (https://portals.broadinstitute.org/gpp/public/gene/details?geneId=16596, 5′-TGAGACTGTCTTACCCTCCAT-3′, 5′-CCGGCGAACTTTGGCACCTAA -3′), then cloned into plko.1 vector. shRNA vector and packaging plasmid were transfected into 293T cells. After 48-h culturing, virus particles in the medium were harvested to infect KO Lin− BM cells, followed by culturing for 3 days in StemSpan™ SFEM with SCF (50ng/mL) and EPO (2 U/mL). RNA were extracted to detect the expression of Klf1, Hbb-b1, and Hba-a1.
Colony-forming unit assay
Clonogenic progenitors were determined in methylcellulose colony assay medium (MethoCult GF M3434, Stem Cell Technologies). In brief, 1 × 104 total BM cells from WT and KO mice were plated in 35-mm tissue culture dishes containing colony assay medium. After 7 days of incubation at 37 °C in 5% CO2, CFU-erythroid colonies (E), granulocyte colonies (G), macrophage colonies (M), granulocyte-macrophage colonies (GM), and GEMM colonies were counted with an inverted microscope. 1 × 104 cells from primary CFU were replated for serial colony formation, 7 days later, colonies were counted with an inverted microscope to evaluate the proliferation and differentiation ability of WT and KO HSPCs, FLT3−LSK and FLT3+LSK cells were sorted by BD FACSAria™ III (BD Biosciences). A total of 300 cells of each group were plated in M3434 for first CFU assay, then 1 × 104 cells from primary CFU were replated for secondary CFU assay. All colonies were counted after 7 days incubation. MEP cells were also sorted, 1000 cells of each group were plated in M3434 for CFU assay, and colonies were counted after 7 days. 1 × 104 per group sorted mcherry-Rrm2b overexpressed cells were also plated in M3434 for CFU assay, and colonies were counted after 7 days.
Cell apoptosis, cell cycle, and BrdU staining of HSPCs
BM cells were harvested from KO mice and stained with HSPC and MEP surface markers including lineage, anti-c-Kit, anti-Sca1, anti-Flt3, anti-CD34, anti-CD16/32. For HSPC apoptosis assay, 5 µL of Annexin V (BD Biosciences, 556421) and 5 µL of 7-AAD (BD Biosciences, 559925) were added to BM cells stained with HSPC surface markers. Gently vortex and incubate cells at RT (25 °C) for 15 min followed by adding 400 µL of 1× binding buffer to each tube and immediately analyzing cells by flow cytometry. For HSPC and MEP cell cycle analysis, HSPC and MEP surface marker-stained BM cells were fixed with BD Cytofix/Cytoperm™ Fixation (554715) for 1 h at 4 °C in the dark, then washed with BD Perm/Wash™ buffer for two times. 2 µL anti-Ki67 antibodies (Biolegnd, 652404) were added to each tube and cells were incubated at 4 °C in the dark. 30 min later, 5 µL Hoechst 33342 (10 mg/mL, Life Technologies) were added to each tube and cells were immediately analyze by flow cytometry. For HSPC BrdU incorporation assay, mice were intraperitoneally (i.p.) injected with BrdU (10 mg/mL, 100 µL, Sigma), 16 h later, BM cells were isolated and stained with HSPC surface markers. Cells were then fixed in the same way as in anti-Ki67 staining, subjected to determination of BrdU incorporation using BrdU Flow Kit (BD Biosciences) according to the manufacturer’s instructions. Flow cytometry was performed on BD LSRFortessa™ X-20 (BD Biosciences) and analyzed with FlowJo Software.
Intracellular staining of ROS
BM cells were harvested from WT and KO mice and stained with HSPCs surface markers including lineage, anti-c-Kit, anti-Sca1, then incubated with Cellrox deep red reagent (Sigma) for 1 h at 37℃ followed by FACS analysis.
5-fluorouracil treatment
WT and KO mice were i.p.-injected with 150 mg/kg 5-fluorouracil (5-FU; Sigma-Aldrich) once. After 20 days, BM cells were analyzed for HSPC proportion and BrdU incorporation.
BM transplantation assay
For cBMT assay, Lin− BM cells of CD45.2 mice were isolated by micro-beads (Lineage cell depletion kit, 130-090-858, Miltenyi Biotec) at 4 weeks after pI–pC induction, then stained with lineage cocktail (B220, CD3e, Ter119, CD11b, Gr1, all from Ebioscience) and sorted by FACS. 1 × 105 Lin− cells per mice mixed with 1 × 106 cells from CD45.1 mice were transplanted into lethally irradiated CD45.1 through lateral tail vein injection. For ncBMT assay, total BM cells were isolated from 5-week-old uninduced CD45.2 mice. 2 × 106 cells per mice were then transplanted into lethally irradiated CD45.1 mice. 5 weeks later, three times of pI–pC treatments were administrated every other day to recipient mice. 16 weeks after pI–pC induction, 1 × 106 total BM cells per mice from primary ncBMT mice were secondary transplanted into lethally irradiated recipient mice. For both cBMT and ncBMT assays, donor-derived cells were monitored by flow cytometry every 4 weeks. Mice were killed for analysis at 16 weeks after transplantation.
Homing assay
Each lethally irradiated CD45.1 mouse was transplanted with 2 × 107 total BM cells from CD45.2 mice at 4 weeks after pI–pC induction. Donor-derived cells were detected by flow cytometry at 6 h and 18 h after transplantation.
Hematologic blood parameters
Blood was collected from the orbital vein, blood routine examinations were performed with pocH-100. Blood smears of WT and KO mice at 8 months after last pI–pC injection were prepared and stained with Wright’s stain solution.
Histological section analysis
Spleen and liver tissues of WT and KO mice at 8 months after last pI–pC injection were dissected, fixed with 37% neutral formaldehyde and then subjected to paraffin embedding. Serial sections (4 μm) were cut, deparaffinized, and stained with hematoxylin and eosin. Freshly dissected long bones were also fixed with 37% neutral formaldehyde, subjected to decalcify. Paraffin sections were then made like spleen and liver tissue. Reticulin fibers were revealed by silver staining according to Gordon-Sweet. Images were obtained with an Olympus DP71 Observer microscope.
Immunofluorescence and immunohistochemistry staining
The Sorted BM HSPCs were fixed with 4% paraformaldehyde for 10 min, followed by 0.5% Triton-X 100 in PBS for 5 min. Cells were incubated at 4 °C overnight with rabbit anti-H3K36me3 antibody (Cell Signaling Technology), then incubated with AlexaFluor 594-goat anti-rabbit IgG[H + L] antibody (Invitrogen) for 1 h. Hoechst 33342 staining was conducted subsequently. Liver sections were deparaffinized followed by an overnight incubation at 4 °C with anti-mouse CD45 antibody. CD45-positive cells were then identified with an HRP Detection Kit (BD Bioscience).
Western blot analysis
An aliquot 2 × 105 sorted LSK cells of WT and KO BM mice were lysed with 1 × SDS sample buffer (Beyotime), followed by an overnight incubation at 4 °C with anti-H3 (Abcam, ab1791) and anti-H3K36me3 (Abcam, ab9050) antibodies, respectively.
RNA-seq and quantitative PCR
Total RNAs from sorted LSK cells of WT and KO mice at 5 weeks and 8 months old after pI–pC induction were extracted with Trizol (Invitrogen), then purified by RNeasy Micro Kit (Cat#74004, QIAGEN). First, rRNA was removed using Ribo-Zero rRNA Removal Kits (Epicentre, MRZMB126). Subsequently, rRNA-depleted samples were fragmented, then subjected to first and second strand cDNA synthetization. cDNA was ligated with sequencing adapters according to standard illumine Hiseq 2000/2500 protocols. The sequencing reads were mapped to the mouse genome (mm10) using Hisat2 (version: 2.0.4). Stringtie (version: 1.3.0) was used to calculate fragments per kilobase of exon model per million mapped reads (FPKM) values. edgeR was used to analyze differential genes based on twofold changes and q ≤ 0.05. GO enrichment analyses were performed with DAVID software (http://david.abcc.ncifcrf.gov/). For real-time PCR, total RNA was reverse-transcripted with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed with the SYBR Premix Ex Taq Kit (TAKARA). The relative gene expression was calculated by the ΔΔCt method, Gapdh was used as internal control.
Gene set enrichment analysis
Normalized RNA-seq data were rank-ordered by fold change. Gene sets from the Molecular Signatures Database (MsigDB, hallmark gene sets and curated gene sets) were used to perform GSEA analysis. Besides, manually constructed gene sets were also used, which was composed of downregulated and upregulated genes in HSPCs and LSKs of varied KO mice from GEO Datasets. GSEA was performed using GSEA v2.2.3 software.
Whole-genome sequencing
DNA was extracted from KO total BM cells at 8 months after pI–pC induction, the paired mouse tail DNA was used as the germline reference for whole-genome sequencing (WGS) analysis. DNA were sheared by sonication for 300 bp and adapters were ligated to the fragments. Fragments were then amplified by ligation-mediated PCR, purified and subjected to DNA sequencing on the Illumina Hiseq 2500 platform. Sequence data were mapped to mm10 by bwa software (version 0.7.12). The average sequencing depth was ×35. Small variations including SNVs and INDELs were called with the Mutect2 and filtered with recommended threshold (quality ≥ 30 and at least four reads covered) for cases. CNVs were called with CNVnator_v0.3 and calculated with Fermikit.
Whole-genome bisulfate sequencing
About 100 ng genomic DNA isolated from BM LSKs were fragmented to 200–300 bp by sonication using a Covaris sonication system (Covaris S220). Lambda DNA was used as a negative control. Fragments were treated with bisulfate using Accel-NGS Methyl-seq DNA library Kit (Swift Biosciences). After that, DNA libraries were constructed using the TruSeq DNA sample preparation kit (Illumina), followed by quality control. DNA libraries were then sequenced with Illumina HiSeq sequencing system, and adapters and low-quality reads were trimmed using trimmomatic software. The average depth of total genome sequencing was ×20, and the sequencing depth for C was ×8. Paired-end sequencing reads were aligned to the mm10 mouse genome using Bismark software, DNA methylation level of C with at least five reads was extracted to calculate the methylation ratio calling [0, 1] by Bismark. To compare the distribution of DNA methylation level between WT and KO LSK cells, the mouse genome was segmented into eight categories: CGI, CGI_shore, promoter (the region 2 kb upstream of the TSS), 5′ UTR, exon, intron, 3′ UTR, and repeats. CpG island shores were defined as regions 0–2 kb away from CpG islands. CpG islands were downloaded from the UCSC Genome Browser. DSS software was used to identify DMRs with parameters as follows: smoothing = TRUE, smoothing.span = 200, delta = 0, p.threshold = 1e-05, minlen = 50, minCG = 3, dis.merge = 100 and pct.sig = 0.5.
Statistical analysis
Student unpaired two-tailed t-test was used for most statistical analyses. Differences between groups were considered significant when P < 0.05, and indicated with a single asterisk (P < 0.05), double asterisks (P < 0.01), triple asterisks (P < 0.001), or quadruple asterisks (P < 0.0001). All statistical analyses were performed with GraphPad Prism software (GraphPad Software, San Diego, CA).
Accession numbers
RNA-seq and whole-genome bisulfate seq data have been deposited to the NCBI Gene Expression Omnibus under accession number GSE108617.
Electronic supplementary material
Acknowledgments
We thank Prof. Bo Zhou from Shanghai Institute of Biochemistry and Cell Biology for his constructive comments. We thank Shu-Min Xiong, Min Zhang, Jing Xie and other colleagues for their help and support. This work was supported by National Natural Science Foundation of China (81500080, 81670149); National Key Basic Research Project (973, 2013CB966801); 1000 Talents Program for Young Scholars; Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152506); and Samuel Waxman Cancer Research Foundation.
Author contributions
Z.Y.L., H.Q.H., S.X.J., C.S.J., and C.Z. designed the experiments and wrote the manuscript. Z.Y.L., S.J.W., X.Y.Y., Z.Y., S.J.C., L.P., L.D., and X.A.N. performed all experiments. X.C.H. performed bioinformatics analysis. All authors were involved in the analysis and interpretation of the data.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yuan-Liang Zhang, Jie-Wen Sun, Yin-Yin Xie and Yan Zhou.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0015-9.
Contributor Information
Zhu Chen, Email: zchen@stn.sh.cn.
Sai-Juan Chen, Email: sjchen@stn.sh.cn.
Xiao-Jian Sun, Email: xjsun@sibs.ac.cn.
Qiu-Hua Huang, Email: hqh10632@rjh.com.cn.
References
- 1.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 2.Cullen SM, Mayle A, Rossi L, Goodell MA. Hematopoietic stem cell development: an epigenetic journey. Curr. Top. Dev. Biol. 2014;107:39–75. doi: 10.1016/B978-0-12-416022-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi L, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell. Stem. Cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdel-Wahab O, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue D, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J. Clin. Invest. 2013;123:4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey A, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dameshek W. Riddle: what do aplastic anemia, paroxysmal nocturnal hemoglobinuria (PNH) and “hypoplastic” leukemia have in common? Blood. 1967;30:251–254. [PubMed] [Google Scholar]
- 12.Zhu X, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 2014;46:287–293. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mar BG, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat. Commun. 2014;5:3469. doi: 10.1038/ncomms4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker H, et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia. 2016;30:2179–2186. doi: 10.1038/leu.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu M, et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc. Natl. Acad. Sci. USA. 2010;107:2956–2961. doi: 10.1073/pnas.0915033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun XJ, et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 20.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. Embo. J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baubec T, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 22.Morselli M, et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. Elife. 2015;4:e06205. doi: 10.7554/eLife.06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesh S, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 24.Li F, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Neri F, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 28.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 30.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park IY, et al. Dual chromatin and cytoskeletal remodeling by SETD2. Cell. 2016;166:950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep. 2014;8:1989–2002. doi: 10.1016/j.celrep.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 33.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanu N, et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34:5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfister SX, et al. Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell. 2015;28:557–568. doi: 10.1016/j.ccell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell. Stem. Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kogan SC, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.V100.1.238. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou T, Kinney MC, Scott LM, Zinkel SS, Rebel VI. Revisiting the case for genetically engineered mouse models in human myelodysplastic syndrome research. Blood. 2015;126:1057–1068. doi: 10.1182/blood-2015-01-624239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos MA, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takubo K, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Kong LJ, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- 45.Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 47.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 48.Xu L, et al. Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers. Proc. Natl Acad. Sci. USA. 2014;111:8589–8594. doi: 10.1073/pnas.1407688111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfeifer GP, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 50.Hatano Y, et al. Molecular heterogeneity of the NUP98/HOXA9 fusion transcript in myelodysplastic syndromes associated with t(7;11)(p15; p15) Br. J. Haematol. 1999;107:600–604. doi: 10.1046/j.1365-2141.1999.01754.x. [DOI] [PubMed] [Google Scholar]
- 51.Pellagatti A, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon−stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108:337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- 52.De Keersmaecker K, Sulima SO, Dinman JD. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood. 2015;125:1377–1382. doi: 10.1182/blood-2014-10-569616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaFave LM, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med. 2015;21:1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111:4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 56.Della Porta MG, et al. Clinical relevance of bone marrow fibrosis and CD34-positive cell clusters in primary myelodysplastic syndromes. J. Clin. Oncol. 2009;27:754–762. doi: 10.1200/JCO.2008.18.2246. [DOI] [PubMed] [Google Scholar]
- 57.Tefferi A. Myelofibrosis with myeloid metaplasia. N. Engl. J. Med. 2000;342:1255–1265. doi: 10.1056/NEJM200004273421706. [DOI] [PubMed] [Google Scholar]
- 58.Carvalho S, et al. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41:2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.