Significance
Cell instructive biomaterial cues are a major topic of interest in both basic and applied research. In this work, we clarify how surface energy of soft biomaterials can dramatically affect mesenchymal stem cell receptor recruitment and downstream signaling related to cell fate. We elucidate how surface protein self-assembly and the resulting surface topology can act to steer mechanotransduction and related biological response of attached cells. These findings fill a critical gap in our basic understanding of cell–biomaterial interaction and highlight soft biomaterial surface energy as a dominant design factor that should not be neglected.
Keywords: mechanobiology, stem cell, PDMS, surface energy, ligand assembly
Abstract
Although mechanisms of cell–material interaction and cellular mechanotransduction are increasingly understood, the mechanical insensitivity of mesenchymal cells to certain soft amorphous biomaterial substrates has remained largely unexplained. We reveal that surface energy-driven supramolecular ligand assembly can regulate mesenchymal stem cell (MSC) sensing of substrate mechanical compliance and subsequent cell fate. Human MSCs were cultured on collagen-coated hydrophobic polydimethylsiloxane (PDMS) and hydrophilic polyethylene-oxide-PDMS (PEO-PDMS) of a range of stiffnesses. Although cell contractility was similarly diminished on soft substrates of both types, cell spreading and osteogenic differentiation occurred only on soft PDMS and not hydrophilic PEO-PDMS (elastic modulus <1 kPa). Substrate surface energy yields distinct ligand topologies with accordingly distinct profiles of recruited transmembrane cell receptors and related focal adhesion signaling. These differences did not differentially regulate Rho-associated kinase activity, but nonetheless regulated both cell spreading and downstream differentiation.
Studies of stem cell behavior on soft biomaterials have typically employed 2D platforms of synthetic polymers coated with monomeric protein ligands (1). Such studies (2–4) have demonstrated that modulating the stiffness of porous gels can direct stem cell fate. However, experimental outcomes on amorphous biomaterial substrates vary widely. Beyond the inherent biological variability of eukaryotic cell culture systems, existing models of cell–biomaterial interaction fail to coherently explain divergence of experimental results. For instance, it has until now not been understood why mesenchymal stem cells (MSCs) readily attach and spread on soft elastomeric silicone (3–5), while they tend to not spread on similarly soft substrates such as polyacrylamide that have been coated with similar extracellular matrix ligands (3, 4, 6).
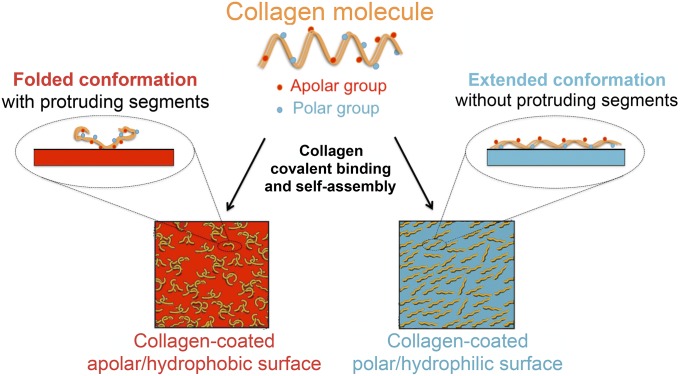
Although cell responses on synthetic hydrogels and elastomers are regularly compared, these materials present very distinct chemical and physical features (7). Among these features, one characteristic that has been widely ignored is the inherent difference in surface energy of these material classes. In the field of biomedical implant design, surface energy has long been recognized to control protein adhesion and downstream cellular reaction (8, 9). The property of biomaterial surface energy can be viewed as the physical work done by intermolecular forces acting to increase phase surface area. As such, surface energy depends on the charge and polarity of the outermost functional groups of the biomaterial. Surface energy can be increased by the presence of polar functional groups, with higher energy substrates having more polar groups yielding a more hydrophilic surface (10, 11). Monomeric type I collagen is a widely used model extracellular matrix ligand comprising both polar and apolar amino acid residues (Fig. 1). We have shown previously that biomaterial surface energy plays a dominant role in determining which groups are exposed after deposition, with a downstream influence on the supramolecular organization of adsorbed collagen layers (12). In this earlier work, we demonstrated that surface energy on stiff (2.15–2.40 MPa), atomically flat substrates steers cell–material interactions and promotes osteogenic MSC differentiation by regulating the topography of the adsorbed ECM protein layer presented to the cells. To achieve these insights, we designed a polydimethylsiloxane (PDMS)-based platform in which stiffness and surface energy can be independently tailored in a straightforward manner by addition of surfactant in small quantities. The chief technical challenge was to rigorously control for, and prevent, potential confounding effects of any divergent chemical, physical, and mechanical properties at the cell–material interface.
Fig. 1.
Schematic of surface energy-driven assembly of collagen I. Collagen I molecules containing polar and apolar amino acid residues covalently bind and self-assemble to exhibit different conformations, topologies, and functionalities when coated on substrates of different surface energy. Adapted from ref. 12.
We now adapt this tunable biomaterial system to achieve very soft substrates (elastic modulus <1 kPa) to isolate and investigate the role of surface energy on human MSC mechanosensitivity to substrate stiffness. We specifically focused on collagen I as a model ligand that supports osteogenic, tenogenic, or adipogenic differentiation in a substrate stiffness-dependent manner (6). We found that surface energy indeed regulates cell adhesion and differentiation on soft and hard substrates, with hydrophobic surfaces suppressing cell mechanosensitivity to bulk material stiffness. We reveal how surface energy directs ligand topography to alternately promote or inhibit cell mechanosensitivity and response to soft amorphous substrates. This understanding likely resolves conflicting mechanistic theories on how MSCs sense and react to soft biomaterial substrates (3, 4, 13–15). More broadly, the present study demonstrates biomaterial surface energy as a crucial consideration in soft biomaterial design, and one that cannot be neglected in study of stem cell–biomaterial interaction and cell fate.
Results
To isolate biological effects of surface-driven ligand assembly, we developed a PDMS-based platform that allowed stiffness and surface energy to be varied independently. Polar (hydrophilic) PDMS surfaces were obtained by adding 0.2% PDMS-b-PEO surfactant with a neutral and polar polyether to the standard PDMS (12). We established mechanically equivalant PDMS and PEO-PDMS substrates over a wide stiffness range (elastic moduli from over 2,000 to less than 1 kPA), generated by adjusting base-to-catalyst mixing ratios. As expected (3), softer elastomers yielded more viscous material responses (SI Appendix, Fig. S1). Microscale mechanical surface homogeneity was demonstrated using subcellular-sized indenter tips (10 μm radius) (SI Appendix, Fig. S2). In contrast to previous reports of high microscale elastic moduli of soft PDMS (4), we measured an elastic modulus of less than 1 kPa for 80:1 PDMS at 10% s−1 strain rates (SI Appendix, Fig. S2B), consistent with our previous investigations on stiff elastomeric substrates (12). Surface treatment with a heterobifunctional protein cross-linker (sulfo-SANPAH) did not alter surface mechanics (SI Appendix, Fig. S2B). Collagen coating only slightly increased nanoscale surface stiffness (SI Appendix, Table S1). A colorimetric micro bicinchoninic acid (microBCA) assay verified effectively equivalent adsorption of collagen on both substrate classes over the full stiffness range. The quantification experiments revealed an approximate protein load of 10 ng/mm2, representing an average of 2 × 104 collagen type I molecules per square micrometer and 167.3 × 104 molecules per square micrometer in the case of the synthetic peptide (SI Appendix, Fig. S3A). This results in a theoretical average distance between molecules of 3.54 nm and 0.39 nm, respectively. Although collagen and GFOGER form quaternary structures that increase the actual intermolecular distance, this remains below the ligand spacing previously shown to affect cell spreading (16). Nevertheless, an effect of different ligand densities at the nanoscale on nucleation and maturation of focal adhesions, and potentially on differentiation, cannot be completely excluded (17).
Consistent with our previously reported observations (12), atomic force microscopy (AFM) imaging analysis of collagen-coated PDMS and PEO-PDMS of different stiffness indicated a clear difference in ligand layer topography between polar and apolar surfaces. On PDMS, all collagen-coated surfaces appeared rough with an average roughness (Ra) between 3.12 and 4.10 nm with more prominent aggregates on stiffer substrates (SI Appendix, Fig. S4). On the other hand, PEO-PDMS surfaces presented a smoother ligand layer with an average roughness between 0.63 and 1.01 nm (SI Appendix, Fig. S4). Thus, collagen conformation and supramolecular organization atop these material surfaces depends on competitive interplay of collagen–substrate and collagen–collagen interactions (Fig. 1). On hydrophobic PDMS, molecules are covalently bound to the surface but do not lie flat. Instead, immobilized monomers adopt a folded conformation that may interact with further collagen molecules that are still in suspension. These intermolecular collagen interactions typically result in the formation of multilayer molecular aggregates. Such aggregation suggests a higher affinity for collagen–collagen interaction than for collagen–surface interaction. On the contrary, collagen molecules seeded onto hydrophilic PEO-PDMS lie flat within a relatively smooth collagen layer. This tendency of collagen to lie in monolayer on the more polar PEO-PDMS indicates that collagen–surface affinity dominates the interactions involved in collagen deposition on this hydrophilic material (18, 19). While the kinetics of collagen and GFOGER attachment to the two material classes is similar (SI Appendix, Fig. S3 E and F) there was approximately threefold increased density of sulfo-SANPAH cross-linker on the hydrophobic PDMS substrates used in these experiments (SI Appendix, Fig. S5). Importantly, this suggests that higher amounts of covalent surface cross-linker cannot overcome the effects of material surface energy in driving biologically relevant differences in ligand supramolecular assembly.
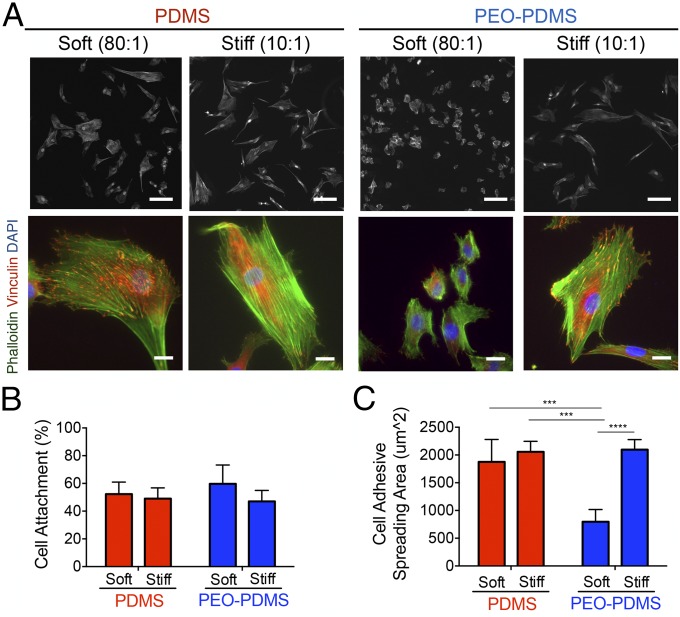
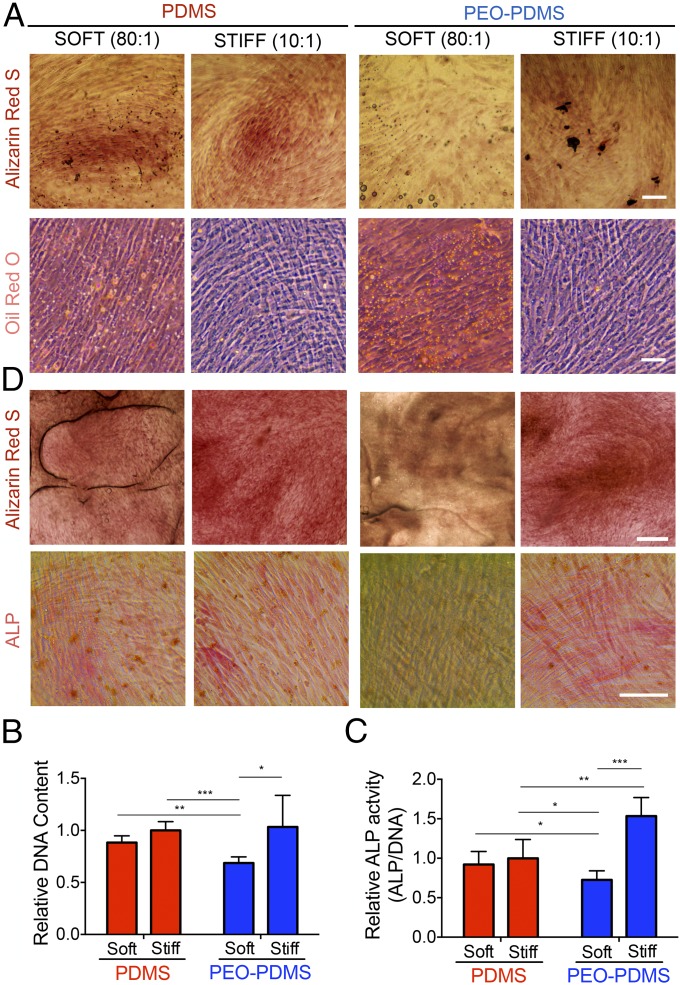
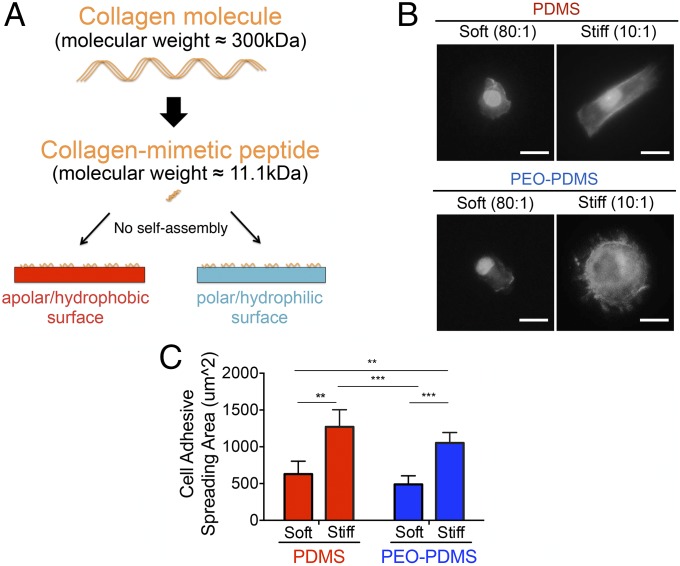
To test whether surface energy affects cell adhesion on elastomers of different stiffness, we cultured human bone marrow mesenchymal stem cells (hBMSCs) on PDMS and PEO-PDMS substrates and measured both cell attachment and spreading. At 1 h after cell seeding at high density (25,000 cells per square centimeter), the percentage of cell attachment evaluated by fluorescent nuclear staining on a large central area of each substrate was found to be similar, with ∼50% of seeded cells attaching to all substrates (Fig. 2B). As expected, cell spreading 24 h after cell seeding was not diminished on low-stiffness PDMS with an apolar, hydrophobic surface (Fig. 2). However, it was markedly diminished on soft hydrophilic elastomeric PEO-PDMS. The attachment footprint of cells on soft PEO-PDMS (80:1) was approximately threefold smaller than that of cells on all other substrates (Fig. 2C). Similarly, Vertelov et al. (15) recently reported reduced cell spreading on soft commercially available silicone gels. While phalloidin staining of cells seeded on stiff PDMS showed a more pronounced actin cytoskeleton with localized focal adhesions (antivinculin immunostaining) at the cell edge, cells on soft PDMS presented more dispersed cytoskeletal elements (Fig. 2A), consistent with previous reports (5). Taken together these results suggest that surface energy affects collagen-driven cell spreading and can dominate cell response to substrate stiffness cues. To evaluate the role of surface energy in modulating stem cell differentiation, we first cultured hBMSCs for 7 d at low density (5,000 cells per square centimeter) in coinduction medium containing osteogenic and adipogenic inducers. Cells were stained with Alizarin Red for calcium deposits, a standard marker of differentiated osteoblasts. Cultures were also stained with Oil Red O for lipid droplets, an indicator of the degree of adipogenesis. In contrast to all of the other substrates, soft PEO-PDMS substrates presented very low calcium deposits and a substantial amount of formed lipid droplets (Fig. 3A). Consistent with previous reports (3), hBMSCs cultured on both soft and stiff PDMS presented similar DNA amounts and alkaline phosphatase (ALP) activity (Fig. 3 B and C); however, the amount of DNA on soft PEO-PDMS substrates was significantly lower compared with the other substrates, suggesting reduced proliferation (20) (Fig. 3B). Quantification of ALP activity revealed a threefold lower expression of this osteogenic marker on soft PEO-PDMS compared with the stiff material (Fig. 3C).
Fig. 2.
Cell spreading is affected by the surface energy on elastomer substrates of different stiffness. (A) Morphology of hBMSCs on functionalized substrates seeded at 5,000 cells per square centimeter after 24-h culture on PDMS and PEO-PDMS substrates of different stiffness (soft, 0.07–0.10 kPa; stiff, 2.15–2.40 MPa). Cells were immunostained with an antibody against vinculin (red), Alexa-488 phalloidin (green), and DAPI (blue). (Upper row scale bar, 100 μm; Lower row scale bar, 20 μm.) Images of cells on stiff substrates were reported from our previous publication (12). (B) Attachment of hBMSCs on PDMS and PEO-PDMS substrates when seeded at 25,000 cells per square centimeter after 1-h culture (n = 3). Data on stiff substrates were reported from our previous publication (12). (C) hBMSCs spreading area on PDMS and PEO-PDMS when seeded at 5,000 cells per square centimeter after 24-h culture (n = 4; number of cells ≥2,400). Data on stiff substrates were taken from our previous publication (12). Data are represented as mean ± SD. Significance is indicated for P ≤ 0.05 (***P ≤ 0 0.001, ****P ≤ 0.0001).
Fig. 3.
Surface energy directs osteogenic stem cell differentiation independently of bulk substrate stiffness. hBMSCs after 7-d culture in mixed-induction medium on PDMS and PEO-PDMS substrates of different stiffness (soft, 0.07–0.10 kPa; stiff, 2.15–2.40 MPa) seeded at 5,000 cells per square centimeter. (A) Staining with Alizarin Red for calcium deposit (Scale bar, 100 μm.) and with Oil Red O for lipid droplets (Scale bar, 200 μm.). (B) Total DNA content. Data on stiff substrates were adapted from our previous studies (12) (n = 4–5). (C) Total ALP activity per DNA normalized by the mean value of the stiff PDMS. Data on stiff substrates were adapted from our previous publication (12) (n = 4–5). (D) Staining of hBMSCs after 14-d culture in basal growth medium on PDMS and PEO-PDMS substrates of different stiffness (soft, 0.07–0.10 kPa; stiff, 2.15–2.40 MPa) seeded at 5,000 cells per square centimeter with Alizarin Red (Scale bar, 500 μm.) and for alkaline phosphatase (ALP) detection with a fast red violet solution. (Scale bar, 100 μm.) Data are represented as mean ± SD. Significance is indicated for P ≤ 0.05 (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
We further investigated the hBMSC differentiation in basal growth medium for 14 d at low seeding density (5,000 cells per square centimeter) by staining for ALP and calcium deposition. Previous studies (4, 12) have reported a tendency for differentiation toward osteogenic lineages when stem cells are cultured on PDMS substrates independently of their stiffness. Consistent with this observation, hBMSCs cultured on all tested substrates, except for soft PEO-PDMS, exhibited a positive staining for ALP and a high calcium deposit (Fig. 3D). Collectively, our data suggest that surface energy is a key factor in stem cell differentiation as driven by substrate stiffness.
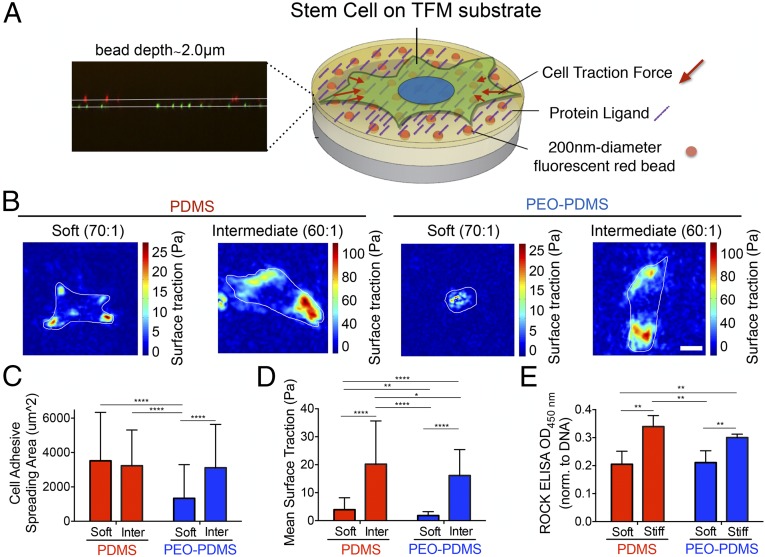
A unique traction force microscopy (TFM) approach was implemented in a manner that avoids confounding effects of bead-coating chemistry and topography that accompanies standard use of this assay. Fluorescent beads were covalently attached to a substrate of appropriate stiffness before addition of a 2-μm spincoat layer of bulk material (Fig. 4A and SI Appendix, Fig. S6). Cells were cultured for 16 h at low seeding density (650 cells per square centimeter), before measuring traction with an optimized tracking algorithm (21) (see SI Appendix for further details). Mean surface traction stress exerted by the cells on PDMS of either a soft (0.2–0.3 kPa) or an intermediate stiffness (5–6 kPa) was significantly higher than for cells on PEO-PDMS (Fig. 4 B–D and SI Appendix, Fig. S7). Substrates with an intermediate stiffness (60:1) were used in place of the stiffer substrates (10:1) to allow cell substrate deformations that could be sensitively resolved by light microscopy; cells on the intermediate substrates (60:1 and 70:1) adopted rounded cell morphologies and displayed adipogenic differentiation potential consistent with the substrates that they were intended to represent (Fig. 4C and SI Appendix, Fig. S8). Interestingly, although spreading was equivalent, mean surface traction stress of cells cultured on soft PDMS was fivefold lower than the cells cultured on intermediate stiffness PDMS (Fig. 4D). To further assess the level of cellular contractility on the various substrates, we cultured cells for 24 h at low seeding density (2,500 cells per square centimeter) and determined the level of ROCK phosphorylation, a key regulator of the cytoskeleton and cellular contraction (22). ROCK phosphorylation was found to be significantly higher on both stiff PDMS and PEO-PDMS than on soft substrates (Fig. 4E). We thus conclude that cell spreading is least partly decoupled from ROCK-mediated cellular contractility. To evaluate downstream effects of surface energy on collagen-binding receptors and focal adhesion components, we analyzed gene expression by quantitative PCR (qPCR) after 24 h. Different cell response on PDMS compared with PEO-PDMS was evident. Substantially diminished signaling related to focal adhesion maturation, including diminished integrin alpha 1, integrin alpha 2, vinculin, paxillin, and focal adhesion kinase expression on the softer PEO-PDMS substrates. This trend was reversed on PDMS, with signaling related to most of these elements increasing on soft PDMS substrates compared with the stiffer material (SI Appendix, Fig. S9). The discoidin domain receptors, including DDR1 and DDR2, which are also activated by collagen, were up-regulated on both soft materials, but more significantly on (apolar) PDMS (SI Appendix, Fig. S9). Taken together, the results suggest that integrin and DDR pathways are differently regulated by collagen when coated on PDMS and PEO-PDMS. As previously described (12, 23), DDRs may be involved in the recognition of a differential spatial collagen organization and lead to the activation of further downstream signaling.
Fig. 4.
Traction force microscopy indicates cells spread on soft PDMS without strongly contracting, and is not predominantly mediated by Rho-associated kinase (ROCK) activity. (A) Schematic for PDMS-based TFM platform with embedded 200-nm-diameter fluorescent trackers at a depth of 2.0 μm. (B) Snapshots of traction stress map with color values corresponding to different stress values (see corresponding axis) generated by hBMSCs on PDMS and PEO-PDMS of different stiffness (soft, 0.22–0.35 kPa; intermediate, 5–6 kPa) seeded at 625 cells per square centimeter after 16 h in culture. (Scale bar, 25 μm.) (C) Quantification of the corresponding cell spreading areas with the fluorescent live-cell nucleic acid Syto-13 stain. (n = 56–90). (D) Quantification of the corresponding mean surface traction stresses (see SI Appendix for details about data processing) (n = 56–90). (E) Semiquantification of phosphorylated ROCK by immune-sandwiched enzyme-linked immunosorbent assay when seeded at 5,000 cells per square centimeter after 24-h culture on PDMS and PEO-PDMS substrates of different stiffness (soft, 0.07–0.10 kPa; stiff, 2.15–2.40 MPa) (n = 4–5). Data are represented as mean ± SD. Significance is indicated for P ≤ 0.05 (*P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001).
To test whether the observed differences in cell behavior could be attributed to surface energy-driven differences in collagen self-assembly, we employed a well-described collagen mimetic peptide containing the minimal GFOGER cell-binding sequence that binds the α2β1 integrin receptor (24). This model ligand does not self-assemble into larger structures, a process that in the native collagen molecule depends on specific amino acid sequences that are absent from the synthetic peptide (25). Additionally, the GFOGER peptide has a comparatively small molecular weight of 11.1 kDa compared with the full-length collagen molecule with a mass of 300 kDa (Fig. 5A). As in all experiments, we first ensured that PDMS and PEO-PDMS presented similar amounts of ligand by adjusting the molarity of the peptide solutions adsorbed to the surface (SI Appendix, Fig. S3B). In contrast to experiments using the native collagen molecule, MSCs plated on soft PDMS did not fully spread, adopting a smaller, rounded shape on both soft PDMS and soft PEO-PDMS (Fig. 5B). In contrast, cell spreading on stiff PDMS and PEO-PDMS substrates was more than twofold higher than on the softer material variants (Fig. 5C). These results suggest that collagen deposition directed by surface energy (12) overrides mechanically driven MSC response to soft PDMS substrates.
Fig. 5.
Inactivation of collagen self-assembly promotes cell spreading according to PDMS stiffness. (A) Schematic for inactivation of collagen self-assembly by employing a collagen-mimetic (GFOGER) peptide. (B) Morphology of hBMSCs on PDMS and PEO-PDMS substrates of different stiffness (soft, 0.07–0.10 kPa; stiff, 2.15–2.40 MPa) when seeded at 2,500 cells per square centimeter after 24-h staining with Alexa-488 phalloidin and DAPI. (Scale bar, 50 μm.) (C) Quantification of the cell spreading area on the corresponding substrates (n = 4–5; number of cells ≥500). Data are represented as mean ± SD. Significance is indicated for P ≤ 0.05 (**P ≤ 0.01, ***P ≤ 0.001).
Discussion
Understanding cell–material interaction is essential for biomaterial design. Although mechanics and biochemistry of cellular attachment points are important, the activity state of a given ligand may be adsorption dependent and can be affected by various physical factors (26, 27). We have shown previously (12) that surface energy-driven ligand assembly and the resulting surface nanotopography on rigid elastomeric bulk material can strongly affect osteogenic stem cell signaling. We extended these studies to soft substrates aiming to potentially resolve the large body of conflicting evidence regarding stem cell sensitivity, or rather insensitivity, to soft PDMS (3, 4, 15). We hypothesized a potentially critical role of surface-driven ligand topography in regulating mesenchymal cells’ detection of and response to mechanical cues at the cell–material interface.
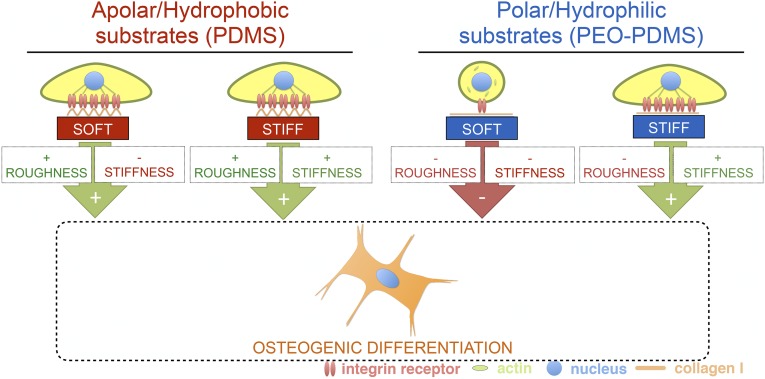
We developed a PDMS-based platform that can be mechanically tuned within a wide range of potential stiffness (from 70 Pa to 2.3 MPa) and with a range of surface energies that enable the creation of hydrophilic and hydrophobic variants of a given material stiffness, without otherwise affecting baseline physical properties of the substrate surface—most critically, collagen topology. This system allows one to limit variation in topology as a key confounding factor that often plagues parametric study of cell–biomaterial interaction. Multiscale mechanical characterization demonstrated consistent mechanical properties across size scales. This contrasts a recent study reporting inconsistent mechanical properties of PDMS across metric scales (4), a discrepancy we attribute to deformation rates. Because viscoelastic effects can be large in these materials (28, 29), we probed mechanical properties within a range of physiological strain rates (up to 10% s−1). We also considered probe fouling by soft PDMS, which can lead to dramatic stiffness overestimation at micro- and nanoscales (30). Using this well-controlled material platform, we pinpointed surface energy as a fundamental material property that can significantly impact stem cell fate on soft biomaterials. We demonstrate that ligand topology driven by surface energy can override adherent cell response to material stiffness, even though substrate stiffness is well described as a dominant contextual cue for stem cells in culture (2, 6). We show that collagen monomer assembly into rough nanotopography on hydrophobic surfaces (12) allows stem cells to spread and osteogenically differentiate on soft PDMS (Fig. 6). As previously reported (31), the presence of nanofeatures can push a cell to elongate, contract, and eventually undergo osteogenic differentiation. We show that using nonaggregating minimal peptides on soft hydrophobic PDMS can rescue the ability of stem cells to sense and react to soft substrates, chiefly in the form of cell rounding. Although studies have investigated the effects of nanotopography on stem cell behavior on rigid substrates (32), the present study is to our knowledge unique in demonstrating a dominant effect of nanotopography on very soft substrates. This information has eluded detection until now, mostly due to the substantial technological challenges involved in fabricating soft structured substrates or characterizing very soft substrates with regard to nanoscale topology and multiscale mechanics. On stiff substrates that are substantially easier to handle, it is well described that nanoscale disorder strongly induces osteogenic differentiation in the absence of chemical supplements (33). Our data indicate that this relationship between nanoscale roughness and osteogenic signaling extends to soft substrates as well. We propose that stochastic ligand assembly driven by an apolar biomaterial surface yields sufficiently rough ligand networks (12) exhibiting lateral and vertical disorder of cell-binding sites that drive bone differentiation independently of substrate rigidity. Dependency of cell behavior on material stiffness can be rescued by using a nonaggregating synthetic collagen peptide, demonstrating that in the absence of adequately rough ligand self-assembly, MSCs can detect and react to the stiffness of a soft or hard PDMS substrate with rounding or spreading, respectively. These experiments add essential mechanistic support to previous reports of stem cell sensitivity to soft substrates being modulated by cell-scale patterning and/or PDMS surface chemistry (15, 34).
Fig. 6.
Schematic of the interplay of matrix stiffness and surface energy-driven ligand topography in osteogenic stem cell differentiation.
Previous studies have demonstrated that the distribution and magnitude of cytoskeletal tension regulate ultimate differentiation of hBMSCs (2, 34–36). The fact that cells with severely diminished mechanical tension and ROCK activity can nonetheless spread on very soft hydrophobic substrates, suggests that cytoskeletal tension can be a secondary factor to topology in determining cell fate. This finding echoes observations by Chaudhuri et al. (14), in which substrate stress relaxation and decreased cytoskeletal tension enhanced cell spreading on soft substrates. The present study shows that spread morphology on even soft substrates functionalized with a suitable extracellular matrix ligand is sufficient to direct MSCs to an osteogenic fate. Conversely, previous studies have demonstrated that stiff substrates which confine cell spreading on micropatterned surfaces can promote adipogenic differentiation (35, 36). Thus, spread cell morphology, rather than the development of cytoskeletal contractility per se, seems to be a necessary condition to determine cell fate in the 2D culture conditions that form the majority basis of our understanding on stem cell mechanosensitivity. This conclusion contrasts with recent experiments with 3D platforms that reported cell fate to be independent of cell morphology but rather more traction dependent (37, 38). Still, 3D culture systems have their own disadvantages, including relative lack of control over hydrostatic stresses and osmotic gradients, and further work is required to determine how ROCK-mediated contractility and spread cell morphology potentially interact to regulate cell signaling or act as a central checkpoint in determining cell fate.
Collectively these results are striking, with potentially critical implications for research employing type I collagen as a “standard” 2D cell culture reagent. We demonstrate that stochastic collagen self-assembly translates to large potential for experimental variability, and/or systematically biased biological outcomes. Other ECM ligands, such as fibronectin, can behave quite differently from collagen (e.g., SI Appendix, Fig. S10), and work is required to characterize which combinations of biomaterials, interface chemistry, extracellular protein milieu, and cells yield reliably emergent system behavior.
In conclusion, we have demonstrated that surface energy-driven ligand self-assembly (12) can steer a cell to very different fates on soft substrates. Controlling for surface energy enables stem cells to spread and differentiate according to PDMS stiffness. These findings fill an important gap in our collective understanding (3, 4), explaining why stem cells spread and undergo osteogenic differentiation on soft apolar silicone when coated with collagen compared with rounding on soft polar substrates such as polyacrylamide. Although thoroughly described in the field of rigid biomaterials used in implants (8, 9), effects of surface energy on very soft substrates are difficult to control, and as such have been widely ignored in opinion-leading papers on stem cell–matrix interaction (3, 4). We suggest that surface energy is nonetheless a major biomaterial design factor that must be considered when designing cell-instructive biomaterials.
Methods
Tunable Surface Energy PDMS Substrate Preparation.
The 12- to 25-mm-diameter glass coverslips were cleaned with milli-Q H2O and ethanol (12). The PDMS base was mixed first with or without 0.2% (wt/wttotal) the surfactant polydimethylsiloxane-b-ethylene oxide. The PDMS cross-linker was then mixed in different ratios ranging from 80:1 to 10:1 base:cross-linker, spread on the glass coverslips.
Additional Methods.
The preparation of the various biocompatible surfaces, and the methods used, are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Barbara Niederöst for technical contribution to this work. This work was generously supported by the Swiss National Science Foundation (Grants 138221 and 205321).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704543115/-/DCSupplemental.
References
- 1.MacQueen L, Sun Y, Simmons CA. Mesenchymal stem cell mechanobiology and emerging experimental platforms. J R Soc Interface. 2013;10:20130179. doi: 10.1098/rsif.2013.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 4.Wen JH, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prager-Khoutorsky M, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 6.Sharma RI, Snedeker JG. Biochemical and biomechanical gradients for directed bone marrow stromal cell differentiation toward tendon and bone. Biomaterials. 2010;31:7695–7704. doi: 10.1016/j.biomaterials.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi A, Ling Q-D, Chang Y, Hsu S-T, Umezawa A. Physical cues of biomaterials guide stem cell differentiation fate. Chem Rev. 2013;113:3297–3328. doi: 10.1021/cr300426x. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, et al. Influence of substratum surface chemistry/energy and topography on the human fetal osteoblastic cell line hFOB 1.19: Phenotypic and genotypic responses observed in vitro. Biomaterials. 2007;28:4535–4550. doi: 10.1016/j.biomaterials.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mager MD, LaPointe V, Stevens MM. Exploring and exploiting chemistry at the cell surface. Nat Chem. 2011;3:582–589. doi: 10.1038/nchem.1090. [DOI] [PubMed] [Google Scholar]
- 10.Ramos SC, et al. Influence of polar groups on the wetting properties of vertically aligned multiwalled carbon nanotube surfaces. Theor Chem Acc. 2011;130:1061–1069. [Google Scholar]
- 11.de Gennes PG. Wetting: Statics and dynamics. Rev Mod Phys. 1985;57:827–863. [Google Scholar]
- 12.Razafiarison T, Silván U, Meier D, Snedeker JG. Surface-driven collagen self-assembly affects early osteogenic stem cell signaling. Adv Healthc Mater. 2016;5:1481–1492. doi: 10.1002/adhm.201600128. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Han D, Zhao Y-P. Kinetic behaviour of the cells touching substrate: The interfacial stiffness guides cell spreading. Sci Rep. 2014;4:3910. doi: 10.1038/srep03910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri O, et al. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vertelov G, et al. Rigidity of silicone substrates controls cell spreading and stem cell differentiation. Sci Rep. 2016;6:33411. doi: 10.1038/srep33411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalcanti-Adam EA, et al. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilian KA, Mrksich M. Directing stem cell fate by controlling the affinity and density of ligand-receptor interactions at the biomaterials interface. Angew Chem Int Ed Engl. 2012;51:4891–4895. doi: 10.1002/anie.201108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont-Gillain CC, Jacquemart I, Rouxhet PG. Influence of the aggregation state in solution on the supramolecular organization of adsorbed type I collagen layers. Colloids Surf B Biointerfaces. 2005;43:179–186. doi: 10.1016/j.colsurfb.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Narayanan B, Gilmer GH, Tao J, De Yoreo JJ, Ciobanu CV. Self-assembly of collagen on flat surfaces: The interplay of collagen-collagen and collagen-substrate interactions. Langmuir. 2014;30:1343–1350. doi: 10.1021/la4043364. [DOI] [PubMed] [Google Scholar]
- 20.Mih JD, et al. A multiwell platform for studying stiffness-dependent cell biology. PLoS One. 2011;6:e19929. doi: 10.1371/journal.pone.0019929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holenstein CN, Silvan U, Snedeker JG. High-resolution traction force microscopy on small focal adhesions: Improved accuracy through optimal marker distribution and optical flow tracking. Sci Rep. 2017;7:41633. doi: 10.1038/srep41633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riveline D, et al. Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund AW, Stegemann JP, Plopper GE. Mesenchymal stem cells sense three dimensional type I collagen through discoidin domain receptor 1. Open Stem Cell J. 2009;1:40–53. doi: 10.2174/1876893800901010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojtowicz AM, et al. Coating of biomaterial scaffolds with the collagen-mimetic peptide GFOGER for bone defect repair. Biomaterials. 2010;31:2574–2582. doi: 10.1016/j.biomaterials.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prockop DJ, Fertala A. Inhibition of the self-assembly of collagen I into fibrils with synthetic peptides. Demonstration that assembly is driven by specific binding sites on the monomers. J Biol Chem. 1998;273:15598–15604. doi: 10.1074/jbc.273.25.15598. [DOI] [PubMed] [Google Scholar]
- 26.Arima Y, Iwata H. Effect of wettability and surface functional groups on protein adsorption and cell adhesion using well-defined mixed self-assembled monolayers. Biomaterials. 2007;28:3074–3082. doi: 10.1016/j.biomaterials.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Schaap-Oziemlak AM, Kühn PT, van Kooten TG, van Rijn P. Biomaterial–stem cell interactions and their impact on stem cell response. RSC Adv. 2014;4:53307–53320. [Google Scholar]
- 28.Lakes RS. Viscoelastic measurement techniques. Rev Sci Instrum. 2004;75:797–810. [Google Scholar]
- 29.Nordin M, Frankel VH. Basic Biomechanics of the Musculoskeletal System. Lippincott Williams & Wilkins; Baltimore: 2013. [Google Scholar]
- 30.Meli F, Küng A. AFM investigation on surface damage caused by mechanical probing with small ruby spheres. Meas Sci Technol. 2007;18:496–502. [Google Scholar]
- 31.Oh S, et al. Stem cell fate dictated solely by altered nanotube dimension. Proc Natl Acad Sci USA. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalby MJ, Gadegaard N, Oreffo ROC. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 33.Dalby MJ, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 34.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci USA. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 37.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]