Significance
Here we describe a fully functional gene drive system constructed in a major worldwide crop pest, Drosophila suzukii. This system is composed of a synthetic Medea drive with a maternal miRNA “toxin” and a zygotic “antidote,” and we demonstrate that it can bias inheritance with 100% efficiency and can persist in a population given high release frequencies. We discuss how such a system may be used to suppress D. suzukii populations or render them harmless to target crops.
Keywords: Medea, gene drive, Drosophila suzukii
Abstract
Synthetic gene drive systems possess enormous potential to replace, alter, or suppress wild populations of significant disease vectors and crop pests; however, their utility in diverse populations remains to be demonstrated. Here, we report the creation of a synthetic Medea gene drive system in a major worldwide crop pest, Drosophila suzukii. We demonstrate that this drive system, based on an engineered maternal “toxin” coupled with a linked embryonic “antidote,” is capable of biasing Mendelian inheritance rates with up to 100% efficiency. However, we find that drive resistance, resulting from naturally occurring genetic variation and associated fitness costs, can be selected for and hinder the spread of such a drive. Despite this, our results suggest that this gene drive could maintain itself at high frequencies in a wild population and spread to fixation if either its fitness costs or toxin resistance were reduced, providing a clear path forward for developing future such systems in this pest.
Spotted wing Drosophila, Drosophila suzukii, is a major worldwide crop pest of various soft-skinned fruits (1). Unlike other drosophilids that prefer to oviposit on overripe fruits, D. suzukii utilizes its serrated ovipositor to lay eggs inside ripening fruits, causing significant crop losses (1–3). Found only in Japan before the 1930s (4), in the last several decades D. suzukii has spread invasively to every continent except Antarctica (1, 2). In the United States, for example, D. suzukii was initially discovered in Santa Cruz, CA, in 2008 and since then has rapidly invaded many states and is a significant threat to fruit industries across the country (2). For example, between 2009 and 2014, D. suzukii caused an estimated $39.8 million in revenue losses for the California raspberry industry alone (5) and is responsible for 20–80% crop losses in other fruit production areas (1, 3, 4, 5). Current methods to control D. suzukii rely considerably on the use of broad-spectrum insecticides (e.g., malathion), which have variable efficacy (2), are difficult to use due to timing of fruit infestation (6), and face the risk of D. suzukii’s evolving resistance (7). While other forms of control may be possible [e.g., the use of recently identified natural predators (8) or oral delivery of dsRNA by microbes (9)], these approaches have not been widely adopted (10, 11). Therefore, given the rapid worldwide spread and potential economic impact of D. suzukii, novel effective control measures are urgently needed.
An alternative approach that would complement existing control methods would be the use of engineered D. suzukii as a genetic-based control strategy (12). Use of genetically modified insects for wild population manipulation was first suggested over half a century ago (13–15) and has garnered considerable interest in recent years (16–18). In fact, one method of using genetically modified insects for population control, a system called RIDL (Release of Insects carrying a Dominant Lethal) (19–21), where males carrying a repressible dominant lethal transgene are released to mate with wild females and produce nonviable progeny, has recently been implemented in the field. Although this strategy has been shown to be effective in reducing insect populations (22–24), it requires continuous rearing and ongoing inundative releases of large numbers of individuals, making it rather costly and labor-intensive; furthermore, it has not been developed for D. suzukii.
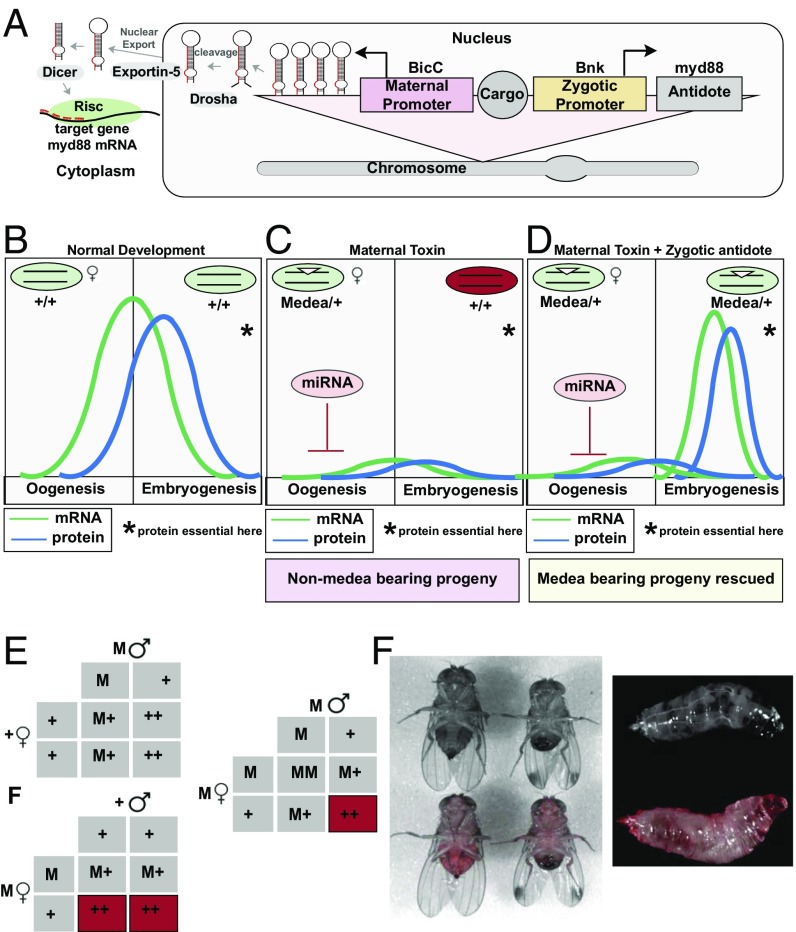
Other proposed methods of using genetically modified organisms for population control rely on engineered gene drive systems that function in a non-Mendelian fashion, allowing the drives to increase in frequency with each generation even without conferring fitness advantages to their host (16, 20, 25). Such methods could be utilized to spread desirable genes through populations or even to suppress target populations (26) and are promising self-sustaining tools for various applications where manipulation of wild populations may be desirable (17, 27). A number of engineered gene drive mechanisms have been proposed (16, 17, 20, 25, 26, 28) and tested in the laboratory (29–33); however, to date, only Medea (Maternal Effect Dominant Embryonic Arrest) and an underdominance-based approach have been demonstrated to bring about robust population replacement in WT laboratory populations (34–36). Specifically, Medea systems rely on expression of a toxin–antidote combination, such as a microRNA (miRNA) toxin that is expressed during oogenesis in Medea-bearing mothers, and a tightly linked antidote expressed early during embryogenesis in Medea-bearing progeny (Fig. 1A). The toxin is inherited by all progeny from a Medea-bearing mother, resulting in miRNA-mediated suppression of an essential embryonic gene that causes disruption of normal development during embryogenesis (Fig. 1 A–C). Offspring that inherit Medea receive a tightly linked antidote, consisting of a zygotically active miRNA-resistant copy of the targeted essential gene, that allows for restoration of normal development (Fig. 1D); non-Medea-bearing progeny from Medea-bearing mothers lack this antidote and perish (Fig. 1E). Due to this biased inheritance, Medea is predicted to rapidly spread itself, and any linked cargo genes, through a target population (34–36).
Fig. 1.
A synthetic Medea system in D. suzukii. A D. suzukii Medea transgene was generated to comprise an miRNA “toxin” targeting the 5′UTR of D. suzukii myd88 expressed under the predicted D. suzukii female germline–specific BicC promoter, an “antidote” consisting of D. suzukii myd88 coding region driven by the predicted D. suzukii early embryo-specific bnk promoter, and two separate transformation markers, eGFP under control of the eye-specific 3xP3 promoter and dsRed under control of the ubiquitous hr5-IE1 promoter (A). During normal development maternal myd88 is deposited into the embryo, where it is required for normal development (B). The Medea miRNA toxin targets myd88 mRNA during oogenesis, preventing proper deposition into the embryo and causing embryonic lethality in progeny that lack the Medea system (C). In embryos that possess a copy of the Medea system, a version of myd88 that is insensitive to the miRNA toxin is expressed during early embryogenesis, rescuing miRNA-induced lethality (D). When heterozygous Medea males are crossed out to WT females, all progeny survive since the maternal toxin is not expressed; however, when heterozygous Medea females are crossed to WT males, 50% of the progeny, the ones that fail to inherit Medea, perish. When heterozygous females are crossed to heterozygous males, 75% of the progeny inherit Medea, either from the mother or the father, and survive, while those that fail to inherit a Medea system perish (E). The hr5-IE1 promotes robust expression of dsRed in both D. suzukii adults and larvae, allowing for facile identification of Medea-bearing individuals (F).
For D. suzukii, since suppression of the pest population is ultimately desired, a synthetic Medea could be used to achieve suppression by spreading a cargo gene proffering susceptibility to a particular chemical, by driving in a conditional lethal gene activated by an environmental cue such as temperature or diapause (36), or by driving a synthetic target site for a population-suppressing homing gene drive system. Although the diapause approach has not been practically demonstrated, mathematical modeling has indicated that this is theoretically possible (36), and the increasing amount of information regarding D. suzukii life history (e.g., refs. 37 and 38) may make it more practically feasible. Therefore, given the potential utility of a Medea system in D. suzukii, we leveraged the limited genetic tools and techniques available in this nonmodel organism, for example the draft genome assembly (39) and transgenesis (40), to engineer a Medea-based population control technology. As D. suzukii is generally poorly genetically characterized, we had to resolve multiple independent issues to accomplish this feat (e.g., identifying and testing necessary components such as maternal and zygotic promoters and demonstrating the ability to engineer miRNAs that target desired sequences). Notwithstanding, we overcame these challenges and herein describe the successful development of a potent Medea system in D. suzukii. We demonstrate that this system is capable of drastic biased inheritance to achieve non-Mendelian transmission frequencies of up to 100% in many geographically distinct populations. This represents an example of a synthetic gene drive mechanism developed in a major crop pest.
Results
Generation of a Synthetic Medea Gene Drive.
To create a synthetic Medea gene drive in D. suzukii, we engineered a piggyBac vector comprising a miRNA toxin coupled with a toxin-resistant antidote, inspired by the architectures used to generate previous Medea systems in Drosophila melanogaster (36, 34). We designed synthetic miRNAs to target D. suzukii myd88, a highly conserved gene shown to be maternally deposited and required for dorsal–ventral patterning in the early embryo in D. melanogaster (41). We used the predicted D. suzukii female germline-specific bicoid (BicC) promoter to drive expression of a “toxin” consisting of a polycistronic array of four synthetic miRNAs each designed to target the 5′UTR of D. suzukii myd88 (Fig. 1A). Importantly, to ensure these miRNAs could target the desired sequence, we performed genomic DNA sequencing of the myd88 5′UTR target region in our reference D. suzukii strain (collected from Corvallis, OR) and designed the miRNAs against this sequence (Fig. S1). This Medea drive also contained an “antidote” consisting of the D. suzukii myd88 coding region, insensitive to the miRNAs as it did not contain the miRNA-targeted 5′UTR, driven by the predicted D. suzukii early embryo-specific bottleneck (bnk) promoter, and two separate transformation markers: eGFP driven by the eye-specific 3xP3 promoter (42) and dsRed driven by the ubiquitous hr5-IE1 promoter (43).
Characterization of Medea Genetic Behavior.
Following microinjection of the Medea transgene into D. suzukii embryos, a single G1 transformant male was recovered, as identified by ubiquitous hr5-IE1–driven expression of dsRed (Fig. 1F), and weak eye-specific 3xP3-driven eGFP. When outcrossed to several WT (non-Medea-bearing, +/+) females, this male produced roughly ∼50% Medea-bearing and ∼50% WT offspring, as would be expected from standard Mendelian segregation without biased inheritance (Table 1). Resulting heterozygous G2 Medea-bearing progeny were individually outcrossed to WT individuals of the opposite sex to determine inheritance patterns, and these individual outcrosses were continued for six generations (Table 1). Remarkably, until the G5 generation, all heterozygous Medea/+ mothers (n = 91) produced 100% Medea-bearing progeny (n = 1,028), while heterozygous Medea/+ fathers (n = 16) produced ∼50% Medea-bearing progeny (n = 268). While the majority of heterozygous Medea/+ G5 (23/31) and G6 (16/25) generation females also produced 100% Medea-bearing progeny, some heterozygous G5 (8/31), and G6 (9/25) females unexpectedly produced a small yet notable number (52/1,219) of WT offspring. Although the exact reason for the difference is unclear, later analysis suggested that resistance to the miRNA toxin might explain this unexpected observation. Notwithstanding, individually these G5 and G6 heterozygous Medea/+ females displayed significantly biased inheritance rates ranging from 76–96%, with an average rate of 86.4%. Overall, in six generations of individual female outcrosses, the percentage of Medea-bearing progeny borne by single heterozygous Medea/+ mothers (n = 147) was 97.7% (2,195/2,247; Table 1) as opposed to the 50% that would be expected with standard Mendelian segregation, indicating that the Medea drive system is extremely functional at biasing inheritance.
Table 1.
D. suzukii Medea shows predicted genetic behavior
| Generation | Sex (no. crossed) | No. of progeny | Average percent Medea, % |
| G1 | ♂ (1) | 22 | 54.5 |
| G2 | ♀ (9) | 126 | 100 |
| G2 | ♂ (3) | 45 | 43.3 |
| G3 | ♀ (32) | 299 | 100 |
| G3 | ♂ (12) | 201 | 48.8 |
| G4 | ♀ (50) | 603 | 100 |
| G5 | ♀ (31) | 785 | 96.8 |
| G6 | ♀ (25) | 434 | 93.8 |
| Medea+/total individuals from females (147) | 2,195/2,247 | 97.7 | |
Results of heterozygous Medea D. suzukii individual fly outcrosses to WT D. suzukii. G1 indicates the offspring from injected G0 individuals, with subsequent numbers (G2–G6) indicating subsequent generations.
D. suzukii Medea Exhibits Maternal-Effect Lethality and Zygotic Rescue.
To further characterize the genetics behind the highly biased inheritance patterns described above, additional crosses between individuals of various Medea genotypes were performed and confirmed that Medea exhibits maternal-effect lethality and zygotic rescue (Table 2). For example, matings between heterozygous Medea/+ mothers and WT fathers resulted in 55.63 ± 0.76% total embryo survival with 94.20 ± 1.33% of the progeny being Medea-bearing, while matings between heterozygous Medea/+ mothers and heterozygous Medea/+ fathers yielded 79.11 ± 3.95% total embryo survival with 94.12 ± 0.67% of the progeny being Medea-bearing. The higher-than-expected embryo survival is consistent with the observation that not all heterozygous Medea/+ mothers give rise to 100% Medea-bearing progeny, indicating that not all WT progeny from a heterozygous Medea/+ mother perish.
Table 2.
D. suzukii Medea chromosomes show maternal-effect lethality and zygotic rescue
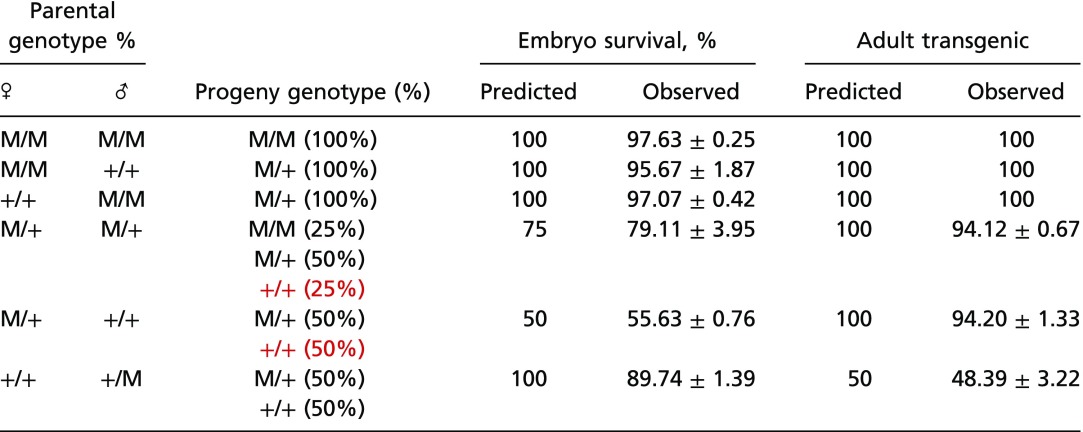 |
Crosses between parents of specific genotypes (indicated in the two leftmost columns) were carried out, and progeny survival to crawling first-instar larvae was quantified (% of surviving progeny and SD, third column from right). M indicates Medea, + indicates WT, and red text indicates genotypes expected to be inviable. The percentage of transgenic adults resulting from each cross type, together with the SD, was quantified (rightmost column).
Medea Functionality in Geographically Distinct Populations.
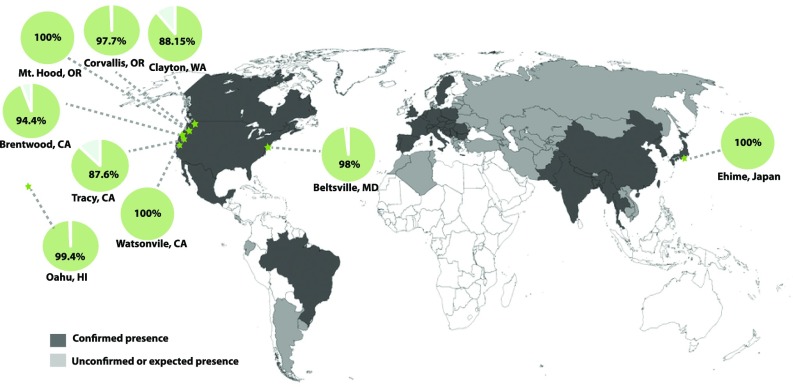
To assess whether the D. suzukii Medea could function in geographically distinct populations that possibly harbor genetic variability in regions that canonically have less conservation such as the 5′UTR, heterozygous Medea/+ flies were tested in eight additional D. suzukii strain backgrounds. These strains were collected from various locations around the world, including Mt. Hood, OR; Clayton, WA; Brentwood, CA; Tracy, CA; Watsonville, CA; Oahu, HI; Beltsville, MD; and Ehime, Japan. Interestingly, for three of eight strains the Medea inheritance rate from heterozygous Medea/+ mothers was 100%, while from five of nine strains the inheritance rate ranged from 87.6 to 99.4%, with an overall transmission rate of 94.2% (Fig. 2). These results strongly demonstrate that the Medea drive described here can dominantly bias transmission in diverse D. suzukii populations.
Fig. 2.
Medea functions in diverse populations of D. suzukii. Heterozygous Medea/+ individuals were crossed with eight geographically distinct D. suzukii populations and Medea inheritance was measured. Overall, Medea biased inheritance with rates ranging from 87.6 to 100%, suggesting that a Medea system generated in the laboratory could be utilized to manipulate some, but not all, diverse wild populations of D. suzukii. Green stars indicate the collection locations of the flies tested, green pie charts indicate the percentage Medea inheritance observed from heterozygous Medea/+ females, and shaded areas on the map indicate locations where D. suzukii populations have been confirmed. The Corvallis, OR strain was our reference D. suzukii strain used to engineer the Medea.
Long-Term Population Cage Experiments.
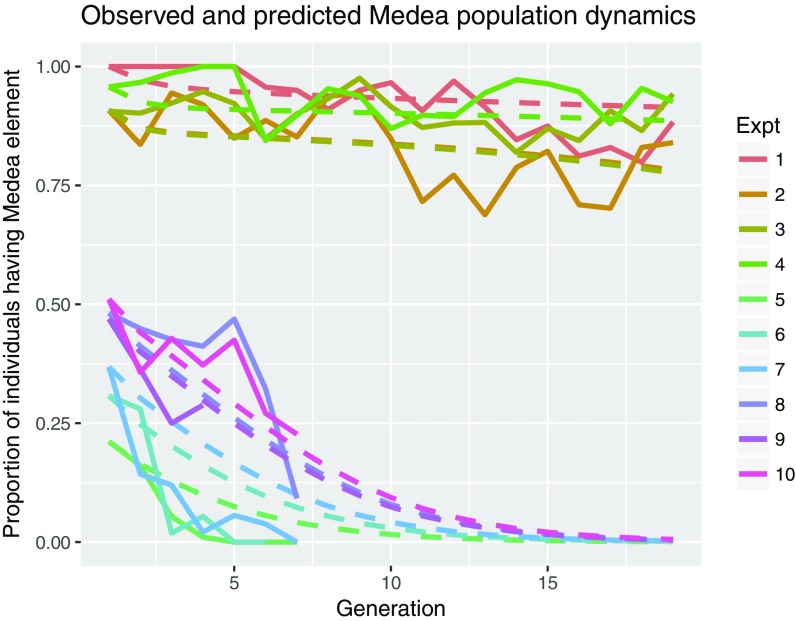
The above observations suggested that D. suzukii Medea should be able to drive robust population replacement. To test this prediction, we performed several long-term multigenerational population cage experiments specifically challenging the Medea drive with a WT strain that harbored preexisting resistance (Corvallis, OR). We set up these population cage studies after maintaining this population for approximately 10 generations; we mated Medea-bearing fathers to WT Corvallis, OR strain mothers at three distinct introduction (G0) frequencies: low frequency (25 heterozygous Medea/+ and 25 WT +/+ males mated to 50 WT +/+ virgins, Medea allele frequency of ∼12.5% and genotype frequency of ∼25%), medium frequency (50 heterozygous Medea/+ males mated to 50 WT +/+ virgins, Medea allele frequency of ∼25% and genotype frequency of ∼50%), and high frequency (50 homozygous Medea/Medea males mated to 50 WT +/+ virgins, Medea allele frequency of ∼50% and genotype frequency of ∼50%). These experiments were conducted in separate bottles in biological triplicate for the low- and medium-threshold drives and quadruplicate for the high-threshold drives, producing 10 distinct populations with G1 Medea allele frequencies ranging from ∼12.5–50% and genotype frequencies ranging from ∼25–100%. Altogether, these population cage experiments were followed for nine generations (for lower-allele-frequency populations, as the Medea allele disappeared from the population by that time) or 19 generations (for higher-allele-frequency populations), counting the number of Medea-bearing adults in each generation to determine the genotype frequency, as described previously (34, 36). Interestingly, the observed changes in Medea frequency over time indicated that, for release proportions (defined as the genotype frequency in the G1 population) of 50% or smaller, the D. suzukii Medea drive was unable to drive into the WT population, likely because of selected drive resistance combined with high fitness costs outweighing the effect of drive. However, at higher release proportions of >90%, similar to classical chromosomal rearrangement thresholds (44), the drive largely compensated for the fitness cost, allowing the drive to remain in the population at high frequencies for the duration of the experiment (19 generations; Fig. 3). Although unintended, the self-limiting dynamics of the generated Medea system may be useful in achieving a transient population transformation of the type associated with other proposed gene drives (e.g., ref. 45).
Fig. 3.
Observed and predicted dynamics of the D. suzukii Medea drive system. Population cage experiments were set up by mating WT (+/+) and heterozygous Medea (Medea/+) or homozygous Medea males (Medea/Medea) with WT (+/+) females, producing a frequency of heterozygotes (Medea/+) in the first generation of 25–100%. Population counts were monitored over 19 generations. Results from these experiments are shown as solid lines, with fitted model predictions shown as dashed lines. Observed data are consistent with a toxin efficiency of 100% in Medea-susceptible mothers, 93% in Medea-resistant mothers (95% CrI: 90–95%), a heterozygote fitness cost of 28% (95% CrI: 27–30%), a homozygote fitness cost of 65% (95% CrI: 62–67%), and an initial resistant allele population frequency of 78% (95% CrI: 57–97%). For high initial heterozygote frequencies (90–100%), the drive is capable of manipulating inheritance in its favor to maintain its presence at high population frequencies, despite a fitness cost. For lower initial heterozygote frequencies (∼50% or less), the drive is eliminated from the population.
Molecular Characterization of Resistance.
To understand whether resistance of the target mRNA to the toxin played a role in observed Medea inheritance rates of <100%, we performed genomic DNA sequencing of the myd88 5′UTR miRNA target region from randomly selected Medea/+ and +/+ progeny from generation 19 of the highest-threshold drive experiments described above to determine whether the miRNA target sites contained any mutations compared with our reference strain (against which the miRNAs were designed). Genomic sequence analysis revealed that, out of four miRNA target sites, one or two sites were perfectly conserved in Medea/+ individuals (site 4 or sites 1 and 4, depending on the individual), while only one (site 4) was perfectly conserved in +/+ individuals (Fig. S1). Additionally, for sites that had mutations, some of the mutation types were found in both Medea/+ and +/+ flies (an A → T mutation at position 7 of site 1, a C → A mutation at position 3 of site 2, and an A → T mutation at position 6 of site 3), while one mutation (an addition of an A after position 15 for site 1) was only found in +/+ flies, which may be indicative of its role in creating resistance.
To further this analysis, we also sequenced +/+ individuals from all of the geographically distinct populations tested for Medea functionality (shown in Fig. 2) and discovered a similar trend. In particular, only one of the four miRNA target sites was perfectly conserved (4), two others (2 and 3) had the same mutations in all strains (and the same mutations found in the Medea/+ and +/+ individuals described above), and a third site (1) had variable mutations (either deletion of the A at position 15 or an A → T mutation at position 7) that may correlate with Medea efficiency, since all strains with the deletion showed inheritance rates of <100%, while all strains with the A → T mutation had inheritance rates of 100%. Together, these observations indicate that the nature of mutations differed between genetic backgrounds with different observed Medea inheritance rates, and that mutations in the 3′ end of target site 1 (in the approximate region of the miRNA seed sequence) in particular seemed to correlate with inheritance rates of <100%. This suggests that the efficiency of the miRNA “toxin” is likely influenced by resistance alleles, which reduce Medea transmission; the exact effect of these alleles is worth further investigation, given the ability of such alleles to potentially block diverse types of drive systems in the wild.
Mathematical Modeling.
To characterize the population dynamics observed in the above cage experiments, we fitted a mathematical model to the observed data in which the Medea drive had an associated fitness cost in heterozygotes and homozygotes and there was a Medea-resistant allele present in the population that reduced toxin efficiency. For the fitted model, the Medea drive was estimated to have a toxin efficiency of 93% in individuals homozygous for the resistant allele [95% credible interval (CrI): 90–95%] and was assumed to have a toxin efficiency of 100% in individuals lacking the resistant allele. The Medea drive was estimated to confer a large fitness cost on its host—28% in heterozygotes (95% CrI: 27–30%) and 65% in homozygotes (95% CrI: 62–67%)—and the resistant allele was estimated to have an initial allele frequency of 78% in the population (95% CrI: 57–97%).
Predictive mathematical modeling based on these parameter estimates suggests that the Medea drive would spread to fixation in the absence of toxin resistance if released above a threshold frequency of 79% (Fig. S2A). Spread to fixation would also be expected if the fitness costs of the generated Medea drive were halved (Fig. S2C), even if all individuals in the population were homozygous for the Medea-resistant allele (Fig. S2D), provided the drive was released above a threshold frequency of ∼25–27%. Consistent with the experimental results (Fig. 3), a Medea drive with a large fitness cost in a Medea-resistant population is expected to be maintained at high frequencies through its drive; however, its eventual elimination is inevitable unless supplemental releases are carried out. However, for high release frequencies (90–95%), the drive may be maintained at high frequencies (>75%) for ∼20 generations (Fig. S2B), which likely exceeds the duration required for agricultural impact. Of note, the ability of the drive to counteract large fitness costs is significant, as demonstrated by comparison with nondriving alleles with analogous fitness costs that rapidly decline in frequency following a 95% release (black lines in Fig. S2 A and C).
Prior mathematical modeling demonstrated the potential for Medea to induce population suppression by spreading a conditional lethal gene into a population (36). In Fig. S3, we illustrate that a Medea drive may also be used to drive a synthetic target site for a population-suppressing homing gene drive system. If the Medea drive has negligible fitness costs, then the WT allele can be driven out of a population. The Medea allele then acts as a target site for a population-suppressing homing construct, which can cause a population crash provided that it carries the zygotic antidote to the Medea toxin in its cargo (Fig. S3D). Such a two-phased approach may be preferred over the use of a homing-based drive by itself due to the invasiveness of homing-based gene drive systems (46); however, it should be noted that the WT allele acts as a homing-resistant allele following release of the homing construct and hence must first be eliminated entirely. Such approaches require further modeling in finite, structured populations.
Discussion
This study represents a comprehensive characterization of a fully functional Medea-based gene drive being challenged with preexisting resistance in long-term, multigenerational population cage experiments (19 generations). The synthetic Medea drive described here showed maximal levels of biased inheritance, up to 100% in some populations, but <100% inheritance bias in other populations, and <100% inheritance bias in later generation outcrosses performed with the original population. Although it is not entirely clear why <100% inheritance bias appeared in the patterns observed (e.g., in G5 and G6, but not previous generation outcrosses), we hypothesized that, in general, this difference in biased inheritance rates could be attributed to the presence of resistance arising from naturally occurring genetic variation that rendered certain embryos immune to the miRNA toxin. This hypothesis is supported by the sequencing data, as many of the sequenced miRNA target sites contained mutations that likely affected miRNA function and lowered toxin efficiency. Although we did not attempt to measure individual miRNA efficiency, it is possible that not all of the miRNAs are effective at target gene knockdown, and that particular target site mutations reduce toxin efficiency significantly enough to allow survival of a few WT individuals. This is further supported by sequencing data collected from the eight distinct geographic populations, which suggest that certain target site mutations may be correlated with incomplete Medea-biased inheritance patterns, although such patterns may also be explained by variable penetrance, and warrants further investigation, as it may have practical implications for the ability to use multiplexing to overcome drive resistance in general (47).
The above observations highlight the importance of resistance as a possible impediment to the use of many kinds of gene drives, including toxin–antidote drive systems, in the field (17, 25, 27). Multiple recent studies have highlighted resistance as a major obstacle to gene drive utility, mostly in the context of homing-based CRISPR/Cas9 drives (47–51). Although a Medea drive system may be less prone to resistance-associated spread impediment because, unlike homing-based drive, its mechanism of action is not likely to generate resistant alleles (17, 47), it will face preexisting resistant alleles given the natural genetic diversity found in wild populations. Furthermore, such mutations would be expected to face strong positive selection in the presence of the drive and increase in frequency over time, which would likely expand their effect and mitigate drive spread. Therefore, any meaningful attempt at generating a Medea-based gene drive system capable of manipulating diverse wild populations must plan for, and mitigate the effects associated with, both resistant alleles and fitness costs, which may be achieved in several ways.
To reduce the chances of resistance acting as an impediment to spread, sequencing-based characterization of naturally occurring genetic variation in geographically distinct target populations can be used to help guide selection of target sites that are well conserved across all populations in which the drive is intended to function. Additionally, miRNA target site selection could be limited to the coding DNA sequence regions of a genome, which tend to be strongly conserved, as opposed to regions such as the 5′UTR, which canonically have higher tolerance for sequence variation. Finally, the choice of multiple target sites that have been independently validated to achieve knockdown and the creation of a polycistronic “toxin,” perhaps consisting of eight miRNAs as opposed to the currently utilized four, may ensure that toxin efficiency is maximally high and unlikely to be deactivated by a single target site mutation. To decrease fitness costs, verification that the “antidote” components (e.g., the recoded targeted gene and promoter used to express said gene) function efficiently enough to restore WT function can ensure that an imperfect antidote does not impose fitness costs. Additionally, reducing the expression of the marker gene to a specific tissue type will likely reduce some of any possible fitness costs associated with high ubiquitous overexpression of an exogenous gene.
Moreover, modeling results suggest that a Medea drive having a high fitness cost and high (though imperfect) toxin efficiency may be capable of maintaining itself in a population for a period of several years following a series of large-scale releases of homozygous males. Either decreasing the fitness cost of the drive or minimizing resistance to the toxin is expected to enable the drive to spread to fixation above a release threshold of ∼25–79% (the lower bound corresponds to halved fitness costs). While the stated release thresholds are high, they may be achievable given multiple successive releases and are well below releases associated with the successful sterile insect technique for the Mediterranean fruit fly (52). This may be desirable for biosafety considerations, as Medea drive with significant fitness costs is less likely to spread widely (52). Medea also has the added benefit that, if fitness costs decline over time, its drive is frequency-dependent and hence large, and intentional releases are more likely to lead to spread than small, unintentional ones (45). That said, the potential and implications of fitness costs’ evolving should be further investigated.
Materials and Methods
Construct Assembly.
To generate plasmid OA-961B, components were cloned into the piggyBac plasmid pBac[3xP3-EGFP afm] (53) using Gibson assembly/EA cloning (54). Specifically, the predicted D. suzukii bottleneck (bnk) promoter was amplified from D. suzukii genomic DNA using primers 961B.5 and 961B.6, the predicted D. suzukii myd88 coding region was amplified from D. suzukii genomic DNA using primers 961B.3 and 961B.4, and the SV40 3′UTR fragment was amplified from template pWalium20-10XUAS-3XFLAG-dCas9-VPR (Addgene plasmid 78897) using primers 961B.1 and 961B.2. The pBac[3xP3-EGFP afm] plasmid was digested with AscI and FseI, and the above three fragments were cloned in via EA cloning. The resulting plasmid was then digested with PmeI, and the following fragments were cloned in via EA cloning: the predicted D. suzukii Bicaudal-C (BicC) promoter region amplified with primers 961B.7 and 961B.8 from D. suzukii genomic DNA, the SV40 3′UTR fragment amplified with primers 961B.9 and 961B.10 from template pWalium20-10XUAS-3XFLAG-dCas9-VPR (Addgene plasmid 78897), the hr5-IE1 promoter region (43) amplified from vector pIEx-4 (Novagen plasmid 71235-3) using primers 961B.11 and 961B.12, and the dsRed-SV40 3′UTR fragment amplified from template pScarlessHD-DsRed (Addgene plasmid 64703) with primers 961B.13 and 961B.14. Assembled miRNAs were then subcloned into final plasmid OA-961B using PacI and FseI. A list of primer sequences used in the above construct assembly can be found in Table S3; the D. suzukii myd88 coding region sequence, bnk promoter region sequence, and BicC promoter region sequence can be found in Table S1. The D. suzukii myd88, BicC, and bnk gene orthologs were identified using the Augustus gene prediction tool (55). The full sequence of final plasmid OA-961B is available on Addgene (plasmid 104967).
Fly Culture and Strains.
D. suzukii WT flies from Corvallis, OR, were a kind gift of P. Shearer, Oregon State University, Corvallis, OR and were maintained under standard conditions described in detail in Supporting Information, which also describes the specific schemes utilized to assess Medea genetic behavior. To prevent any unintentional release of the D. suzukii Medea-bearing flies into the environment we have undertaken stringent precautions including the following measures: (i) shatterproof polypropylene plastic vials were used, (ii) boxes with triple-contained flies were kept in locked facilities at all times in an institutional biosafety committee-approved BSL2 insectary, and (iii) only a single highly expert investigator handled the flies. We reasoned that these precautions are sufficient given that spread of this Medea drive requires a significant introduction threshold.
Mathematical Modeling.
Model fitting was carried out using Bayesian Markov chain Monte Carlo (MCMC) methods in which parameters describing the population dynamics of the Medea drive were estimated, including 95% CrIs. Estimated parameters include fitness costs associated with being heterozygous, sHet, or homozygous, sHom, for the Medea drive, and the reduced maternal toxin efficiency associated with the Medea-resistant allele, eR, present in the population at a given initial frequency, pR. Prior information on the parameter eR was inferred from G5 and G6 outcrosses in which heterozygous Medea females were mated with WT males and the proportion of WT offspring was nonzero. A simplified version of the fitted model was used to infer the expected dynamics of the generated Medea drive and one with its fitness costs halved in both a fully Medea-susceptible population and a fully Medea-resistant population. It was also extended to model the use of Medea to introduce a target site for a population-suppressing homing construct. The modeling framework is described in Supporting Information.
Supplementary Material
Acknowledgments
We thank Joanna Chiu (University of California, Davis) for providing the various D. suzukii strains used in this study. The molecular work was supported by a grant from the California Cherry Board (to O.S.A.). The modeling work was supported by a grant from the Innovative Genomics Institute, University of California, Berkeley/San Francisco (to J.M.M.).
Footnotes
Conflict of interest statement: A.B. and O.S.A. have filed a patent on the Suzukii Medea drive system.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713139115/-/DCSupplemental.
References
- 1.Walsh DB, et al. Drosophila suzukii (Diptera: Drosophilidae): Invasive pest of ripening soft fruit expanding its geographic range and damage potential. J Integr Pest Manag. 2011;2:G1–G7. [Google Scholar]
- 2.Asplen MK, et al. Invasion biology of spotted wing Drosophila (Drosophila suzukii): A global perspective and future priorities. J Pest Sci. 2015;88:469–494. [Google Scholar]
- 3.Goodhue RE, et al. Spotted wing drosophila infestation of California strawberries and raspberries: Economic analysis of potential revenue losses and control costs. Pest Manag Sci. 2011;67:1396–1402. doi: 10.1002/ps.2259. [DOI] [PubMed] [Google Scholar]
- 4.Langille AB, Arteca EM, Newman JA. The impacts of climate change on the abundance and distribution of the spotted wing Drosophila (Drosophila suzukii) in the United States and Canada. PeerJ. 2017;5:e3192. doi: 10.7717/peerj.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farnsworth D, et al. Economic analysis of revenue losses and control costs associated with the spotted wing drosophila (Drosophila suzukii (Matsumura)) in the California raspberry industry. Pest Manag Sci. 2017;73:1083–1090. doi: 10.1002/ps.4497. [DOI] [PubMed] [Google Scholar]
- 6.Cini A, Ioriatti C, Anfora G. A review of the invasion of Drosophila suzukii in Europe and a draft research agenda for integrated pest management. Bull Insectol. 2012;65:149–160. [Google Scholar]
- 7.Smirle MJ, Zurowski CL, Ayyanath M-M, Scott IM, MacKenzie KE. Laboratory studies of insecticide efficacy and resistance in Drosophila suzukii (Matsumura) (Diptera: Drosophilidae) populations from British Columbia, Canada. Pest Manag Sci. 2017;73:130–137. doi: 10.1002/ps.4310. [DOI] [PubMed] [Google Scholar]
- 8.Gabarra R, Riudavets J, Rodríguez GA, Pujade-Villar J, Arnó J. Prospects for the biological control of Drosophila suzukii. BioControl. 2015;60:331–339. [Google Scholar]
- 9.Murphy KA, Tabuloc CA, Cervantes KR, Chiu JC. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci Rep. 2016;6:22587. doi: 10.1038/srep22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woltz JM, Donahue KM, Bruck DJ, Lee JC. Efficacy of commercially available predators, nematodes and fungal entomopathogens for augmentative control of Drosophila suzukii. J Appl Entomol. 2015;139:759–770. [Google Scholar]
- 11.Abrieux A, Chiu JC. Oral delivery of dsRNA by microbes: Beyond pest control. Commun Integr Biol. 2016;9:e1236163. doi: 10.1080/19420889.2016.1236163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rota-Stabelli O, Blaxter M, Anfora G. Drosophila suzukii. Curr Biol. 2013;23:r8–r9. doi: 10.1016/j.cub.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Curtis CF. Possible use of translocations to fix desirable genes in insect pest populations. Nature. 1968;218:368–369. doi: 10.1038/218368a0. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton WD. Extraordinary sex ratios. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- 15.Serebrovskii AS. On the possibility of a new method for the control of insect pests. Zool Zh. 1940;19:618–630. [Google Scholar]
- 16.Burt A. Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130432. doi: 10.1098/rstb.2013.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17:146–159. doi: 10.1038/nrg.2015.34. [DOI] [PubMed] [Google Scholar]
- 18.National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Life Sciences; Committee on Gene Drive Research in Non-Human Organisms: Recommendations for Responsible Conduct . Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values. National Academies; Washington, DC: 2016. [PubMed] [Google Scholar]
- 19.Thomas DD, Donnelly CA, Wood RJ, Alphey LS. Insect population control using a dominant, repressible, lethal genetic system. Science. 2000;287:2474–2476. doi: 10.1126/science.287.5462.2474. [DOI] [PubMed] [Google Scholar]
- 20.Alphey L, et al. Genetic control of Aedes mosquitoes. Pathog Glob Health. 2013;107:170–179. doi: 10.1179/2047773213Y.0000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alphey L, et al. Sterile-insect methods for control of mosquito-borne diseases: An analysis. Vector Borne Zoonotic Dis. 2010;10:295–311. doi: 10.1089/vbz.2009.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey-Samuel T, et al. Pest control and resistance management through release of insects carrying a male-selecting transgene. BMC Biol. 2015;13:49. doi: 10.1186/s12915-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris AF, et al. Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes. Nat Biotechnol. 2012;30:828–830. doi: 10.1038/nbt.2350. [DOI] [PubMed] [Google Scholar]
- 24.Harris AF, et al. Field performance of engineered male mosquitoes. Nat Biotechnol. 2011;29:1034–1037. doi: 10.1038/nbt.2019. [DOI] [PubMed] [Google Scholar]
- 25.Bull JJ. Evolutionary decay and the prospects for long-term disease intervention using engineered insect vectors. Evol Med Public Health. 2015;2015:152–166. doi: 10.1093/emph/eov013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- 27.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan G, Braig H. Genetically Engineered Organisms: Assessing Environmental and Human Health Effects. CRC; Boca Raton, FL: 2001. The spread of genetic constructs in natural insect populations; pp. 251–314. [Google Scholar]
- 29.Windbichler N, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves RG, Bryk J, Altrock PM, Denton JA, Reed FA. First steps towards underdominant genetic transformation of insect populations. PLoS One. 2014;9:e97557. doi: 10.1371/journal.pone.0097557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33:1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantz VM, Bier E. Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112:E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C-H, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 35.Akbari OS, et al. A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr Biol. 2013;23:671–677. doi: 10.1016/j.cub.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbari OS, et al. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth Biol. 2014;3:915–928. doi: 10.1021/sb300079h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shearer PW, et al. Seasonal cues induce phenotypic plasticity of Drosophila suzukii to enhance winter survival. BMC Ecol. 2016;16:11. doi: 10.1186/s12898-016-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiman NG, et al. Drosophila suzukii population response to environment and management strategies. J Pest Sci. 2016;89:653–665. doi: 10.1007/s10340-016-0757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu JC, et al. Genome of Drosophila suzukii, the spotted wing drosophila. G3 (Bethesda) 2013;3:2257–2271. doi: 10.1534/g3.113.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schetelig MF, Handler AM. Germline transformation of the spotted wing drosophilid, Drosophila suzukii, with a piggyBac transposon vector. Genetica. 2013;141:189–193. doi: 10.1007/s10709-013-9717-6. [DOI] [PubMed] [Google Scholar]
- 41.Kambris Z, et al. DmMyD88 controls dorsoventral patterning of the Drosophila embryo. EMBO Rep. 2003;4:64–69. doi: 10.1038/sj.embor.embor714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berghammer AJ, Klingler M, Wimmer EA. Genetic techniques: A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 43.Ren L, et al. Comparative analysis of the activity of two promoters in insect cells. Afr J Biotechnol. 2011;10:8930–8941. [Google Scholar]
- 44.Foster GG, Whitten MJ, Prout T, Gill R. Chromosome rearrangements for the control of insect pests. Science. 1972;176:875–880. doi: 10.1126/science.176.4037.875. [DOI] [PubMed] [Google Scholar]
- 45.Gould F, Huang Y, Legros M, Lloyd AL. A killer-rescue system for self-limiting gene drive of anti-pathogen constructs. Proc Biol Sci. 2008;275:2823–2829. doi: 10.1098/rspb.2008.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall JM. The effect of gene drive on containment of transgenic mosquitoes. J Theor Biol. 2009;258:250–265. doi: 10.1016/j.jtbi.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 47.Marshall JM, Buchman A, Sánchez CHM, Akbari OS. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep. 2017;7:3776. doi: 10.1038/s41598-017-02744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unckless RL, Clark AG, Messer PW. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics. 2017;205:827–841. doi: 10.1534/genetics.116.197285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drury DW, Dapper AL, Siniard DJ, Zentner GE, Wade MJ. CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci Adv. 2017;3:e1601910. doi: 10.1126/sciadv.1601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. Evolutionary dynamics of CRISPR gene drives. Sci Adv. 2017;3:e1601964. doi: 10.1126/sciadv.1601964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammond AM, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 2017;13:e1007039. doi: 10.1371/journal.pgen.1007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyck VA, Hendrichs J, Robinson AS. The Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management. Springer; Dordrecht, The Netherlands: 2005. [Google Scholar]
- 53.Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210:630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- 54.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 55.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]