Significance
It is commonly observed that proteasome impairment results in accumulation of ubiquitinated proteins in the cytosol. Even proteins originally located in the nucleus show similar cytosolic accumulation, suggesting that unidentified machinery proactively transports them to the cytosol. Here, we report that a protein complex, UBIN–polyubiquitinated substrate transporter, harboring ubiquitin binding domain and nuclear export signal specifically mediates this process. In addition, their worm homologues showing similar transportation activity are important to maintain the lifespan of worms under natural condition. Our findings provide an answer to the long-standing question of why ubiquitinated proteins are deposited in the cytosol by proteasome impairment; they provide definite identification of underlying molecular machinery and show its essential involvement in the proteostasis in animal cells.
Keywords: protein quality control, ubiquitin, protein aggregation, protein transport
Abstract
Although mechanisms for protein homeostasis in the cytosol have been studied extensively, those in the nucleus remain largely unknown. Here, we identified that a protein complex mediates export of polyubiquitinated proteins from the nucleus to the cytosol. UBIN, a ubiquitin-associated (UBA) domain-containing protein, shuttled between the nucleus and the cytosol in a CRM1-dependent manner, despite the lack of intrinsic nuclear export signal (NES). Instead, the UBIN binding protein polyubiquitinated substrate transporter (POST) harboring an NES shuttled UBIN through nuclear pores. UBIN bound to polyubiquitin chain through its UBA domain, and the UBIN-POST complex exported them from the nucleus to the cytosol. Ubiquitinated proteins accumulated in the cytosol in response to proteasome inhibition, whereas cotreatment with CRM1 inhibitor led to their accumulation in the nucleus. Our results suggest that ubiquitinated proteins are exported from the nucleus to the cytosol in the UBIN-POST complex-dependent manner for the maintenance of nuclear protein homeostasis.
Cellular protein homeostasis, which is primarily mediated by protein folding and degradation, is essential for cell survival. Many molecular chaperones function in protein folding/refolding, prevent protein aggregates formation, and are involved in the degradation of terminally misfolded proteins with potential cytotoxic effects (1). The ubiquitin proteasome system (UPS) and the autophagy lysosome pathway play important roles in the removal of terminally misfolded and aggregated proteins (2). Imbalances in these protein quality control machineries cause the accumulation of misfolded and ubiquitinated proteins, resulting in the formation of cytotoxic aggregates inside and outside the cell. These protein aggregates are implicated in the pathogenesis of neurodegenerative disorders, such as Alzheimer’s disease, amyotrophic lateral sclerosis, and Parkinson’s disease (3, 4).
The isolation and clearance of cytotoxic misfolded proteins can occur across different cellular compartments. The most extensively studied example is the endoplasmic reticulum-associated degradation (ERAD) (5), in which endoplasmic reticulum (ER) resident molecular chaperones selectively recognize terminally misfolded proteins in the ER. These proteins are then translocated from the ER into the cytosol by specialized factors, including p97 ATPase, which provides a major driving force for retrotranslocation, and degraded by the UPS in the cytosol. Mitochondrial proteins are also extracted across the membrane and targeted for proteasomal degradation in the cytosol (6, 7). However, the ER and mitochondria have access to other proteolytic pathways: the ER is connected to the lysosome via a vesicular transport system, and mitochondrial proteins can be degraded by intrinsic proteases (8). Despite the availability of proteolytic systems, these organelles proactively translocate damaged proteins to the cytosol for proteolytic clearance, which could be attributable to the large capacity of the cytosolic protein quality control system. The cytosol contains various molecular chaperones with different substrate specificities, folding enzymes with a broad range of activities, two strong and flexible proteolytic machineries (namely, the UPS and autophagy), and regulated protein deposition systems, such as the aggresome, aggresome-like induced structure (ALIS), insoluble protein deposit (IPOD), and juxta nuclear quality control compartment (JUNQ), which sequester damaged proteins (9–11). Thus, the cytosol constitutes a robust system for maintaining protein homeostasis that extends to distinct organelles within the cytosol.
Nuclear protein homeostasis is maintained by similar systems to those operating in the cytosol. The nucleus and the cytosol are considered topologically identical, because they are connected by a large nuclear pore that allows bidirectional free diffusion of small biomolecules in addition to the proactive transport of macromolecules mediated by a range of mechanisms, including more than 20 different importin, exportin, and NXF proteins (12–15). Among them, CRM1 (also known as exportin-1) is the major protein, which facilitates the export of proteins and RNAs across the nuclear pore (16). Despite such a high degree of connectivity, molecular chaperones are purposely relocated from the cytosol to the nucleus in response to various proteotoxic stresses, including heat shock, suggesting that the mechanisms of protein quality control differ considerably between the nucleus and the cytosol (17–20). In addition to the relocation of molecular chaperones, misfolded proteins in the cytosoland the ER are also imported into the nucleus for degradation by the proteasome as shown recently in yeast (21, 22). However, the nucleus is not the final destination of all ubiquitinated proteins in cells. Ubiquitinated proteins form cytosolic inclusions in the presence of proteasomal inhibition or are proactively sequestered to specialized structures in the cytosol, such as the aggresome, IPOD, and JUNQ (23–25), to prevent cytotoxicity. Therefore, in addition to their transport into the nucleus, ubiquitinated proteins in mammalian cells appear to be relocated from the nucleus to the cytosol to prevent their cytotoxicity, although no molecular machineries mediating such transportation from the nucleus to the cytosol have been identified.
We previously identified UBIN (also referred to as UBQLN4), which harbors ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains (26). This class of proteins transports ubiquitinated proteins to the proteasome; however, the biological role of UBIN remains elusive. Here, we identified a small 15-kDa protein termed polyubiquitinated substrate transporter (POST)/DeSI1, which contained a nuclear export signal (NES), as a binding partner of UBIN. We showed that POST and the Ran-dependent nuclear export mediator CRM1 cooperatively export UBIN bound to polyubiquitinated proteins from the nucleus to the cytosol. The UBIN-POST system is a pathway that actively removes polyubiquitinated proteins from the nucleus and may contribute to nuclear protein homeostasis.
Results
POST Is the UBIN Binding Partner and Is Exported from the Nucleus to the Cytosol by CRM1.
To explore the biological role of UBIN, UBIN binding proteins were identified by interactome analysis using MS with HEK293 cells, in which UBIN with an FLAG epitope tag at the C terminus (UBIN-FLAG) served as bait. Most of the UBIN binding proteins identified were 19S and 20S proteasome subunits (Table S1), indicating the functional importance of the UBL domain of UBIN, which is recognized by 26S proteasome (27). In addition, one protein of 15 kDa was identified as a UBIN-interacting protein and was later named POST. MS analysis using POST harboring an FLAG tag at its N terminus (FLAG-POST) as bait conversely identified UBIN and its homologs, UBQLN1 and UBQLN2 (Table S2), as POST binding proteins.
The interaction of UBIN with POST was confirmed by coprecipitation analysis with HEK293 cells transfected with UBIN-Flag and HA-POST (Fig. 1A). As shown, HA-POST was coimmunoprecipitated with UBIN-Flag (Fig. 1A, Lower, lane 9) in contrast to the negative controls (Fig. 1A, lanes 7 and 8). Consistently, UBIN-Flag was coimmunoprecipitated with HA-POST (Fig. 1A, Upper, lane 12) in contrast to the negative control (Fig. 1A, lanes 10 and 11). Thus, HA-POST and UBIN-Flag were coimmunoprecipitated with each other.
Fig. 1.
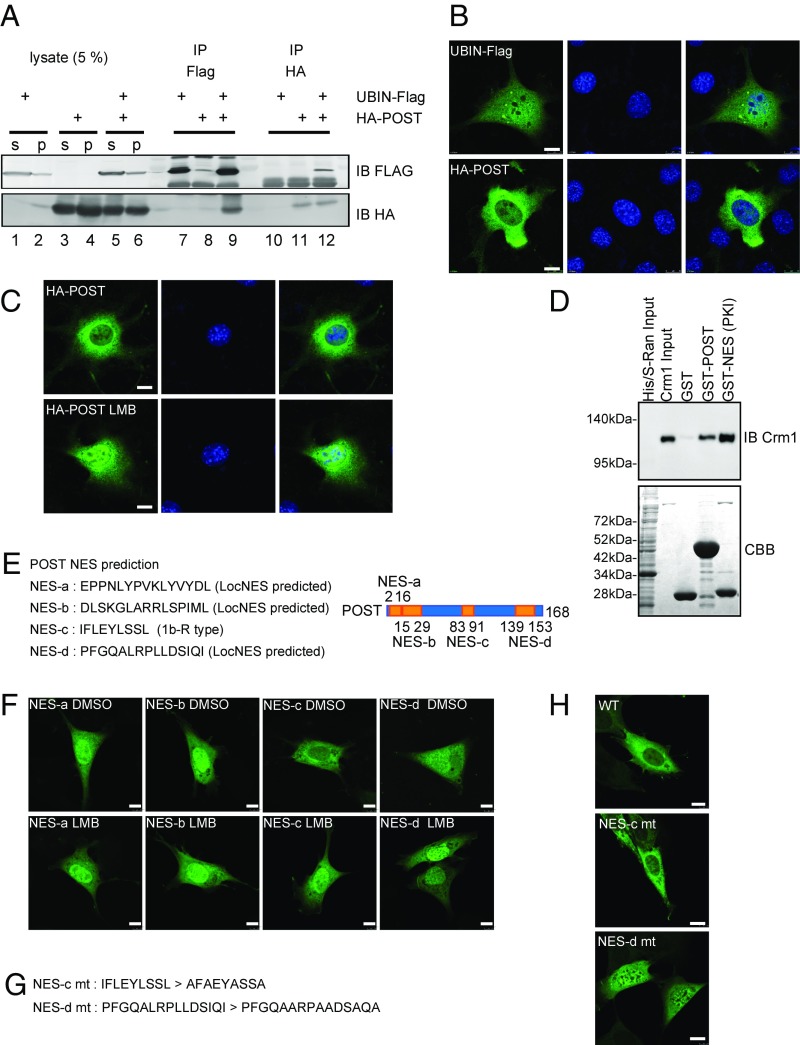
The UBIN binding protein POST localizes to the cytosol in a CRM1-dependent manner. (A) Physical binding between UBIN and POST. HEK293 cells cultured on 35-mm dishes were transfected with 0.8 μg of pcDNA3.1-UBIN-Flag (lanes 1, 2, 5, and 6) or 0.2 μg of pCAG-HA-POST (lanes 3–6); s means the supernatant and p means the precipitates after 15,000 × g centrifugation for 15 min. The UBIN-expressing cell lysate (lane 1) was immunoprecipitated with anti-Flag M2 antibody (lane 7) or anti-HA antibody (5D8; lane 10). The POST-expressing cell lysate (lane 3) was immunoprecipitated with anti-Flag M2 antibody (lane 8) or anti-HA antibody (lane 11). The UBIN- and POST-expressing cell lysate (lane 5) was immunoprecipitated with anti-Flag M2 antibody (lane 9) or anti-HA antibody (lane 12). Lysates and immunoprecipitates were subjected to immunoblotting (IB) with anti-Flag or anti-HA antibody as shown. IP, immunoprecipitation. (B) POST predominantly localizes to the cytosol despite the distribution of its binding partner UBIN throughout the cell. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.5 μg of UBIN-Flag or 0.1 μg of HA-POST. The cells were immunostained with anti-Flag M2 or anti-HA (12CA5) antibody (green). Nuclei were stained with DAPI (blue). (Scale bar: 10 μm.) (C) Involvement of CRM1 in the cytosolic distribution of POST. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.1 μg of HA-POST. In Lower, the cells were treated with 10 nM LMB for 12 h. The cells were immunostained with anti-HA (12CA5) antibody (green). (Scale bar: 10 μm.) (D) Pulldown assay to analyze the binding between POST and CRM1. GST-POST binds with CRM1 in the presence of RanGTP. Immobilized GST, GST-POST, and GST-NES (PKI) as a positive control were incubated with CRM1 (18 μg) and RanGTP. Bound proteins were subjected to SDS/PAGE and analyzed by Coomassie staining (CCB; Lower) or IB with anti-CRM1 antibody (Upper). (E) Prediction of NESs in POST and their positions in POST. NES-a, -b, and -d were predicted by LocNES program. NES-c was found by similarity with reverse-type NES-like sequence of class 1b (1b-R). (F) Subcellular localization of Venus-conjugated NES-a, -b, -c, and -d. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.6 μg of Venus-NES-a, -b, -c, and -d. The cells were treated with 10 nM LMB for 12 h (Lower). (Scale bar: 10 μm.) (G) Sequences of NES-c and -d mutants. Hydrophobic residues (Φ) in potential NES were replaced by alanine. (H) Involvement of the NES-d in the cytosolic distribution of POST. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.6 μg of HA-POST (WT), HA-tagged NES-c mt POST, or HA-tagged NES-d mt POST. The cells were immunostained with anti-HA antibody. (Scale bar: 10 μm.)
To examine the subcellular localization of these proteins, NIH 3T3 cells were analyzed by immunostaining with an anti-FLAG antibody (FLAG-M2). The results showed that UBIN-Flag localized to the cytosol and the nucleus, whereas HA-POST predominantly localized to the cytosol (Fig. 1B), indicating a possibility that POST is proactively retained in the cytosol by any mechanism.
CRM1 is the major receptor for protein nuclear export. CRM1 recognizes NESs of cargo proteins and exports them from the nucleus to the cytosol in a RanGTP-dependent manner (28). Leptomycin B (LMB) blocks the interaction between CRM1 and cargo proteins and inhibits the cargo export. We examined whether POST is exported by CRM1 or not. We treated the cells that express HA-POST with LMB and found that the treatment resulted in nuclear localization of HA-POST (Fig. 1C). These observations suggest that POST was located in the cytosol by its proactive export from the nucleus by CRM1.
Next, we examined direct interaction between GST-POST and CRM1 in the presence of RanGTP by GST pulldown analysis in vitro. GST as a negative control, GST-NES (SGNSNELALKLAGLDINKT) of protein kinase inhibitor (PKI) as a positive control, or GST-POST was immobilized on glutathione Sepharose and incubated with purified CRM1, His/S-Ran expressing bacteria lysate, and 0.5 mM GTP. CRM1 bound to GST-POST and GST-NES of PKI but not to GST (Fig. 1D), clearly showing direct interaction between POST and CRM1. These results suggested that POST is exported from the nucleus through direct binding to CRM1.
We surveyed NES responsible for the CRM1 binding in POST utilizing an NES prediction program LocNES (29) (prodata.swmed.edu/LocNES/LocNES.php), which predicted three potential NESs: EPPNLYPVKLYVYDL (NES-a), DLSKGLARRLSPIML (NES-b), and PFGQALRPLLDSIQI (NES-d) (Fig. 1E). In addition, we found a reverse-type NES-like sequence of class 1b (Φ1xxΦ2xxΦ3xΦ4; Φ is the hydrophobic residue) (29) IFLEYLSSL (NES-c) in POST (Fig. 1E). To determine which potential NES functions in cells, NES-a, -b, -c, and -d were each fused to the C terminus of a fluorescent protein Venus and expressed in NIH 3T3 cells. Venus-NES-a and NES-b predominantly localized to the nucleus, and LMB treatment did not affect the localization of Venus-NES-a and NES-b at all (Fig. 1F). Whereas Venus-NES-c showed localization to the cytosol, Venus-NES-d localized throughout the cytosol and the nucleus (Fig. 1F). LMB treatment biased these distributions of Venus-NES-c and NES-d toward the nucleus (Fig. 1F). These results suggested that NES-c and -d are potential functional NESs in POST.
To examine functional involvement of the potential NESs (c and d), the essential hydrophobic residues of NES-c or NES-d of POST were substituted for alanine residues to abolish the recognition by CRM1 (Fig. 1G). HA-tagged full-length WT-POST, NES-c mutant (NES-c mt) POST, or NES-d mutant (NES-d mt) POST was expressed in NIH 3T3 cells and immunostained with HA antibody. WT-POST and NES-c mt POST localized in the cytosol, whereas a considerable proportion of NES-d mt POST was detectable in the nucleus (Fig. 1H). From these results, we concluded that NES-d is the major NES for the CRM1-dependent export of POST from the nucleus (Fig. 1H).
UBIN Is Exported from the Nucleus to the Cytosol by POST in a CRM1-Dependent Manner.
Although UBIN was localized in the cytosol and the nucleus, its binding partner POST was predominantly localized in the cytosol through the action of its intrinsic NES (Fig. 1 B, G, and H). Therefore, we examined the effects of WT-POST and NES-d mt POST on the subcellular distribution of UBIN. Cotransfection with WT-POST led to the predominant cytosolic localization of UBIN, whereas NES-d mt POST did not affect the UBIN subcellular localization (Fig. 2A). These results indicated that POST mediates the export of UBIN from the nucleus to the cytosol in an NES-dependent manner.
Fig. 2.
UBIN is exported from the nucleus to the cytosol in a POST- and CRM1-dependent manner. (A) POST alters the subcellular distribution of UBIN from the nucleus to the cytosol. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.5 μg of UBIN-Flag only (Top), 0.5 μg of UBIN-Flag and 0.1 μg of HA-POST (Middle), or 0.5 μg of UBIN-Flag and 0.1 μg of HA-POST NES-d mt (Bottom). The cells were immunostained with anti-Flag antibody (green) and anti-HA antibody (red). (Scale bar: 10 μm.) (B) The cytosolic localization of UBIN is affected by the CRM1 inhibitor LMB. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.5 μg of pEGFP-UBIN-Flag. The cells were treated with vehicle (Left) or 10 nM LMB for 12 h (Right). (Scale bar: 10 μm.) (C) FLIP analysis of EGFP-UBIN. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.5 μg of pEGFP-UBIN-Flag and treated with vehicle (Upper) or 10 nM LMB (Lower) for 2 h. The red-circled areas in the cytosol were continuously bleached. The fluorescence intensity of the nucleus was measured after the cytoplasmic area was mostly bleached. (Scale bar: 10 μm.) (D) Quantification of the FLIP analysis shown in C. The average of three independent experiments is shown with SD (n = 3).
To confirm the CRM1-dependent export of UBIN, NIH 3T3 cells were transfected with UBIN harboring GFP at its N terminus (GFP-UBIN) and treated with LMB. The results showed nuclear accumulation of GFP-UBIN (Fig. 2B). Fluorescence loss in photobleaching (FLIP) analysis was performed to observe the dynamic transport of GFP-UBIN between the nucleus and the cytosol under the same conditions. In Fig. 2C, the area surrounded by the red circle in the cytosol was continuously quenched, and the change in the intensity of GFP in the nucleus was observed (Fig. 2C). The fluorescence intensity of GFP-UBIN initially disappeared from the cytosol and then from the nucleus, suggesting that UBIN translocates from the nucleus into the cytosol (Fig. 2 C and D). However, this effect was almost completely abolished by the CRM1 inhibitor LMB (Fig. 2 C and D). Taken together, these results suggested that UBIN is exported from the nucleus to the cytosol in a POST- and CRM1-dependent manner.
Proteasome Inhibition Leads to the Accumulation of Polyubiquitinated Proteins in the Cytosol.
As UBIN was exported from the nucleus by a POST-CRM1 complex, we next questioned whether ubiquitinated proteins bound to UBIN via its UBA domain in the nucleus can be exported from the nucleus by POST and CRM1. A previous study showed that stably expressing GFP-ubiquitin in Mel JuSo cells predominantly localized to the nucleus, and this localization shifted to the cytosol in the presence of the proteasome inhibitor MG-132; most of the nuclear ubiquitin existed in a complex with histone as monoubiquitinated histone, and treatment with proteasome inhibitors caused the deubiquitination of this complex and the translocation of ubiquitin into the cytosol (30). In this study, myc-ubiquitin was also detected in the nucleus as previously reported (30), and myc-ubiquitin accumulated and stained with aggregates in the cytosol in cells treated with proteasome inhibitor epoxomycin (EPX). Treatment with EPX and LMB resulted in the accumulation and aggregation formation of myc-ubiquitin in the nucleus (Fig. 3A) and accumulated in the cytosol, partly forming aggregates on EPX treatment, whereas combined treatment with EPX and LMB resulted in its accumulation and aggregation formation in the nucleus (Fig. 3A). Similarly, CRM1 knockdown by siRNA in conjugation with EPX treatment led to ubiquitin accumulation and aggregation formation in the nucleus (Fig. 3A). The knockdown efficiency of CRM1 was confirmed by real-time PCR (Fig. S1). These observations suggested that a portion of myc-ubiquitin bound to nuclear proteins as reported, and the ubiquitinated proteins in the nucleus were actively exported to the cytosol primarily by CRM1 under the proteasome-inhibited condition.
Fig. 3.
Proteasome inhibition leads to the accumulation of polyubiquitinated proteins in the cytosol in a CRM1-dependent manner. (A) Ubiquitinated proteins accumulate in the cytosol on proteasome inhibition and are relocated to the nucleus in response to inhibition of nuclear export. NIH 3T3 cells were transfected with 0.5 μg of HA-ubiquitin and treated with DMSO, 0.5 mM EPX for 8 h, 0.5 mM EPX and 10 nM LMB for 8 h, or CRM1 knockdown for 72 h and 0.5 mM EPX for the last 8 h. The cells were immunostained with anti-HA (12CA5) antibody (green). The white circles indicated by dashed lines indicate the nuclear region. (Scale bar: 10 μm.) (B) Detection of the nuclear export of endogenous ubiquitinated proteins using antiubiquitin antibody. NIH 3T3 cells were treated with DMSO, 0.5 mM EPX for 8 h, 0.5 mM EPX and 10 nM LMB for 8 h, or CRM1 knockdown for 72 h and 0.5 mM EPX for the last 8 h. The cells were immunostained with antiubiquitin (Apu2) antibody (red). The white circles indicated by dashed lines indicate the nuclear region. (Scale bar: 10 μm.) (C) Nuclear export of ubiquitinated proteins in HUVECs. HUVECs were treated with DMSO (Left) or 0.5 mM EPX for 8 h (Right). The cells were immunostained with antiubiquitin (FK2) antibody (green) and antiubiquitin (Apu2) antibody (red). The white circles indicated by dashed lines indicate the nuclear region. (Scale bar: 10 μm.) (D) Quantification of the Apu2 immunostaining of HUVECs (with SD). The total fluorescence intensity of the cytoplasmic region or nuclear region of HUVECs immunostained with Apu2 antibody was quantified (n = 10 experiments; each experiment contains 500 cells). ***P < 0.001.
K48-linked polyubiquitination targets proteins for proteasomal degradation. As immunofluorescence analysis of myc-ubiquitin cannot distinguish different forms of ubiquitination, such as mono-, K48-, and K63-linked ubiquitination, NIH 3T3 cells were stained with an anti–K48-linked polyubiquitin chain antibody (Apu2) (Fig. 3B). The Apu2 antibody detected a weak diffuse staining pattern of K48-linked polyubiquitinated proteins throughout the cell under the control condition, whereas EPX treatment resulted in a strong cytosolic staining pattern, including puncta formation of them. Cotreatment with EPX and LMB (or CRM1 knockdown) resulted in the nuclear accumulation and puncta formation of K48-linked polyubiquitin chains (Fig. 3B), indicating that K48-linked polyubiquitinated proteins are exported from the nucleus to the cytosol in a CRM1-dependent manner.
A similar distribution pattern of ubiquitinated proteins in response to treatment with EPX and LMB was observed in HeLa cells and human umbilical vein epithelial cells (HUVECs), which are human primary cells. Ubiquitin signal was predominantly detected in the nucleus in DMSO treatment (stained with antiubiquitin antibody FK2), whereas proteasome inhibition markedly caused its relocation to the cytosol (Fig. 3C) and formation of ubiquitin-positive aggregate in the cytosol (Fig. S2A). In HeLa cells and HUVECs, LMB treatment led to the nuclear accumulation and aggregate formation of ubiquitinated proteins (Fig. S2). Furthermore, double treatment with EPX and LMB resulted in enhanced nuclear accumulation and aggregate formation of ubiquitinated proteins in contrast to single treatment with a proteasome inhibitor EPX or LMB (Fig. S2). Quantitative analysis showed that EPX treatment increased cytosolic and nuclear ubiquitin accumulation, whereas additional treatment with LMB significantly increased accumulation of ubiquitinated proteins in the nucleus (Fig. 3D). We also investigated the behavior of protein modified with K63-linked polyubiquitin chain in NIH 3T3 cells. As already shown, immunofluorescent signals attributable to total ubiquitin (detected by FK2) and K48-linked chain (detected by Apu2) were dramatically affected by EPX single or EPX/LMB double treatments (Fig. S3A). In particular, significant ubiquitin aggregation was induced in the nucleus by EPX/LMB double treatment, whereas alteration of overall distribution of K63-linked chain was limited (Fig. S3B). Its predominant distribution in the cytosol barely changed on EPX or EPX/LMB treatments. However, the double staining with FK2 and anti-K63 antibodies showed that ubiquitin-positive nuclear aggregate (detected by FK2) contained an aliquot of K63-positive signal (Fig. S3B, Lower Right), suggesting that a CRM1-mediated nuclear export of K63 chain also occurred, although its biological relevance remains elusive.
UBIN Collaborates with POST in the Export of Ubiquitinated Proteins from the Nucleus to the Cytosol.
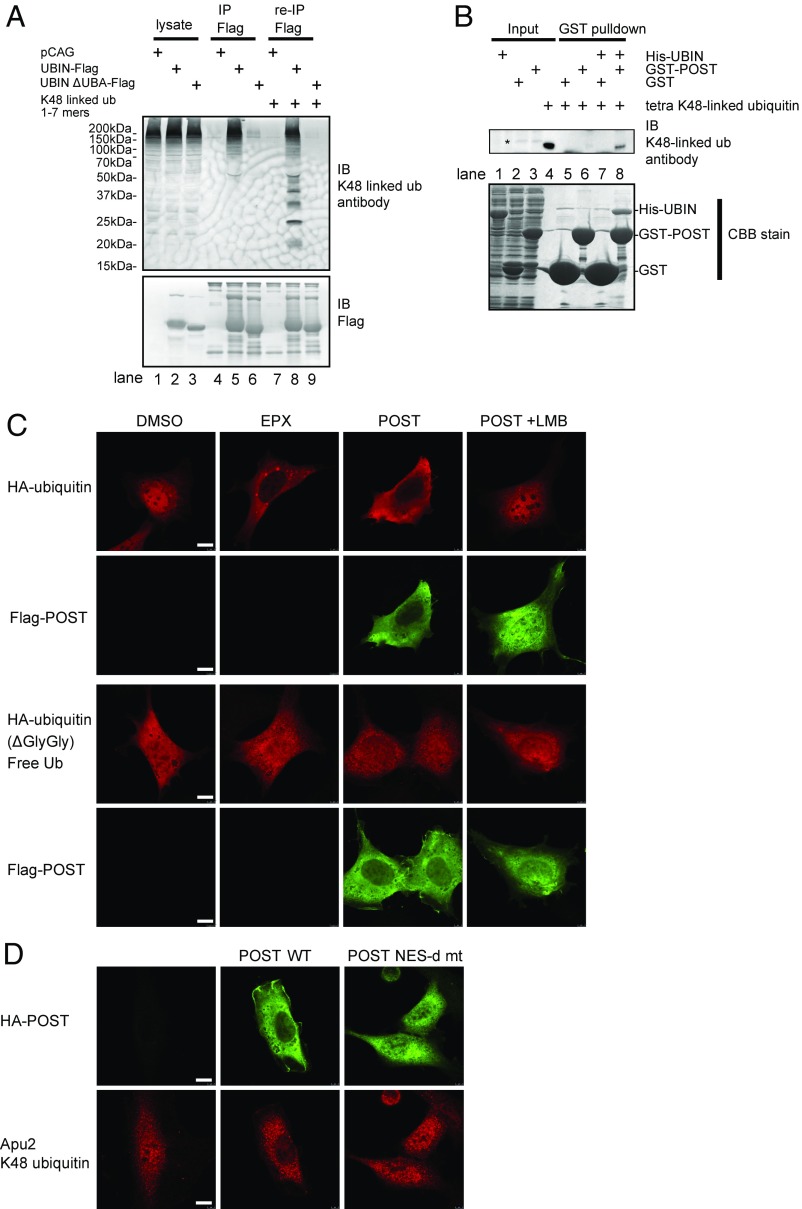
To address the possibility that UBIN exports polyubiquitinated proteins from the nucleus in collaboration with POST and Crm1, we first examined the type of ubiquitination that is recognized by UBIN. UBIN-Flag was transfected into HEK293 cells, and UBIN binding proteins were isolated from cell lysates with Flag-affinity beads (M2). K48-linked ubiquitinated proteins were effectively coprecipitated with UBIN but not with its ΔUBA mutant (Fig. 4A, lanes 5 and 6). Furthermore, the immunoprecipitated UBIN-Flag was incubated with a synthetic K48-ubiquitin chain mixture containing oligoubiquitin 1- to 7-mers for 1 h at 4 °C. Then, oligoubiquitin associated with the UBIN-Flag conjugated onto the Flag-affinity beads was analyzed by immunoblotting (Fig. 4A). The 2–7 K48-linked ubiquitin oligomers, but not the ubiquitin monomer, were successfully coimmnoprecipitated with UBIN-Flag (Fig. 4A, lane 8). UBIN lacking the intrinsic UBA domain (ΔUBA UBIN) binds to neither cellular K48-linked ubiquitinated proteins nor synthetic K48-linked polyubiquitin chains (Fig. 4A, lanes 6 and 9). Thus, UBIN recognized K48-linked polyubiquitinated proteins but not monomer ubiquitin through its UBA domain.
Fig. 4.
UBIN-POST exports ubiquitinated proteins from the nucleus to the cytosol. (A) UBIN physically binds to K48-linked polyubiquitin chains through UBA domain. HEK293T cells cultured on 35-mm dishes were transfected with 1.0 μg of pcDNA3.1-UBIN-Flag or 1.0 μg of pcDNA3.1-UBIN (ΔUBA)-Flag. WT and mutant UBIN-Flag were immunoprecipitated by Flag-M2 beads (lanes 4–6). The UBIN-Flag immobilized on the beads was incubated with recombinant K48-linked oligoubiquitin chains (1- to 7-mers; UC-240) for 1 h at 4 °C, and the precipitated oligoubiquitin chains were analyzed by immunoblotting (IB) (lanes 7–9). The cell lysate (lanes 1–3), proteins immunoprecipitated with UBIN-Flag (lanes 4–6), and coprecipitated oligoubiquitin chains (lanes 7–9) were analyzed by IB with antiubiquitin (Apu2) or anti-Flag antibody. IP, immunoprecipitation. (B) POST indirectly binds with K48-linked ubiquitin chain through UBIN. Immobilized GST or GST-POST on glutathione Sepharose 4B was incubated with (lanes 7 and 8) or without (lanes 5 and 6) His-UBIN for 1 h at 4 °C. After washing with PBS, GST or GST-POST immobilized beads with or without UBIN were incubated with 0.5 μg of K48-linked tetraubiquitin chain. Bound proteins were subjected to SDS/PAGE and analyzed by Coomassie staining (CBB; Lower) or IB with Apu2 antibody (Upper). *Nonspecific band. (C) POST mediates the nuclear export of ubiquitinated proteins but not that of free ubiquitin. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.4 μg of pcDNA3.1-HA-ubiquitin or pcDNA3.1-HA-ubiquitin (ΔGlyGly) with or without 0.1 μg of Flag-POST. The cells were treated with vehicle, 0.5 mM EPX, and/or 10 nM LMB for 8 h. The cells were immunostained with anti-Flag (rabbit) antibody (green) and anti-HA (12CA5) antibody (red). (Scale bar: 10 μm.) (D) The POST NES-d mt did not mediate the nuclear export of polyubiquitinated proteins. NIH 3T3 cells cultured on 35-mm dishes were transfected with 0.1 μg of pCAG-HA-POST or pCAG-HA-POST (NES-d mt). The cells were immunostained with anti-HA (12CA5) antibody and antiubiquitin (Apu2) antibody. (Scale bar: 10 μm.)
We further investigated binding affinity of UBIN to a range of ubiquitin chains, including minor chains. UBIN showed slight preference for K48-linked ubiquitin dimers compared with K63-linked ubiquitin chain in vitro (Fig. S4 A and B). However, UBIN broadly recognized other types of tetraubiquitin chains, including those K9-, K11-, K33-, and K48-linked (Fig. S4C), while biological relevance of the recognition of these minor chains by UBIN currently remains unknown.
We also tested an alternative possibility that POST directly binds to ubiquitin chain without help of UBIN. GST or GST-POST purified from Escherichia coli was immobilized on glutathione beads and incubated with K48-linked tetraubiquitin chain. GST and GST-POST did not solely bind with K48-linked tetraubiquitin chain (Fig. 4B, Upper, lanes 5 and 6), whereas GST-POST bound with K48-linked tetraubiquitin chain only in the presence of UBIN (Fig. 4B, Upper, lane 8). These results suggested that POST directly interacted with UBIN and indirectly bound with ubiquitin chain through the UBA domain of UBIN.
To determine whether UBIN and POST export ubiquitinated proteins from the nucleus, HEK293 cells were cotransfected with HA-ubiquitin and POST. HA-ubiquitin was predominantly detected in the nucleus in the control and accumulated in the cytosol in response to proteasome inhibition (Fig. 4C). Similar distribution pattern was observed in cells overexpressing POST in the absence of proteasome inhibition, suggesting that POST mediates the nuclear export of polyubiquitinated proteins (Fig. 4C). However, LMB treatment canceled such the effect of POST (Fig. 4C), suggesting that POST mediated the nuclear export of polyubiquitinated proteins in a CRM1-dependent manner. Of note, ubiquitin lacking the two C-terminal glycine residues (ΔGlyGly), which loses the C-terminal substrate conjugation site and remains as a monomer, was not exported into the cytosol in response to POST overexpression (Fig. 4C).
Finally, immunostaining with the anti–K48-linked polyubiquitin chain antibody (Apu2) confirmed that K48-linked polyubiquitinated nuclear proteins were predominantly observed in the cytosol in cells overexpressing POST, whereas overexpression of the NES-deficient POST mutant failed to export K48-linked polyubiquitinated proteins to the cytosol (Fig. 4D). These results strongly suggest that UBIN and POST cooperatively export K48-linked ubiquitinated proteins from the nucleus to the cytosol in an NES/CRM1-dependent manner.
Depletion of UBIN and POST Led to the Nuclear Accumulation of Ubiquitinated Proteins and a Shorter Lifespan in Caenorhabditis elegans.
The UBIN and POST proteins are conserved among metazoans. Therefore, the evolutionary conservation and in vivo relevance of the UBIN/POST system was examined in C. elegans. The expression patterns of ubql-1 and F36D4.5, which are the nematode homologs of UBIN and POST, respectively, were examined. The upstream regions of ubql-1 and F36D4.5, including regulatory elements, were cloned into the pPD95.75 vector, with GFP fused downstream of each regulatory element (Fig. S5 A–D). We found that the regulatory region of ubql-1 actively drives the expression of GFP in the pharynx, hypodermis, intestine, and head neurons (Fig. S5 A and B), while that of F36D4.5 is activated in the pharynx, hypodermis, intestine, head neuron, and tail neuron (Fig. S5 C and D). Therefore, ubql-1 and F36D4.5 were simultaneously expressed in the pharynx, intestine, and head neurons. We selected intestinal tissues to examine the cooperative nuclear export of ubiquitinated proteins mediated by ubql-1 and F36D4.5.
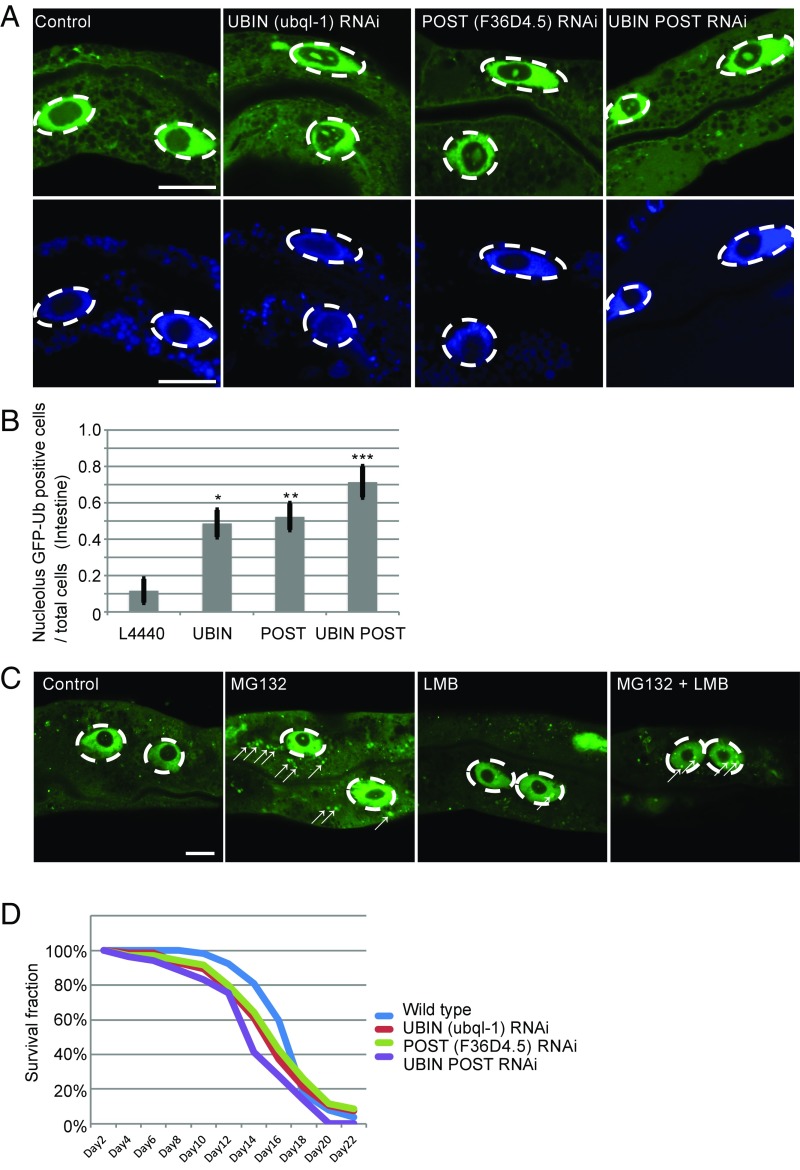
The localization of ubiquitinated proteins was examined in nematodes expressing GFP-tagged ubiquitin (GFP::Ub) under Ubql-1 (UBIN) and/or F36D4.5. (POST) knockdown condition. Knockdown of UBIN and/or POST led to the accumulation of ubiquitinated proteins in the nucleolus of intestinal cells (Fig. 5 A and B) (note that the nucleus of intestinal cells in worm typically harbors a single nucleolus as shown in Fig. S5E) (31, 32), suggesting that the UBIN/POST system is also responsible for the export of ubiquitinated proteins from the nucleus in the nematode.
Fig. 5.
Depletion of UBIN and POST led to nuclear accumulation of ubiquitinated proteins and a shorter lifespan in C. elegans. (A) Abnormal nucleolar deposition of ubiquitinated proteins in response to ubql-1 (UBIN) and F36D4.5 (POST) knockdown. Ubiquitinated protein deposition was observed with GFP-tagged ubiquitin (GFP::Ub; green; Upper). The nematodes were treated with ubql-1 RNAi, F36D4.5 RNAi, or both ubql-1 and F36D4.5 RNAi. The nucleus was stained with Hoechst-33342 (blue; Lower). The white circles indicated by dashed lines indicate the nuclear region, and the medial unstained region is the nucleolus. GFP-Ub was not distributed in the nucleolus under normal conditions (green). (Scale bar: 10 μm.) (B) Quantification of the data shown in A. The rate of the cells that showed nucleolar GFP signal per total cells is shown. Statistical significance was tested by one-way ANOVA and Tukey’s multiple comparison test. Multiple cells of five worms were analyzed for each treatment. Three independent trials were performed (thus, a total of 15 worms were analyzed for each treatment). *P < 0.05; **P < 0.01; ***P < 0.001. (C) Subcellular localization of ubiquitinated proteins. Fluorescence of GFP-Ub was observed by confocal microscopy. Nematodes were treated with MG-132 and/or LMB. White arrows indicate GFP-Ub–positive aggregation. The dashed white circles indicate the nuclear region. (Scale bar: 10 μm.) (D) Lifespan assays in control (L4440; blue line), ubql-1 knockdown (red line), F36D4.5 knockdown (green line), or ubql-1 and F36D4.5 double-knockdown nematodes (purple line). Mean median survival times (± SEM) of L4440, ubql-1 knockdown, F36D4.5 knockdown, and the double knockdown were 18 ± 0.67, 16.5 ± 1.00, 16.5 ± 1.00, and 14.75 ± 0.63 (n = 4). Each group in each trial included 70 worms. Only the double knockdown showed significantly less median survival time than that of control (P < 0.05; tested by Tukey’s multiple comparison test).
The localization of ubiquitinated proteins was also observed in nematodes expressing GFP-Ub (GFP::Ub) in the presence or absence of the proteasome inhibitor MG-132 (Fig. 5C). The GFP signal was predominantly observed in the nucleus and faintly observed in the cytosol. Such an increase in cytosolic GFP signal was not observed with the presence of LMB (Fig. 5C). We did not take into account these very small dots less than 1 μm in diameter, because they were detectable even in the control cells (e.g., the left control cell includes a few faint dots in the nucleolus). These findings were consistent with the results in mammalian cells. Treatment with MG-132 induced the formation of cytosolic aggregates, whereas simultaneous treatment with MG-132 and LMB resulted in the accumulation of aggregates in the nucleoplasm (Fig. 5C, arrows).
Careful observation revealed that GFP-Ub accumulated in the nucleolus in the ubql-1 (UBIN) and/or POST knockdown nematodes (Fig. 5A), which differed from the pattern in mammalian cells; however, the underlying mechanism remains unclear. Such a difference may be attributable to differences in intranuclear morphology: in nematodes, a single large nucleolus occupies a large part of the nucleus and stains negative for Hoechst-33342 under steady-state conditions (Fig. S5E). As previously reported (Discussion), CRM1 may be involved in the transport of ubiquitinated proteins to the nucleolus or cytosol from the nucleoplasm, and transport to the cytosol by CRM1 may be dependent on UBIN/POST. This possibility should be explored in future studies.
Finally, we performed lifespan assay under UBIN and/or POST knockdown conditions in nematode. Double knockdown of ubql-1 (UBIN) and F36D4.5 (POST) significantly shortened the lifespan of nematodes. Although single knockdown of them commonly showed similar tendencies, they were not statistically significant (Fig. 5D). This suggests that the UBIN/POST system is functional under physiological conditions and involved in the maintenance of protein homeostasis in the nematode.
Discussion
Here, we showed that the UBL-UBA protein UBIN and the small NES harboring protein POST form a nucleus–cytosol shuttling complex that mediates the nuclear export of ubiquitinated proteins. UBIN broadly binds to various types of ubiquitin chain, including the K48-linked chain via the intrinsic UBA domain, and POST mediates their export in cooperation with CRM1 through its intrinsic NES sequence, potentially supporting nuclear protein homeostasis, particularly when the protein degradation capacity is saturated in the nucleus. POST contains four potential NESs, whereas our data consistently suggested that just one of them (referred to as NES-d) is functional in cells. A reported crystal structure of POST (Protein Data Bank ID code 3EBQ) showed that the leucine residues of NES-d are exposed on the molecular surface of POST, while others are buried and most likely not accessible by CRM1, supporting this model. Depletion of UBIN and/or POST led to the nucleolar accumulation of ubiquitinated proteins in C. elegans and shortened its lifespan, suggesting that the UBIN/POST system contributes to the maintenance of cellular/organismal homeostasis.
These results indicated that polyubiquitinated proteins can be exported from the nucleus to the cytosol via the UBIN/POST system, presumably for degradation by the ubiquitin–proteasome system. Previous study suggested that the nuclear proteasome degrades the ubiquitinated proteins in the nucleus in yeast (33). In addition, the transport of misfolded proteins from the cytosol to the nucleus was reported in yeast and mammalian cells and suggested to mediate the sequestration of cytotoxic misfolded proteins (22, 34). However, treatment with proteasome inhibitors triggers the formation of insoluble cytosolic inclusions, such as aggresomes, ALIS, and the predominant accumulation of ubiquitinated proteins in the cytosol of mammalian cells. Thus, the accumulation of ubiquitinated proteins and their degradation in the nucleus or in the cytosol may be determined by the efficiency and capacity of each compartment for the elimination of those proteins.
The cytosol has a higher capacity for the elimination of unwanted proteins, such as misfolded, aggregate-prone proteins, than the nucleus, because autophagy as a bulk degradation system can degrade large molecules, such as aggregates. p62-mediated autophagy selectively eliminates ubiquitinated proteins, and the regulated protein sequestration systems, such as the aggresome, ALIS, and IPOD, prevent the cellular toxicity caused by the production of aggregates-prone proteins. These machineries are not available in the nucleus. Therefore, the capacity for protein quality control is greater in the cytosol than in the nucleus, despite the existence of nuclear proteasomes. ERAD consists of the retrograde translocation of misfolded proteins from the ER to the cytosol through the ER membrane. The degradation of misfolded mitochondrial proteins is mediated by their transport out of mitochondria before degradation. These findings support the notion that the cytosol is more suitable for the degradation of misfolded proteins. In addition, aggregates formed within the nucleus are difficult to eliminate, whereas aggregates formed in the ER and mitochondria may be eliminated by ER-phagy and mitophagy, respectively (35–38). The existence of nucleophagy was suggested by several reports, although data are limited (36, 39).
Knockdown of UBIN and/or POST led to inclusion body formation in the nucleolus of the nematode. This could indicate the existence of an alternate cargo transportation system in the nucleus. CRM1 mediates the bidirectional transportation of the cargo protein p53 between the nucleolus and the nucleoplasm, which is sensitive to LMB. A similar system mediates the localization of cytoplasmic polyadenylation element binding protein 1 (CPEB1). CPEB1 localizes to the cytosol and the CRM1-nucleolar bodies in the nucleolus; however, LMB treatment leads to the nucleoplasmic localization of CPEB1 (40). Similarly, ubiquitinated proteins were possibly exported from the nucleoplasm to the cytosol and nucleolus. Its nucleolar accumulation by ubql-1 or F36D4.5 knockdown suggested that they participate in nucleoplasm–cytosol shuttling but not in nucleolus–nucleoplasm shuttling. Although the molecular mechanism underlying this pathway and its applicability to other substrate proteins remain to be fully elucidated, our observation suggests that the nucleolus–nucleoplasm pathway is more active in the nematode than in the mammalian cell.
UBL-UBA proteins physically link ubiquitinated proteins to the proteasome and function as ubiquitin receptors, mediating the effective proteasomal degradation of misfolded proteins. This class of ubiquitin receptor often shows a preference for aggregation-prone proteins with exposed hydrophobic moieties on their molecular surfaces. The best studied member of the UBL-UBA protein family, Dsk2, a yeast homolog of UBIN, is involved in inclusion body formation of the mutant huntingtin (Htt) protein and its vacuolar degradation in yeast (41–43). Dsk2 also shows activity as a ubiquitin receptor for proteasomal degradation. Mammalian UBQLN family proteins (UBIN is the fourth of the UBQLN1 to -5 proteins) show preference for binding to aggregation-prone proteins; therefore, UBQLN maintains the solubility of mitochondrial membrane proteins, mediates the clearance of newly synthesized transmembrane ER proteins, promotes autophagic degradation of aggregated proteins in the cytosol, and promotes clearance of aggregated proteins by the proteasome in an autophagy-independent manner (44–48). Hjerpe et al. (48) recently showed that UBQLN2 mediates the proteasomal degradation of aggregated proteins, such as the mutant Htt protein, through hsp110-mediated disaggregation in the nucleus. Loss of the UBIN/POST system led to enhanced aggregates formation in the nucleus (or nucleolus in the nematode), suggesting that this system preferably acts on aggregation-prone species of ubiquitinated proteins. The correlation between POST and other UBL-UBA proteins, including UBQLN2, should also be addressed in future studies.
Materials and Methods
Expression Vectors and siRNAs.
Human cDNAs encoding UBIN and POST were amplified from reverse-transcribed mRNA from HeLa cells by PCR. Human POST cDNAs with mutant NESs were produced by site-directed mutagenesis. Human Flag-tagged UBIN was subcloned into the mammalian expression vector pcDNA3.1. Human HA- or Flag-tagged POST was subcloned into the mammalian expression vector pCAGGS. siCRM1-8 (CCUGGAAAUCCAGUUAACAACCAAA) and siCRM1-9 (UCAGCCUCAGCAGGUUCAUACAUUU) were purchased from Thermo Fisher Scientific.
Cell Culture and Transfection.
NIH 3T3, HEK293, and HeLa cells were grown in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin streptomycin, and 1% l-glutamine at 37 °C in a 5% CO2 humidified incubator. NIH 3T3 cells were transfected using Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions. For 3.5-cm dish subconfluent NIH 3T3 cells, 1 μg of DNA was incubated with 6.5 μL of enhancer in 100 μL of DNA-condensation buffer (buffer EC, which is included in the Effectene transfection kit) for 3 min at room temperature, and 10 μL of Effectene reagent was added and incubated for 5 min at room temperature. This transfection DNA solution was added to a 3.5-cm dish directly and incubated until the next experiment. HEK293 cells were transfected using Lipofectamine LTX reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. For 3.5-cm dish subconfluent HEK293 cells, 1 μg of DNA was incubated with 2 μL of PLUS reagent in 100 μL of DMEM for 3 min at room temperature, and 3 μL of Lipofectamine LTX reagent was added and incubated for 20 min at room temperature. This transfection DNA solution was added to a 3.5-cm dish directly and incubated until the next experiment.
HUVECs were cultured in HUVEC growth medium endothelial basal medium-2 with growth supplements (Lonza) at 37 °C in a 5% CO2 humidified incubator.
Immunoprecipitation.
Cells were washed with PBS and lysed with lysis buffer (40 mM Hepes-KOH, pH 7.5, 100 mM NaCl, 5 mM EDTA, 0.2% Triton X-100), and anti-FLAG M2 Affinity Gel (Sigma) was added. After a wash step with lysis buffer, proteins bound to the gel were eluted and separated by SDS/PAGE.
Immunofluorescence Microscopy.
Cells were fixed in 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 in PBS at room temperature. After permeabilization, the cells were blocked with blocking solution (1% goat serum, 1% BSA, 1% glycerol, 0.2% Triton X-100 in PBS) for 1 h. The cells were incubated with primary antibodies in blocking solution for 2 h followed by incubation for 1 h with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific) and 500 ng/mL DAPI, and they were mounted with ProLong Diamond (Thermo Fisher Scientific). Images were acquired with a TCS SP8 confocal microscope (Leica Microsystems).
FLIP Analysis.
After image acquisition in five frames using a 488-nm diode laser at 1.5% power, the cytoplasmic region of EGFP-UBIN–expressing cells was bleached in 57 iterations (5 min) using a 488-nm diode laser source set at 80% power with a TCS SP8 confocal microscope with an HC PL APO 40×/0.85 dry objective (Leica Microsystems) at 30 °C. The scanning and bleaching cycles were repeated for 2 min. Fluorescence signals were collected using a DD488/552 excitation dichroic mirror and prism spectrometer. The pinhole size corresponded to a 3.00 airy unit. One frame was scanned at 1,024 × 1,024 pixels at a 200-Hz scanning speed, and the zoom factor was four. The mean value of the fluorescent intensity of the nuclear EGFP-UBIN was measured by Las X software (Leica Microsystems) every 2 min for 31 samples. The measured fluorescence intensity was normalized to the baseline value.
Real-Time PCR.
Expression levels of CRM1 were determined by real-time PCR. Total RNA was isolated from NIH 3T3 cells treated with siRNA for 72 h using the High Pure RNA Isolation Kit (Roche) and reverse-transcribed using ReverTra Ace (TOYOBO). qRT-PCR was performed using Thunderbird Probe qPCR Mix (TOYOBO) and Universal Probe Library probes (Roche) on a LightCycler 480 (Roche). The sequences of primers used were as follows: CRM1, 5′-ccctaatcaagtgtgggacag-3′ and 5′-tgactgtttcaggatctttcagg-3′; and GusB, 5′-ctctggtggccttacctgat-3′ and 5′-cagttgttgtcaccttcacctc-3′. GusB was used as the internal control.
Quantification of Fluorescent Intensity.
HUVECs were plated onto 96-well clear-bottom plates (Greiner Bio-One GmbH). After treatment with vehicle (DMSO) or the reagents, the cells were fixed, immunostained for ubiquitin, and counterstained with DAPI as described in Immunofluorescence Microscopy. The images were obtained automatically focusing on the DAPI staining at 552 × 552 pixels by the CellInsight NXT high-content screening platform (Thermo Fisher Scientific) with the UPlanFL N 10×/0.30 dry objective lens (Olympus). The individual cells were determined based on the DAPI staining, and the sum total fluorescent intensities of immunostained ubiquitin were quantified in the DAPI-positive nuclear region or the cytosolic region without DAPI signal in individual cells by HCS studio 2.0 cell analysis software. Each fluorescent intensity was normalized by the cytosolic sum total fluorescent intensity of DMSO-treated cells; 500 cells were observed in 1 well of the 96-well plate.
Construction of Bacteria Expression Vectors.
UBIN and POST genes were subcloned into pET28a vector. pET-CRM1 was provided by Kumiko Ui-Tei, University of Tokyo, Tokyo, Japan, and the CRM1 genes were subcloned into pGEX6p-1 vector (49). pET30-His/S-Ran (canine) was provided by Yoshiyuki Matsuura, Nagoya University, Nagoya, Japan (50, 51).
Expression and Purification of Recombinant CRM1.
CRM1 was purified as previously reported (50, 51). Briefly, CRM1 was expressed in E. coli strain BL21 (DE3) RIL with addition of 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 20 °C for 16 h and immobilized onto glutathione Sepharose 4B with buffer M containing 10 mM Tris⋅HCl, pH 7.5, 0.2 M NaCl, 5 mM MgCl2, 0.05% Tween20, and 7 mM 2-mercaptoethanol. After washing the resin with buffer M, CRM1 was eluted by incubation with GST-PreScission protease (GE Healthcare) for removal of GST tag.
GST Pulldown Assay with CRM1.
Pulldown assays were performed in buffer M using the modified protocol as described (50, 51). GST fusion proteins were immobilized on 20 μL of glutathione Sepharose 4B and incubated with 700 μL of His/S-Ran expressing E. coli lysate in buffer M, 18 μg of CRM1, and 0.5 mM GTP at 4 °C for 2 h. Beads were washed twice with 1 mL of buffer M containing 0.1 mM GTP, and bound proteins were eluted with SDS sample buffer. These samples were subjected to SDS/PAGE and analyzed by Coomassie staining or immunoblotting.
GST Pulldown Assay with K48-Linked Tetraubiquitin Chain.
GST, GST-POST, and His-UBIN were expressed in E. coli strain BL21 (DE3) RIL with addition of 0.3 mM IPTG at 30 °C for 6 h. GST, GST-POST, and His-UBIN expressing E. coli were recovered from 50 mL of Luria Broth and lysed with 10 mL of PBS containing 0.2% Triton X-100. After centrifugation at 20,000 × g for 15 min, GST and GST-POST in 1.4 mL supernatant were immobilized onto 20 μL of glutathione Sepharose 4B. Next, GST and GST-POST immobilized glutathione 4B were incubated with or without 1.4 mL of His-UBIN expressing E. coli lysate. After washing, GST and GST-POST immobilized glutathione 4B were incubated with 0.5 μg of K48-linked tetraubiquitin chains in 300 μL of PBS containing 0.2% Triton X-100. Beads were washed twice with 1 mL of PBS, and bound proteins were eluted with SDS sample buffer. These samples were subjected to SDS/PAGE and analyzed by Coomassie staining or immunoblotting.
C. elegans Culture.
C. elegans Bristol (N2) strain nematodes were cultured using standard techniques (52). All strains were grown at 20 °C.
Transgenic C. elegans Strains.
The putative regulatory regions of ubql-1 (1,194 bp upstream of the start codon) and F36D4.5 (1,000 bp upstream of the start codon) were cloned into the PstI/BamHI site of pRPD95.75. These vectors were introduced into nematodes by gonad microinjection. The ERT 261 C. elegans strain was kindly provided by Emily R. Troemel, University of California, San Diego, La Jolla, CA (53).
Feeding RNAi Experiments.
Ublq-1 exons 2–4 (543 bp) or F36D4.5 exon 2 (435 bp) were cloned into the NotI/XbaI site of the RNAi feeding vector L4440. Nematodes at the L1 stage were placed on nematode growth media (NGM) agar plates containing 0.4 mM isopropyl-β-d-thiogalactopyranoside, 10 μg/mL tetracycline, 50 μg/mL ampicillin, and E. coli HT115 transformed with L4440 vector for the expression of Ublq-1 or F36D4.5 dsRNA or empty L4440 vector as a negative control.
C. elegans Imaging.
For imaging, adult nematodes were anesthetized with 2 mM levamisole and mounted on a 2% agar pad; images were captured using a laser-scanning confocal microscope LSM700 (Carl Zeiss Microscopy) with a Plan-Apochromat 20×/0.8 M27 dry or Alpha Plan-Apochromat 100×/1.46 oil DIC M27 oil immersion objective lens.
Lifespan Assay.
Ublq-1 or F23D4.5 knockdown L4 nematodes were transferred to fresh plates seeded with the corresponding bacterium on day 0, and the lines were transferred to fresh plates every 3 d. Nematodes were considered as dead when they no longer responded to a light touch on the nose. Mean median survival time (±SEM) was calculated from four independent trials. Statistical significance was tested by Tukey’s multiple comparison test. P < 0.05 was considered significant.
Drug Treatment.
MG-132 and LMB were dissolved in DMSO and ethanol, respectively, and then added to molten NGM agar before pouring into plastic dishes. The final concentration of MG-132 was 50 µM, and the final concentration of LMB was 50 ng/mL. Control NGM plates contained the appropriate solvent. The nematodes were cultured on the NGM agar plates containing drugs for 16 h.
Statistical Analysis.
All of the error bars represent SD. The group comparisons for the quantitative data obtained by immunofluorescence (Fig. 3D) were analyzed using Student’s t test (***P < 0.001). The multiple group comparisons for Fig. 5B were analyzed using one-way ANOVA and Tukey’s multiple comparison test (*P < 0.05; **P < 0.01; ***P < 0.001). The multiple group comparisons for Fig. 5D were analyzed using Tukey’s multiple comparison test (P < 0.05).
Supplementary Material
Acknowledgments
We are grateful to Dr. Kumiko Ui-Tei (University of Tokyo), Dr. Yoshiyuki Matsuura (Nagoya University), Dr. Emily R. Troemel (University of California, San Diego), and Dr. Kazutaka Araki (National Institute of Advanced Industrial Science and Technology) for reagents and kind helps, respectively. We appreciate Dr. Yo-hei Yamamoto (Kyoto Sangyo University) for kind help. This study was supported by Japan Society for the Promotion of Science KAKENHI Grants 24227009 (to K.N.) and 16K20998 (to S.H.) and partly by Takeda Science Foundation (K.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711017115/-/DCSupplemental.
References
- 1.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka K, Matsuda N. Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim Biophys Acta. 2014;1843:197–204. doi: 10.1016/j.bbamcr.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Lee DY, et al. Modulation of SOD1 subcellular localization by transfection with wild- or mutant-type SOD1 in primary neuron and astrocyte cultures from ALS mice. Exp Neurobiol. 2015;24:226–234. doi: 10.5607/en.2015.24.3.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexopoulou Z, et al. Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc Natl Acad Sci USA. 2016;113:E4688–E4697. doi: 10.1073/pnas.1523597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morito D, Nagata K. Pathogenic hijacking of ER-associated degradation: Is ERAD flexible? Mol Cell. 2015;59:335–344. doi: 10.1016/j.molcel.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Bragoszewski P, et al. Retro-translocation of mitochondrial intermembrane space proteins. Proc Natl Acad Sci USA. 2015;112:7713–7718. doi: 10.1073/pnas.1504615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr Opin Cell Biol. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quirós PM, Langer T, López-Otín C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 9.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 10.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 12.Güttler T, Görlich D. Ran-dependent nuclear export mediators: A structural perspective. EMBO J. 2011;30:3457–3474. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal A, Agrawal DK. Importins and exportins regulating allergic immune responses. Mediators Inflamm. 2014;2014:476357. doi: 10.1155/2014/476357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chook YM, Süel KE. Nuclear import by karyopherin-βs: Recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natalizio BJ, Wente SR. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013;23:365–373. doi: 10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung HY, Fu SC, Chook YM. Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. eLife. 2017;6:e23961. doi: 10.7554/eLife.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelham HR. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984;3:3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kampinga HH, Brunsting JF, Stege GJJ, Konings AWT, Landry J. Cells overexpressing Hsp27 show accelerated recovery from heat-induced nuclear protein aggregation. Biochem Biophys Res Commun. 1994;204:1170–1177. doi: 10.1006/bbrc.1994.2586. [DOI] [PubMed] [Google Scholar]
- 19.Kose S, Furuta M, Imamoto N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell. 2012;149:578–589. doi: 10.1016/j.cell.2012.02.058. [DOI] [PubMed] [Google Scholar]
- 20.Bryantsev AL, et al. Regulation of stress-induced intracellular sorting and chaperone function of Hsp27 (HspB1) in mammalian cells. Biochem J. 2007;407:407–417. doi: 10.1042/BJ20070195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sontag EM, Vonk WIM, Frydman J. Sorting out the trash: The spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol. 2014;26:139–146. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bersuker K, Brandeis M, Kopito RR. Protein misfolding specifies recruitment to cytoplasmic inclusion bodies. J Cell Biol. 2016;213:229–241. doi: 10.1083/jcb.201511024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda M, Koide T, Yorihuzi T, Hosokawa N, Nagata K. Molecular cloning of a novel ubiquitin-like protein, UBIN, that binds to ER targeting signal sequences. Biochem Biophys Res Commun. 2001;280:535–540. doi: 10.1006/bbrc.2000.4149. [DOI] [PubMed] [Google Scholar]
- 27.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 28.Fung HY, Chook YM. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin Cancer Biol. 2014;27:52–61. doi: 10.1016/j.semcancer.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fung HY, Fu SC, Brautigam CA, Chook YM. Structural determinants of nuclear export signal orientation in binding to exportin CRM1. eLife. 2015;4:e10034. doi: 10.7554/eLife.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YI, et al. Nucleolar GTPase NOG-1 regulates development, fat storage, and longevity through insulin/IGF signaling in C. elegans. Mol Cells. 2014;37:51–57. doi: 10.14348/molcells.2014.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jedrusik MA, Vogt S, Claus P, Schulze E. A novel linker histone-like protein is associated with cytoplasmic filaments in Caenorhabditis elegans. J Cell Sci. 2002;115:2881–2891. doi: 10.1242/jcs.115.14.2881. [DOI] [PubMed] [Google Scholar]
- 33.Tsuchiya H, Arai N, Tanaka K, Saeki Y. Cytoplasmic proteasomes are not indispensable for cell growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2013;436:372–376. doi: 10.1016/j.bbrc.2013.05.105. [DOI] [PubMed] [Google Scholar]
- 34.Miller SB, et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015;34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. 2014;127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 37.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci. 2012;125:1488–1499. doi: 10.1242/jcs.094110. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto K. Organellophagy: Eliminating cellular building blocks via selective autophagy. J Cell Biol. 2014;205:435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ernoult-Lange M, et al. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell. 2009;20:176–187. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, van Dyk D, Li Y, Andrews B, Rao H. A genome-wide synthetic dosage lethality screen reveals multiple pathways that require the functioning of ubiquitin-binding proteins Rad23 and Dsk2. BMC Biol. 2009;7:75. doi: 10.1186/1741-7007-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chuang KH, Liang F, Higgins R, Wang Y. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol Biol Cell. 2016;27:2025–2036. doi: 10.1091/mbc.E16-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itakura E, et al. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki R, Kawahara H. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep. 2016;17:842–857. doi: 10.15252/embr.201541402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothenberg C, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hjerpe R, et al. UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell. 2016;166:935–949. doi: 10.1016/j.cell.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19:17–35. doi: 10.1261/rna.034769.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koyama M, Matsuura Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010;29:2002–2013. doi: 10.1038/emboj.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 52.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakowski MA, et al. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10:e1004200, and erratum (2014) 10:e1004371. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]