Significance
Endoplasmic reticulum (ER) stress-mediated relocalization of ER chaperones to the cell surface allows cells to expand their functionality beyond the ER, impacting survival, death, migration, and immunity. However, little is known about the underlying mechanisms and the interacting partners on the cell surface. Both protooncogene tyrosine-protein kinase SRC and TGF-β are important players in cell signaling, growth, apoptosis, and survival. Our discoveries that SRC activation is the driving force in the escape of ER luminal proteins to the cell surface via disruption of retrograde transport provide an oncogenic function for SRC and uncover a mechanism for regulation of TGF-β signaling via the GRP78/CD109 partnership, with therapeutic implications in cancer and other human diseases.
Keywords: endoplasmic reticulum stress, SRC protein kinase, retrograde transport, GRP78, TGF-β signaling
Abstract
The discovery that endoplasmic reticulum (ER) luminal chaperones such as GRP78/BiP can escape to the cell surface upon ER stress where they regulate cell signaling, proliferation, apoptosis, and immunity represents a paradigm shift. Toward deciphering the mechanisms, we report here that, upon ER stress, IRE1α binds to and triggers tyrosine kinase SRC activation, leading to ASAP1 phosphorylation and Golgi accumulation of ASAP1 and Arf1-GTP, resulting in KDEL receptor dispersion from the Golgi and suppression of retrograde transport. At the cell surface, GRP78 binds to and acts in concert with a glycosylphosphatidylinositol-anchored protein, CD109, in blocking TGF-β signaling by promoting the routing of the TGF-β receptor to the caveolae, thereby disrupting its binding to and activation of Smad2. Collectively, we uncover a SRC-mediated signaling cascade that leads to the relocalization of ER chaperones to the cell surface and a mechanism whereby GRP78 counteracts the tumor-suppressor effect of TGF-β.
Endoplasmic reticulum (ER) chaperones, originally identified as glucose depletion-inducible proteins and protein foldases, are increasingly recognized as major regulators of cellular homeostasis in health and disease with unexpected roles beyond the ER compartment (1–4). The recent discovery that ER stress not only induces the expression of ER chaperones to cope with ER protein quality control but also actively promotes their relocation to the cell surface represents a paradigm shift for their functions (5). For example, at the cell surface, the 78-kDa glucose-regulated protein (GRP78), also referred to as “BiP/HSPA5,” acts as a receptor regulating signaling pathways as well as viral entry, while other ER chaperones, such as GRP94, calreticulin (CRT), and protein disulfide isomerase (PDI), modulate functions including immune responses and cell–cell adhesion (1, 6, 7). Global profiling of the cell-surface proteome of tumor cells also revealed a relative abundance of cytosolic heat-shock proteins (8). Thus, relocalization of stress-inducible ER and cytosolic chaperones to the cell surface could represent a major common adaptive mechanism for cells to expand the functionality of these proteins in response to proteotoxic stress, turning on signaling pathways distinct from their intracellular functions and thereby impacting both survival and death. This is particularly important in the context of cancer where cell-surface ER chaperones have been reported to regulate oncogenic signaling pathways, proliferation, cell adhesion, and immunity (4, 6, 9).
Despite these advances, how ER stress actively induces cell-surface relocalization of ER chaperones distinct from passive exposure of ER content during cell death and how they exert their biological functions at the cell surface are largely unknown. A general feature among ER-resident proteins is a C-terminal tetrapeptide, Lys-Asp-Glu-Leu (KDEL), or related motifs as the ER-retention signal (10). When the ER proteins arrive at the Golgi, the KDEL receptors (KDELRs) recognize the KDEL and related motifs and package them into coat protein complex I (COPI) vesicles leading to Golgi-to-ER retrograde transport (7, 11, 12). Thus, a potential escape mechanism for the ER chaperones upon ER stress could be through perturbation of the KDELR retrieval machinery, which remains to be established.
SRC is a nonreceptor protein-tyrosine kinase belonging to the SRC family kinases (SFK) which is overexpressed and activated in a large number of human malignancies (13). SRC activation is majorly regulated via two phosphorylation sites including an activating phospho-tyrosine, pY419, within the kinase domain and an inhibitory phospho-tyrosine, pY530, at the regulatory tail. As the most investigated proto-oncogene, SRC is known to play important roles in cell morphology, differentiation, proliferation, invasion, adhesion, and survival (13, 14). Interestingly, active SRC has been reported to localize to the ER and Golgi complex (15), and SRC activation inhibited the KDEL-dependent retrograde transport by dispersing KDELR from the Golgi with no effect on anterograde transport (16). A major regulator of retrograde vesicle assembly is ADP ribosylation factor 1 (Arf1), a member of the Arf family of small GTPases (11). At the cis-Golgi, Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (GBF1) is the GTP exchange factor (GEF) that forms Arf1-GTP, which then recruits effectors to the Golgi to assemble retrograde vesicle (17, 18). Here, using a combination of biochemical, mutational, and imaging approaches, we uncover a signaling cascade triggered by ER stress linking the ER stress sensor IRE1α and SRC to the disruption of the dynamic equilibrium of KDELR in the Golgi and leading to the escape of ER chaperones to the cell surface.
While the role of the ER form of GRP78 as a pivotal regulator of the unfolded protein response (UPR) is well established (6, 19, 20), how cell-surface GRP78 (csGRP78) regulates cell signaling is just emerging. We recently discovered that csGRP78 majorly exists as a peripheral protein on the plasma membrane via interaction with other cell-surface proteins (21). In examining the repertoire of csGRP78-interacting proteins, our proteomic analysis led to the identification of CD109, the expression of which is up-regulated in a wide range of cancers and is associated with poor prognosis (22, 23). CD109, a member of the α-2-Macroglobulin (α2M) family and a glycosylphosphatidylinositol (GPI)-anchored protein, blocks TGF-β signaling by promoting degradation of the TGF-β receptors (TβRs) (24, 25). Interestingly, activation of SRC suppresses Smad2/3 phosphorylation, and ablation of SRC sensitizes TGF-β–induced growth arrest and apoptosis (26, 27), although the mechanism is not known. Here, we uncover a pathway linking IRE1α, SRC, csGRP78, and CD109, leading to the protection of cancer cells from the tumor-suppressor effect of TGF-β.
Results
IRE1α and SRC Regulate Cell-Surface Relocalization of ER Chaperones.
To determine how ER stress actively promotes ER chaperones’ trafficking to the cell surface, HeLa cells with adherent properties well-suited to withstand the experimental protocol were treated with the ER-stress inducer thapsigargin (Tg) for 6 h before treatment with a membrane-impermeable biotinylating reagent to specifically label cell-surface proteins which were purified from intracellular proteins by avidin pull down (5, 21). The ER luminal chaperones being analyzed include GRP78, GRP94, CRT, and PDI, which all contain the KDEL motif, as well as ERp72 containing the KEEL motif (Fig. S1A). EphB4, a transmembrane cell-surface receptor known to traffic to the cell surface via the Golgi, and MHC class I served as the loading controls for the purified cell-surface protein fraction. Upon Tg treatment, SRC activation, as monitored by pY419 level, was readily detected at 2 h and increased 5.5-fold by 6 h (Fig. 1A). SRC activation preceded the increase in the cell-surface form of the ER chaperones. Within the first 6 h, while the UPR was being activated as evidenced by eIF2α phosphorylation and XBP1 splicing, the total ER chaperone and cell-surface EphB4 levels remained constant (Fig. 1A and Fig. S1B). These results showed that the increase in cell-surface ER luminal chaperones is unlikely to be due to saturation of the KDELR or acceleration of anterograde trafficking. Extending the analysis to 24 h showed increases in the levels of cell-surface GRP78, GRP94, CRT, and PDI, all preceding the increase in their total level from the whole-cell lysate (Fig. S1C). Furthermore, knockdown of CRT, which is known to shuttle with ERp57 to the cell surface (28), had no effect on csGRP78 expression (Fig. S1D).
Fig. 1.
IRE1α and SRC regulate cell-surface relocalization of ER chaperones. (A) HeLa cells were treated with Tg for the indicated times. The indicated proteins from whole-cell lysate (WCL) and the cell surface (CS) were analyzed by Western blot with GAPDH and EphB4 serving as loading controls for whole-cell lysate and cell-surface proteins, respectively. In all panels, “pSRC” indicates pSRC(Y419). The band intensities for pSRC and csGRP78 were quantified from three experiments and graphed. (B) As in A, except stable HeLa cell lines expressing shSRC or control shRNA (−) were treated as indicated. (C) As in A, except HeLa cells were transfected with SRC531 or empty vector (−) and were treated as indicated. (D) As in A, except HeLa cells were transfected with IRE1α shRNA (shIRE1α) or control shRNA (−) and were treated with Tg. (E) Western blot analysis of the indicated proteins in HeLa cells with or without Tg treatment. (F) HeLa cells with or without Tg treatment were subjected to co-IP using anti-IRE1α antibody. The indicated proteins were analyzed by Western blot along with whole-cell lysate. (G) IRE1α-knockout HeLa cells expressing HA-IRE1α were treated or were not treated with Tg and were subjected to immunofluorescent staining for the HA epitope (green) and SRC (red). In the merged image, yellow indicates costaining of the two proteins. (Scale bar, 10 µm.) (H) As in F, except IRE1α-knockout HeLa cells transfected with WT HA-IRE1α or the mutant (ΔP) devoid of amino acids 965–977 were not treated or were treated with Tg and subjected to co-IP. (I) Summary of IRE1α and SRC involvement in ER stress-induced chaperone relocalization. *P < 0.05, **P < 0.01, ***P < 0.005.
To test the involvement of SRC in regulating ER chaperones’ translocation to the cell surface, HeLa cells were pretreated with the SFK inhibitor SU6656 or were subjected to ectopic expression of a dominant-negative mutant of SRC in human breast cancer MCF-7 cells. Both effectively blocked Tg-induced cell-surface expression of all the ER luminal chaperones being tested (Fig. S2 A and B). Previously, we discovered that cancer cells resistant to therapeutic treatment, e.g., the androgen-independent, metastatic prostate cancer cell line C4-2B, intrinsically expressed higher csGRP78 levels than its parental cell line LNCaP (29). In these cells, pSRC(Y419) was also intrinsically elevated compared with LNCaP (Fig. S2C). Upon treatment with SFK inhibitors (SU6656 or PP2), the levels of pSRC(Y419) and cell-surface ER chaperones were all suppressed, while total SRC and EphB4 levels were unaffected (Fig. S2D). The requirement of SRC for this regulation was demonstrated by the nearly complete suppression of cell-surface expression of ER chaperones in SRC-knockdown HeLa and C4-2B cells treated with either Tg or tunicamycin (Tu) (Fig. 1B and Fig. S2 E–G), as well as in HEK293 cells treated with Tg (Fig. S2H). SRC dependence was observed in a wide range of cancer cell lines, including both solid and blood tumors, while SRC independence was observed in the colon cancer cell line HCT116, in which, in contrast to other cells, csGRP78 expression was independent of Golgi integrity (Fig. S2I) (21). Furthermore, overexpression of a constitutively active SRC mutant (SRC531) (30) was more potent than Tg in driving ER luminal chaperones to the cell surface, and the effect was enhanced in combination (Fig. 1C). Collectively, these results demonstrate that SRC is both sufficient and necessary for ER stress-induced relocalization of ER chaperones to the cell surface.
To decipher how ER stress activates SRC, we tested the involvement of the ER transmembrane sensor IRE1α, which is activated upon ER stress. In HeLa cells, knockdown of IRE1α potently suppressed Tg-induced pSRC(Y419) levels and cell-surface relocalization of ER luminal chaperones without affecting EphB4 (Fig. 1D). Dephosphorylation of inhibitory pY530 of SRC is a major mechanism for SRC activation (13). Interestingly, we detected no change in the pSRC(Y530) level in control and Tg-treated cells, while, corresponding with pSRC(Y419) activation, the SRC downstream effectors STAT1 and AKT were activated (Fig. 1E). SRC can be activated in an Src homology 3 (SH3)- or Src homology 2 (SH2)-dependent manner which requires protein–protein interaction (31). Upon Tg treatment, SRC coimmunoprecipitated with IRE1α (Fig. 1F), consistent with the colocalization of the two proteins at the perinuclear region as revealed by confocal microscopy (Fig. 1G). In contrast to IRE1α, csGRP78 expression was not affected by PERK or ATF6 deficiency (Fig. S3A).
Bioinformatic analysis predicted proline-rich motifs at the cytosolic tail of IRE1α which match the class VIII noncanonical SH3-binding motif (Fig. S3B) (32), suggesting a potential SRC-binding site. To test this, we created CRISPR knockout of IRE1α in HeLa cells, which mimicked the effects of IRE1α knockdown (Fig. 1D and Fig. S3C) and showed that reconstitution with WT IRE1α restored Tg-induced SRC binding, activation, and cell-surface expression of ER-resident chaperones; however, the IRE1α deletion mutant lacking the proline-rich motifs (ΔP) was unable to do so (Fig. 1H and Fig. S3D). In contrast, IRE1α mutants defective in dimerization (D123P), kinase (K599A), or RNase activity (K907A), all lacking XBP1-splicing activity (Fig. S3B) (33, 34), were still able to rescue Tg-induced SRC activation and csGRP78 expression (Fig. S3E). Collectively, these results imply that, upon ER stress, SRC is recruited to form a complex with IRE1α dependent on its proline-rich motif at its cytosolic tail and, upon activation, triggers a cascade of events leading to the relocalization of ER luminal chaperones to the cell surface (Fig. 1I).
ASAP1 Is the Downstream Substrate of SRC in Cell-Surface Relocalization of ER Luminal Chaperones.
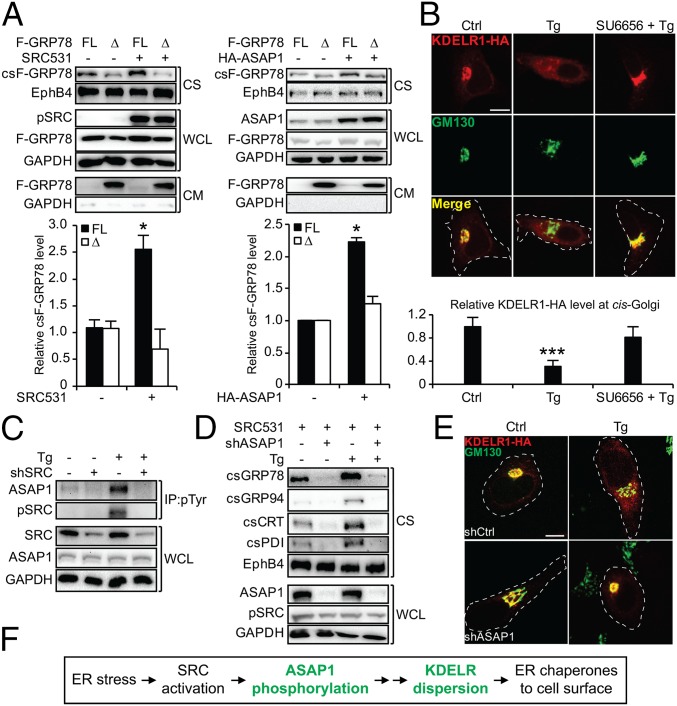
In HeLa cells, SRC531 expression increased cell-surface relocalization of the full-length FLAG-GRP78 (F-GRP78) but not the GRP78 KDEL-deleted mutant (Δ), which was secreted into the medium (Fig. 2A), suggesting that SRC regulation acted through the KDELR retrieval machinery. Among the KDELR isoforms, KDELR1 is the most abundant and exhibits the highest affinity for KDEL (35). To monitor the cellular distribution of KDELR1, we created KDELR1 bearing an HA tag at its C terminus and confirmed by immunofluorescence that it is localized to the cis-Golgi (Fig. 2B). Tg treatment caused about 70% dispersion of KDELR1 from the cis-Golgi in HeLa cells as well as in SK-MEL-28 cells, and this required SRC activation since the effect was blocked by SU6656 (Fig. 2B and Fig. S4A).
Fig. 2.
The SRC substrate ASAP1 mediates ER stress-induced KDELR1 dispersion and ER chaperone relocalization. (A) HeLa cells were transfected with the full-length F-GRP78 (FL) or the KDEL deletion mutant (Δ) in combination with either SRC531 (Left) or HA-ASAP1 (Right). The indicated proteins from the whole-cell lysate, cell surface, and conditioned medium (CM) were analyzed by Western blot with GAPDH and EphB4 serving as loading controls. The lack of GAPDH in the conditioned medium confirmed cell integrity. The relative csF-GRP78 level under each condition was quantified and graphed. (B) HeLa cells were transfected with KDELR1-HA, pretreated with SU6656 followed by Tg, and subjected to immunofluorescent (IF) staining for the HA epitope (red) and GM130 (green), with the latter serving as marker for cis-Golgi. In the merged image, yellow indicates costaining of the two proteins. The white dashed lines outline the cell shape. (Scale bar, 10 µm.) The immunofluorescent intensities of KDELR1-HA at the cis-Golgi were quantified and graphed. (C) HeLa cells expressing shSRC or control shRNA (−) with or without Tg treatment were subjected to immunoprecipitation (IP) using anti–phospho-tyrosine (pTyr) antibody. The indicated proteins were analyzed by Western blot along with whole-cell lysate. (D) As in A, except HeLa cells expressing shASAP1 or control shRNA (−) transfected with the SRC531 expression vector were treated as indicated. (E) As in B, except HeLa cells expressing shASAP1 or control shRNA (shCtrl) were treated as indicated. (Scale bar, 5 µm.) (F) Summary of ASAP1 and KDELR1 involvement in ER stress-induced chaperone relocalization. *P < 0.05; ***P < 0.005.
While SRC531 expression did not lead to tyrosine phosphorylation of KDELR1 (Fig. S4B), in Tg-treated HeLa cells where SRC was activated we observed prominent tyrosine phosphorylation of several protein bands, which could be SRC substrates as their phosphorylation was blocked by the SRC inhibitor dasatinib (Das) currently in clinical use (Fig. S4C) (36). Among these, the 130-kDa band was identified as Arf-GAP with the SH3 domain, ANK repeat, and PH domain-containing protein 1 (ASAP1) reported to be an SRC substrate involved in retrograde transport (Fig. S4C) (37, 38). An in vivo kinase assay confirmed that ASAP1 was phosphorylated by SRC (Fig. S4D), and knockdown of SRC eliminated ASAP1 phosphorylation upon ER stress (Fig. 2C).
ASAP1 is an essential downstream effector for the SRC effect, since SRC531 failed to induce cell-surface relocalization of ER chaperones in HeLa cells with efficient knockdown of ASAP1 by shRNA in either the absence or presence of Tg (Fig. 2D). The dependence on ASAP1 was also observed in MCF-7 cells (Fig. S4E). Similar to SRC531, overexpression of ASAP1 led to the increase of full-length F-GRP78, but not the KDEL-deleted (Δ) mutant, at the cell surface (Fig. 2A). Furthermore, Tg was able to disperse KDELR1 in scramble shRNA-expressing (shCtrl) HeLa cells but not in shASAP1 HeLa cells (Fig. 2E). Other ER stress inducers (DTT and hypoxia) also required the IRE1α/SRC/ASAP1 axis for csGRP78 expression (Fig. S4F), correlating with KDELR dispersion (Fig. S4G). Taken together, these results indicated that ER stress activates SRC, which phosphorylates its substrate ASAP1, leading to KDELR dispersion and cell-surface relocalization of ER chaperones (Fig. 2F).
ER Stress Promotes ASAP1 and GBF1 Complex Formation and Enhances GBF1 GEF Activity.
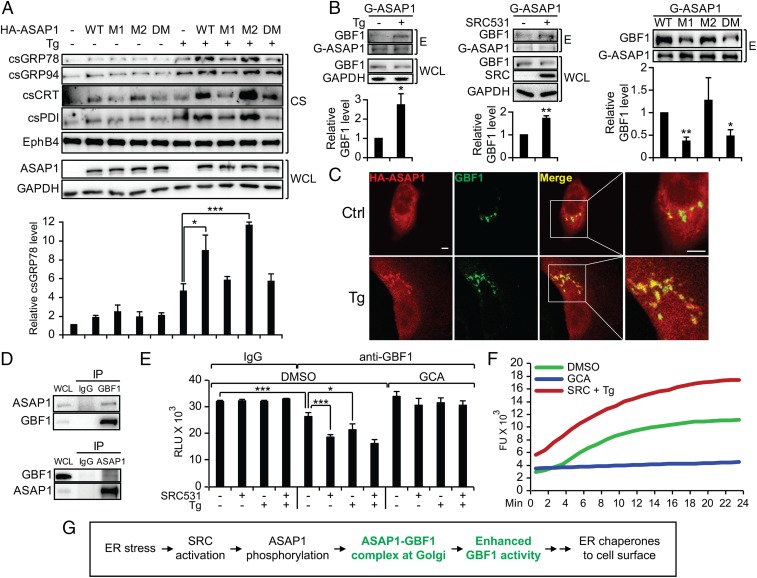
Human ASAP1 contains tyrosine phosphorylation sites at Y312 and Y767 (Fig. S5A) (39). We first determined that SRC531 could form complex and phosphorylate ASAP1 with a mutation at either Y312F or Y767F (referred to as “M1” and “M2,” respectively) but not when both were mutated (DM) (Fig. S5B). Interestingly, only M1 and DM, but not M2, blocked Tg-induced cell-surface relocalization of ER chaperones with no effect on EphB4 (Fig. 3A). Correspondingly, M1 failed to promote SRC531-induced cell-surface relocalization of ER chaperones (Fig. S5C), indicating that Y312 is the critical ASAP1 phosphorylation site to mediate the SRC effect. In probing for downstream targets of ASAP1 using a GST pull-down assay, we uncovered that GBF1, a GEF localized to cis-Golgi and known to regulate retrograde transport (38), could bind to GST-tagged ASAP1 (Fig. S5D). In such an assay, treatment of cells with Tg or expressing SRC531 enhanced the formation of a GBF1 complex with GST-ASAP1, and this interaction was suppressed in the M1 and DM mutants of GST-ASAP1 (Fig. 3B).
Fig. 3.
ER stress promotes ASAP1 and GBF1 complex formation and enhances GBF1 GEF activity. (A) HeLa cells transfected with HA-ASAP1 WT or mutant expression vectors (M1, M2, and DM) were not treated or were treated with Tg. The indicated proteins from whole-cell lysate or the cell-surface preparations were analyzed by Western blots with GAPDH and EphB4 serving as loading controls. The csGRP78 levels were quantified and graphed. (B) HeLa cells were treated with Tg (Left) or were transfected with SRC531 expression vector (Middle) or both (Right). Whole-cell lysate was subjected to a pull-down assay with GST-fused WT or mutant ASAP1 recombinant proteins (G-ASAP1). The indicated proteins from the eluate (E) were analyzed by Western blot. The relative eluted GBF1 level under each condition was quantified and graphed with SD. (C) HeLa cells transfected with the HA-ASAP1 expression vector were not treated (Ctrl) or were treated with Tg and were subjected to immunofluorescent staining for the HA epitope (red) or GBF-1 (green). Costaining of both proteins (yellow) is shown in the merged images and the enlarged views. (Scale bars, 5 µm.) (D) HeLa cells were transfected with the SRC531 expression vector followed by Tg treatment, and the whole-cell lysate was subjected to immunoprecipitation using IgG control, anti-GBF1, or anti-ASAP1 antibodies. The indicated proteins were analyzed by Western blot. (E) HeLa cells were transfected with the SRC531 expression vector and were treated with Tg, DMSO, or GCA as indicated. The whole-cell lysates were subjected to immunoprecipitation using anti-GBF1 or control IgG antibodies, and the immunoprecipitate was subjected to the GTPase-Glo GEF activity assay. (F) As in E, except the GBF1 immunoprecipitates from the indicated treatment conditions were subjected to the GTP exchange assay. (G) Summary of ASAP1 and GBF1 involvement in the ER stress-induced chaperone relocalization. *P < 0.05, **P < 0.01, ***P < 0.005.
GBF1 primarily localizes in the cis-Golgi where it converts Arf1-GDP to Arf1-GTP in regulating retrograde transport (18). Tg treatment showed minimal effect on GBF1 localization (Fig. S5E). In contrast, Tg promoted ASAP1 relocalization from the cytosol to the cis-Golgi, where it showed strong colocalization with GBF1 (Fig. 3C). Coimmunoprecipitation (co-IP) further confirmed complex formation between endogenous ASAP1 and GBF1 in stressed cells expressing activated SRC (Fig. 3D). In testing the effect of Tg and SRC on the GEF activity of GBF1, we utilized the GTPase-Glo GEF activity assay with recombinant Arf1 as substrate and observed that both SRC531-expressing and ER-stressed cells accelerated the GTP-loading reaction of GBF1 isolated from cell lysates, which was blocked by a specific GBF1 inhibitor Golgicide A (GCA) (Fig. 3E) (40). This observation was confirmed using the GTP exchange assay under the same treatment conditions (Fig. 3F). Thus, ER stress induces SRC activation, which phosphorylates a key residue, Y312, on ASAP1 that promotes its interaction with GBF1 in the cis-Golgi, leading to enhanced GBF1 activity and cell-surface relocalization of ER chaperones (Fig. 3G).
GBF1-Mediated Increase of Arf1-GTP and Accumulation in the cis-Golgi Leads to KDELR Dispersion.
In HeLa, MCF-7, and SK-MEL-28 cells, the GBF1 inhibitor GCA potently suppressed Tg-induced cell-surface relocalization of ER chaperones but not EphB4 (Fig. 4A), suggesting that GBF1 is a common and essential component of this process. A major substrate of GBF1 is Arf1, which coordinates the assembly of retrograde vesicle (11). Utilizing the Arf1 activation assay, we determined that, upon Tg treatment or the expression of SRC531 or HA-ASAP1, the level of Arf1-GTP, which is the active form of Arf1, was substantially increased (Fig. 4B). GBF1 was required for the Tg- and SRC531-mediated increase in Arf1-GTP, as this was suppressed by GCA (Fig. 4C). Furthermore, SRC and ASAP1 are both essential for the Tg-induced increase in Arf1-GTP levels, since knockdown of either SRC or ASAP1 abolished the increase (Fig. 4D).
Fig. 4.
GBF1 facilitates the ER stress-induced Arf1-GTP increase at the cis-Golgi. (A) The indicated cancer cell lines were treated with Tg and GCA as indicated. Cell-surface proteins were isolated and probed for the indicated proteins by Western blot with EphB4 serving as loading control. (B) Whole-cell lysate from HeLa cells treated with Tg or transfected with SRC531 or HA-ASAP1 expression vectors were subjected to the Arf1 pull-down activation assay. The indicated proteins from the eluate (E) or whole-cell lysate were analyzed by Western blot. The band intensities for Arf1-GTP were quantified and graphed. (C) As in B, except the cells were treated additionally with GCA as indicated. (D) As in B, except SRC (shSRC)- or ASAP1 (shASAP1)-knockdown stable HeLa cell lines were treated with Tg as indicated. (E) SK-MEL-28 cells were transfected with either WT Arf1-HA or the Q71L mutant expression vectors. The cells were not treated (Ctrl), treated with Tg, or pretreated with SU6656 followed by Tg and were subjected to immunofluorescent staining for the HA epitope (red) or GM130 (green). Yellow indicates costaining of the two proteins in the merged images. The immunofluorescent intensities of Arf1-HA at the cis-Golgi under each condition were quantified and graphed. (Scale bar, 10 µm.) *P < 0.05, ***P < 0.005.
Arf1 binding to GTP causes the exposure and insertion of its N-terminal amphiphilic helix and myristoyl group into the lipid bilayer and stable association with membrane (41). Using confocal microscopy, we observed that, upon Tg stress, the level of Arf1 at the cis-Golgi, as evident by colocalization with GM130, increased by twofold, and this increase was blocked by the SFK inhibitor SU6656 (Fig. 4E). Compared with WT Arf1, a threefold increase in Arf1(Q71L), a constitutively active GTP-locked Arf1 mutant, was detected at the cis-Golgi, as expected.
In HeLa, MCF-7, and SK-MEL-28 cells, Arf1(Q71L) expression alone dramatically increased the cell-surface expression of ER chaperones even in the absence of ER stress, and Tg treatment did not appear to cause any further increase (Fig. 5A). This suggests Arf1 activation is sufficient for the relocalization of KDEL-bearing ER chaperones to the cell surface. In agreement, Arf1(Q71L), known to regulate retrograde but not anterograde transport (42), had no effect on EphB4 levels (Fig. 5A), and the action of Arf1(Q71L) was dependent on the integrity of the KDEL motif (Fig. 5B). Additionally, we demonstrated that expression of SRC531, ASAP1, or Arf1(Q71L) all caused dispersion of KDELR1 from cis-Golgi (Fig. 5C). Collectively, our results demonstrate that ER stress activates SRC via IRE1α, subsequently triggering a signaling cascade linking ASAP1, GBF1, and Arf1 and leading to the disruption of the KDELR retrieval machinery, thus allowing ER chaperones to escape to the cell surface (Fig. 5D).
Fig. 5.
GTP-locked Arf1(Q71L) is sufficient to drive KDELR1 dispersion and ER chaperones to the cell surface. (A) The indicated cancer cell lines were transfected with the Arf1-HA(Q71L) mutant expression vector and were treated with Tg as indicated. The indicated proteins from the whole-cell lysate and the cell surface were analyzed by Western blot with GAPDH and EphB4 serving as loading controls. (B) As in A, except HeLa cells were transfected with the indicated expression vectors, and conditioned medium was collected and assayed for secreted F-GRP78. The band intensities of csF-GRP78 were quantified and graphed. (C) HeLa cells were transfected with the KDELR1-FLAG (KDELR1-F) expression vector alone or in combination with expression vectors for SRC531-His, HA-ASAP1, or Arf1-HA(Q71L), as indicated, and were subjected to immunofluorescent staining for the indicated proteins. GM130 served as the marker for cis-Golgi. (Scale bar, 5 µm.) (D) Summary of the ER stress-signaling cascade leading to the escape of ER luminal chaperones to the cell surface. *P < 0.05.
Identification of CD109 as a csGRP78-Binding Partner in TGF-β Inhibition.
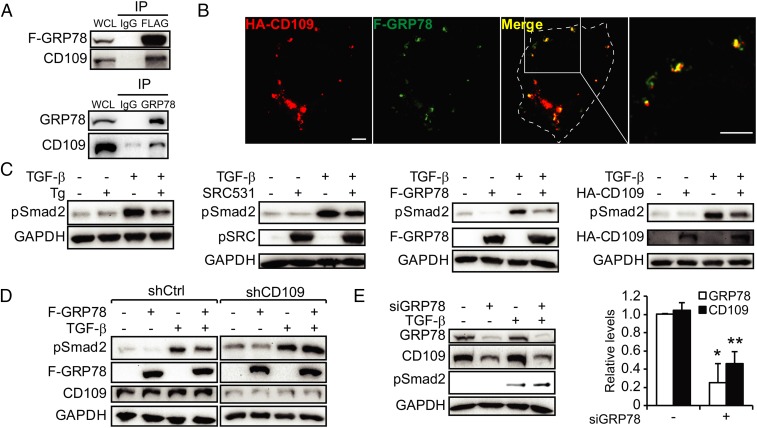
While GRP78 is one of the best-characterized ER chaperones (6, 43), how GRP78 regulates tumor proliferation and survival from the cell surface is not well understood. Taking advantage of the recent finding that a substantial level of csGRP78 interacts with GPI-anchored cell-surface proteins (21), we harvested proteins released from HeLa cells treated with phosphatidylinositol-specific phospholipase C (PI-PLC) and subjected those bound to csGRP78 to LC-MS/MS. This led to the identification of CD109 as a potential binding partner of csGRP78 (Fig. S6A). CD109 is known as a negative regulator of TGF-β signaling via suppressing Smad2/3 phosphorylation and promoting TβR degradation (25). The interaction between endogenous CD109 and F-GRP78 in HeLa cells was confirmed by co-IP (Fig. 6A), in agreement with confocal microscopy showing colocalization of HA-CD109 and F-GRP78 on the surface of nonpermeabilized cells (Fig. 6B). Analysis of the Gene Expression across Normal and Tumor Tissue (GENT) database (44) further revealed that in 1,738 cancer cell lines representing 30 cancer types, CD109 is widely expressed, with the highest levels in skin and cervical cancers (Fig. S6B). Interestingly, the transcript level of Cripto, another TGF-β–inhibitory GPI-anchored protein known to interact with csGRP78 (45), is lower than CD109 in general but is highly expressed in colorectal and rectal cancer cell lines. In the melanoma SK-MEL-28 cells used as a model system in the studies described below, the relative transcript levels of CD109 and Cripto from the GENT database are 1,779 and 57, respectively.
Fig. 6.
csGRP78 partners with CD109 in TGF-β inhibition. (A) Whole-cell lysate from HeLa cells expressing F-GRP78 (Upper) or SK-MEL-28 cells (Lower) were subjected to immunoprecipitation with IgG, anti-FLAG, or anti-GRP78 antibodies as indicated. The immunoprecipitate along with the whole-cell lysate was probed by Western blot for F-GRP78, GRP78, and CD109. (B) HeLa cells were cotransfected with F-GRP78 and HA-CD109 expression vectors and were subjected to immunofluorescent staining for the HA (red) and FLAG (green) epitopes. Costaining of the two proteins is indicated by yellow in the merged image and the enlarged view. (Scale bars, 5 µm.) (C) HeLa cells were treated with Tg or TGF-β and were transfected with SRC531, F-GRP78, or HA-CD109 alone or in combination, as indicated. The indicated proteins were analyzed by Western blot. (D) As in C, except the CD109 (shCD109)- or control shRNA (shCtrl)-knockdown stable HeLa cell line was treated as indicated. (E) As in D, except SK-MEL-28 cells were transfected with siRNA against GRP78 (siGRP78) and were treated with TGF-β as indicated. The band intensities were quantified and graphed. *P < 0.05, **P < 0.01.
Previously we have shown that transfection of cells with plasmid encoding for F-GRP78 leads to its expression at the cell surface (5). In HeLa cells, we observed that Tg treatment and the expression of SRC531, F-GRP78, or HA-CD109 all suppressed TGF-β–induced Smad2 phosphorylation (pSmad2) (Fig. 6C). The GRP78 effect was dependent on CD109, since it was prevented by CD109 knockdown (Fig. 6D). In SK-MEL-28 cells with high CD109 levels, co-IP showed complex formation between endogenous CD109 and GRP78 (Fig. 6A). Knockdown of GRP78 reduced the CD109 level and up-regulated TGF-β–induced pSmad2 (Fig. 6E). Collectively, these results suggest that ER stress activates SRC, leading to an increase in csGRP78, which forms a complex with and stabilizes CD109, thereby suppressing TGF-β signaling mediated by the TβR. To address translational relevance, we treated primary acute myeloid leukemia (AML) cells taken from a patient with relapsed disease with bortezomib, a clinically used proteasome inhibitor known to induce ER stress (46). Bortezomib treatment of AML cells led to an increase in csGRP78 expression, correlating with the suppression of TGF-β–induced Smad2 phosphorylation (Fig. S7).
csGRP78 Suppresses TGF-β Receptor Binding to Smad2 Through Promoting Its Routing to the Caveolae.
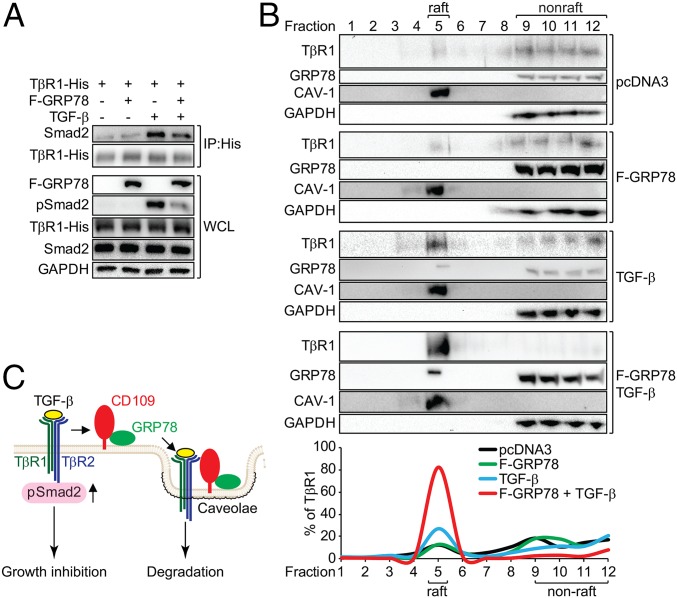
To determine how GRP78 blocks Smad2 phosphorylation, we observed that F-GRP78 expression in SK-MEL-28 cells suppressed TGF-β–induced binding of Smad2 to TGF-β receptor 1 (TβR1), as revealed by co-IP (Fig. 7A). CD109 is reported to regulate TβR endocytosis and degradation to inhibit TGF-β signaling (25). Utilizing sucrose-gradient ultracentrifugation to fractionate the caveolae containing lipid raft associated with receptor degradation from nonraft membranes, which include the plasma membrane, we observed that in control cells and F-GRP78–expressing cells the majority of TβR1 was in the nonraft fraction, with only a minority (12%) in the raft fraction (Fig. 7 B and C). Upon the addition of TGF-β, we detected an increase in TβR1 (27%) in the raft fraction, where some endogenous GRP78 was detected as well. Strikingly, TGF-β treatment of F-GRP78–expressing cells dramatically shifted more TβR1 (80%), as well as GRP78, to the raft fraction (Fig. 7B). This suggests that GRP78, acting in concert with CD109, promotes routing of the TβR to the caveolae, leading to its degradation and the relief of growth inhibition (Fig. 7C).
Fig. 7.
csGRP78 promotes the routing of TβR1 to the caveolae and disrupts its interaction with Smad2. (A) SK-MEL-28 cells transfected with TβR1-His or F-GRP78, alone or in combination were treated with TGF-β as indicated. The cells were subjected to coimmunoprecipitation using anti-His antibody. The indicated proteins were analyzed by Western blot. (B) SK-MEL-28 cells were transfected with pcDNA3 or F-GRP78 or were treated with TGF-β alone or in combination as indicated. The whole-cell lysate was subjected to sucrose-gradient fractionation. The fractions were subjected to Western blot for analysis of the indicated proteins. CAV-1, caveolin 1. The lipid raft was enriched in fraction 5, and nonraft was enriched in fractions 9–12 with CAV-1 and GAPDH serving as markers for the respective fractions. The percentage of TβR1 in each fraction was quantified and graphed. (C) Model of the inhibition of TGF-β signaling by csGRP78/CD109. Upon TGF-β stimulation, csGRP78/CD109 routes the TβR to the caveolae for degradation, disrupting the binding of the receptor to Smad2 and its subsequent activation, thereby blunting TGF-β–mediated growth inhibition.
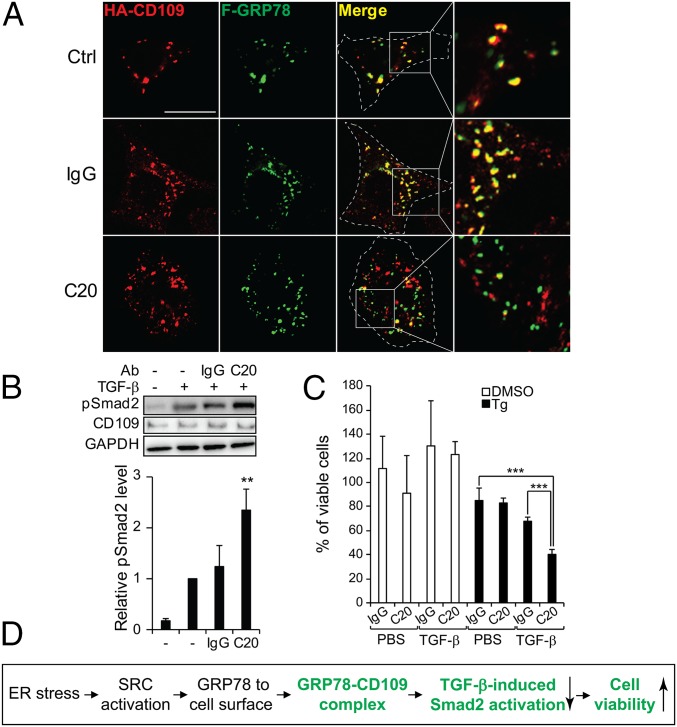
To test directly that csGRP78 regulates CD109/TGF-β signaling, we showed that C20, an anti-GRP78 antibody targeting the C terminus of GRP78, disrupted the cell-surface colocalization of F-GRP78 and HA-CD109 in nonpermeabilized HeLa cells (Fig. 8A). In agreement, upon TGF-β stimulation, C20 treatment resulted in a 2.5-fold increase of Smad2 phosphorylation in SK-MEL-28 cells (Fig. 8B). TGF-β signaling is known to reduce cell viability (47). In the same cells, C20 treatment significantly reduced cell viability in TGF-β–treated cells in combination with Tg but not in control cells treated with DMSO (Fig. 8C). Taken together, these results uncover a pathway whereby ER stress, via SRC activation, promotes ER chaperone relocalization to the cell surface, where GRP78 forms a complex with CD109 and negatively regulates TGF-β signaling to sustain cell viability under stress (Fig. 8D).
Fig. 8.
Effect of antibody targeting GRP78 on the interaction with CD109 and TGF-β signaling. (A) HeLa cells cotransfected with F-GRP78 and HA-CD109 were not treated (Ctrl) or were treated with IgG or the anti-GRP78 antibody C20. The nonpermeabilized cells were subjected to immunofluorescent staining for the HA (red) and FLAG (green) epitopes. The yellow staining in the merged images and enlarged views indicate costaining of the csF-GRP78 with HA-CD109, which was disrupted by C20 treatment. (Scale bar, 10 µm.) (B) SK-MEL-28 cells were treated with IgG, C20, and TGF-β as indicated. The indicated proteins were analyzed by Western blot. The band intensities of pSmad2 were quantified and graphed. (C) As in B, except the cells were treated with DMSO or Tg in combination with the antibodies and then were treated with PBS or TGF-β and subjected to the WST-1 assay. (D) Summary of GRP78/CD109 inhibition of TGF-β signaling and promotion of survival under ER stress. **P < 0.01, ***P < 0.005.
Discussion
In cancer, an adverse tumor microenvironment caused by nutrient deprivation and hypoxia disturbs the protein-folding capacity and creates ER stress (48). The discovery that ER stress actively promotes a process whereby ER chaperones can escape from the ER compartment and relocalize to the cell surface, where they assume regulatory roles impacting cell signaling, proliferation, and survival, raises important questions about how these effects can be achieved. In this study we dissected the fundamental mechanisms and uncovered a number of observations that could have major implications in cancer and other human diseases.
First, our kinetic studies revealed that ER stress rapidly induces cell-surface expression of ER chaperones before an increase in their intracellular protein levels or the onset of apoptosis; thus their escape from the ER is unlikely to be due to over-saturation of the KDELR retrieval machinery or a passive event preceding cell death. In cancer, SRC is well known to play diverse roles in tumorigenesis, proliferation, survival, and metastasis (13). SRC expression and activity, as well as cell-surface expression of ER chaperones, increase as tumor advances (13, 49). Here we provide direct evidence that SRC, in addition to being activated by ER stress (50–52), has a function in actively promoting the cell-surface relocalization of ER chaperones in a wide range of solid and blood cancer cell lines. Most importantly, SRC is both sufficient and necessary for this process. How might ER stress activate SRC? Evidence is emerging that the ER stress sensor IRE1α forms a dynamic scaffold onto which many regulatory components assemble, as exemplified by activated IRE1α binding to TRAF2 and regulating the JNK and NFκB pathways independent of its RNase activity (48, 53). We discovered that, upon ER stress, SRC forms a complex with IRE1α and is activated through Y419 phosphorylation. SFK, including SRC, can be activated through SH3 interactions (54). While the detailed mechanism awaits further investigation, here we determined that the cytosolic tail of IRE1α containing noncanonical SH3-binding proline-rich motifs is critical for ER stress-induced SRC binding and activation and the escape of ER chaperones to the surface. Given that ER luminal GRP78 dissociates from IRE1α upon ER stress (55, 56), this could trigger changes in IRE1α leading to a feed-forward mechanism promoting GRP78 to the cell surface. In a context-dependent manner, other UPR signaling pathways could also contribute to the ER escape mechanism, as it has been reported that CRT exposure at the cell surface is dependent on PERK in immune cells (57).
While the SRC requirement is prevalent in the panel of cell lines that we examined, there are exceptions, such as the human colon cancer cell line HCT116, which utilized a Golgi-independent mechanism for cell-surface expression of GRP78 (21), supporting the notion that the SRC mechanism of action is mediated through the ER–Golgi axis. Thus, one scenario is that, upon activation by ER stress, SRC triggers a signaling cascade partially inhibiting Golgi–ER retrograde trafficking of chaperones bearing KDEL or related motifs that are recognized by the KDELR1 and thereby allowing a subfraction of these ER luminal chaperones to escape to the cell surface. Earlier studies hinted that SRC activation could lead to KDELR dispersion from the Golgi; however, the mechanistic link between SRC activation and KDELR dispersion is unclear. Our studies identified ASAP1 as a key SRC substrate mediating this pathway. ASAP1 regulates recycling of cell-surface receptors (58) and is implicated in tumor invasion, in which phosphorylation of murine ASAP1 by SRC at Y782, corresponding to human ASAP1 at Y767, is critical (37). An ultradeep phosphoproteomic analysis on HeLa cells detects phosphorylation of ASAP1 at Y312 (59). Here we determined that phosphorylation of ASAP1 at Y312 is critical for its binding to GBF1, which subsequently leads to the Arf-GTP increase at the cis-Golgi. While we cannot rule out the involvement of other SRC downstream pathways, the ability of ASAP1 knockdown to block escape of ER chaperones and the potency of the constitutively active Arf1(Q71L) mutant in driving cell-surface expression of ER chaperones in multiple cell model systems imply that the ASAP1/GBF1/Arf1 axis is a major mechanism for ER stress-mediated escape of ER chaperones to the cell surface.
TGF-β signaling transduced by TβR1 and TβR2 plays an important role in cancer and tissue fibrosis. CD109 in skin cells acts as a coreceptor and inhibitor of TGF-β signaling by facilitating TβR degradation in the caveolar compartment (25). Here we identify CD109 as a binding partner of csGRP78 that mediates the anti–TGF-β effect of GRP78. We discovered that GRP78, by promoting the routing of TβR1 to the caveolae for degradation, disrupts the binding of TβR1 to Smad2 and its downstream signaling. Comparison of the transcript levels of CD109 and Cripto, another GPI-anchored protein reported to suppress TGF-β signaling via csGRP78 (45), revealed high-level expression of CD109 in a wide range of cancers, whereas Cripto is notably highly expressed in colorectal and rectal cancer cell lines. Cripto is reported to interact with the N terminus of csGRP78 (60). In our studies, we observed that, among the anti-GRP78 antibodies tested, the one targeting the C terminus of GRP78 is most effective in disrupting CD109 signaling. Future studies will be required to dissect whether csGRP78 coregulates CD109 and Cripto through similar or different mechanisms.
What are the therapeutic implications of our findings? The ER chaperones on the cell surface are emerging as unique targets as well as mediators for antineoplastic treatment and imaging (4, 61–64). On a mechanistic level, csGRP78 promotes PIP3 formation and regulates the PI3K/Akt pathway (29), csGRP94 facilitates HER2 dimerization and promotes cell proliferation in breast cancer (65), and cell-surface PDI mediates integrin disulfide exchange and promotes cell adhesion in glioma (6). Small-molecule SRC inhibitors such as Das are currently in clinical use (36), and IRE1α inhibitors have been identified (20, 53). Thus, inhibition of SRC or its interaction with IRE1α could block the cell-surface expression of ER chaperones and potentially suppress their protumor functions. While this might potentially impede cell-surface CRT expression in immune-mediated toxicity, tumor cells are known to express other compensatory ligands accessible by immune cells (66). In addition to applications in oncology, ER chaperones on the cell-surface function as receptor for pathogen infection. Viruses such as Coxsackie virus, dengue virus, and Borna disease virus and fungi such as Rhizopus oryzae recognize csGRP78 for entry or invasion into the host (2), and the bacterial pathogen Listeria monocytogenes requires csGRP94 to invade (67). The recent discovery that GRP78 autoantibody associates with blood–brain barrier disruption in neuromyelitis optica suggests that csGRP78 is a potential target for promoting the transit of large-molecule therapies for central nervous system diseases (68). Collectively, understanding the biology of cell-surface chaperones could have a major impact in combating cancer, infectious diseases, inflammatory disorders, and neuropathy and warrants vigorous investigation.
Materials and Methods
Cell culture, expression vectors, recombinant protein, site-directed mutagenesis, transfection, biotinylation, pull-down assays, assays for GEF activity, GTP exchange, and cell viability, PI-PLC treatment, mass spectrometry, immunofluorescent staining, immunoprecipitation, immunoblot, sucrose-gradient fractionation, CRISPR/Cas9 knockout, RNA preparation, RT-PCR, and statistical analysis can be found in SI Materials and Methods. Also included are the procedures for the isolation of primary human leukemic cells and flow cytometry. Use of human material was approved by the University of Southern California Institutional Review Board (IRB), and informed consent was obtained for use of the material.
Supplementary Material
Acknowledgments
We thank James Ou, Jain Xu, and Erik Snapp for helpful discussions, Parkash Gill, Albert Koong, and Costas Koumenis for the gift of reagents, the University of Southern California (USC) Proteomics Core for mass spectrometry analysis, and the Cell and Tissue Imaging Core of the USC Research Center for Liver Diseases for confocal microscopy. These studies were supported by NIH Grant R01 CA027607 (to A.S.L.), L. K. Whittier Foundation Grant 003457 (to A.S.L. and K.K.), and NIH/National Cancer Center Core Grants P30 CA014089 to the USC Norris Comprehensive Cancer Center and P30 DK048522 to the USC Research Center for Liver Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714866115/-/DCSupplemental.
References
- 1.Shiu RP, Pouysségur J, Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteinsin Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci USA. 1977;74:3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: Atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434:181–188. doi: 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behnke J, Mann MJ, Scruggs FL, Feige MJ, Hendershot LM. Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol Cell. 2016;63:739–752. doi: 10.1016/j.molcel.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AS. Glucose-regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Liu R, Ni M, Gill P, Lee AS. Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J Biol Chem. 2010;285:15065–15075. doi: 10.1074/jbc.M109.087445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiersma VR, Michalak M, Abdullah TM, Bremer E, Eggleton P. Mechanisms of translocation of ER chaperones to the cell surface and immunomodulatory roles in cancer and autoimmunity. Front Oncol. 2015;5:7. doi: 10.3389/fonc.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin BK, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607–7616. doi: 10.1074/jbc.M210455200. [DOI] [PubMed] [Google Scholar]
- 9.Gopal U, Gonzalez-Gronow M, Pizzo SV. Activated alpha2-macroglobulin regulates transcriptional activation of c-Myc target genes through cell surface GRP78. J Biol Chem. 2016;291:10904–10915. doi: 10.1074/jbc.M115.708131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelham HR. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem Sci. 1990;15:483–486. doi: 10.1016/0968-0004(90)90303-s. [DOI] [PubMed] [Google Scholar]
- 11.D’Souza-Schorey C, Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 12.Raykhel I, et al. A molecular specificity code for the three mammalian KDEL receptors. J Cell Biol. 2007;179:1193–1204. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 14.Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. J Signal Transduct. 2011;2011:865819. doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan PC, Chen HC. p120RasGAP-mediated activation of c-Src is critical for oncogenic Ras to induce tumor invasion. Cancer Res. 2012;72:2405–2415. doi: 10.1158/0008-5472.CAN-11-3078. [DOI] [PubMed] [Google Scholar]
- 16.Bard F, Mazelin L, Péchoux-Longin C, Malhotra V, Jurdic P. Src regulates Golgi structure and KDEL receptor-dependent retrograde transport to the endoplasmic reticulum. J Biol Chem. 2003;278:46601–46606. doi: 10.1074/jbc.M302221200. [DOI] [PubMed] [Google Scholar]
- 17.Spang A. Retrograde traffic from the Golgi to the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5:a013391. doi: 10.1101/cshperspect.a013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casanova JE. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 19.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 21.Tsai YL, et al. Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J Biol Chem. 2015;290:8049–8064. doi: 10.1074/jbc.M114.618736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohshima Y, et al. CD109 expression levels in malignant melanoma. J Dermatol Sci. 2010;57:140–142. doi: 10.1016/j.jdermsci.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Tao J, Li H, Li Q, Yang Y. CD109 is a potential target for triple-negative breast cancer. Tumour Biol. 2014;35:12083–12090. doi: 10.1007/s13277-014-2509-5. [DOI] [PubMed] [Google Scholar]
- 24.Lin M, et al. Cell surface antigen CD109 is a novel member of the alpha(2) macroglobulin/C3, C4, C5 family of thioester-containing proteins. Blood. 2002;99:1683–1691. doi: 10.1182/blood.v99.5.1683. [DOI] [PubMed] [Google Scholar]
- 25.Bizet AA, et al. The TGF-β co-receptor, CD109, promotes internalization and degradation of TGF-β receptors. Biochim Biophys Acta. 2011;1813:742–753. doi: 10.1016/j.bbamcr.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 26.Kathiriya JJ, et al. Presence and utility of intrinsically disordered regions in kinases. Mol Biosyst. 2014;10:2876–2888. doi: 10.1039/c4mb00224e. [DOI] [PubMed] [Google Scholar]
- 27.Ungefroren H, Sebens S, Groth S, Gieseler F, Fändrich F. Differential roles of Src in transforming growth factor-ß regulation of growth arrest, epithelial-to-mesenchymal transition and cell migration in pancreatic ductal adenocarcinoma cells. Int J Oncol. 2011;38:797–805. doi: 10.3892/ijo.2011.897. [DOI] [PubMed] [Google Scholar]
- 28.Panaretakis T, et al. The co-translocation of ERp57 and calreticulin determines the immunogenicity of cell death. Cell Death Differ. 2008;15:1499–1509. doi: 10.1038/cdd.2008.67. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PLoS One. 2013;8:e80071. doi: 10.1371/journal.pone.0080071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irby RB, et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi H, Murata Y, Okazawa H, Matozaki T. Src family kinases: Modulators of neurotransmitter receptor function and behavior. Trends Neurosci. 2011;34:629–637. doi: 10.1016/j.tins.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Teyra J, et al. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure. 2017;25:1598–1610.e3. doi: 10.1016/j.str.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capitani M, Sallese M. The KDEL receptor: New functions for an old protein. FEBS Lett. 2009;583:3863–3871. doi: 10.1016/j.febslet.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 36.Araujo JC, et al. Docetaxel and dasatinib or placebo in men with metastatic castration-resistant prostate cancer (READY): A randomised, double-blind phase 3 trial. Lancet Oncol. 2013;14:1307–1316. doi: 10.1016/S1470-2045(13)70479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bharti S, et al. Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol Cell Biol. 2007;27:8271–8283. doi: 10.1128/MCB.01781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 40.Sáenz JB, et al. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol. 2009;5:157–165. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen LA, Donaldson JG. Analysis of Arf GTP-binding protein function in cells. Curr Protoc Cell Biol. 2010;Chapter 3:Unit 14.12.1–17. doi: 10.1002/0471143030.cb1412s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanoix J, et al. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin G, et al. GENT: Gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shani G, et al. GRP78 and Cripto form a complex at the cell surface and collaborate to inhibit transforming growth factor beta signaling and enhance cell growth. Mol Cell Biol. 2008;28:666–677. doi: 10.1128/MCB.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fels DR, et al. Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res. 2008;68:9323–9330. doi: 10.1158/0008-5472.CAN-08-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 48.Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao X, et al. Cell surface GRP78 accelerated breast cancer cell proliferation and migration by activating STAT3. PLoS One. 2015;10:e0125634. doi: 10.1371/journal.pone.0125634. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Hong M, et al. Transcriptional regulation of the Grp78 promoter by endoplasmic reticulum stress: Role of TFII-I and its tyrosine phosphorylation. J Biol Chem. 2005;280:16821–16828. doi: 10.1074/jbc.M413753200. [DOI] [PubMed] [Google Scholar]
- 51.Zhong Q, et al. Role of endoplasmic reticulum stress in epithelial-mesenchymal transition of alveolar epithelial cells: Effects of misfolded surfactant protein. Am J Respir Cell Mol Biol. 2011;45:498–509. doi: 10.1165/rcmb.2010-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bettaieb A, et al. Differential regulation of endoplasmic reticulum stress by protein tyrosine phosphatase 1B and T cell protein tyrosine phosphatase. J Biol Chem. 2011;286:9225–9235. doi: 10.1074/jbc.M110.186148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moarefi I, et al. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 55.Pincus D, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sepulveda D, et al. Interactome screening identifies the ER luminal chaperone Hsp47 as a regulator of the unfolded protein response transducer IRE1alpha. Mol Cell. 2018;69:238–252.e7. doi: 10.1016/j.molcel.2017.12.028. [DOI] [PubMed] [Google Scholar]
- 57.Panaretakis T, et al. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. EMBO J. 2009;28:578–590. doi: 10.1038/emboj.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie Z, et al. A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr Biol. 2006;16:130–139. doi: 10.1016/j.cub.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 59.Sharma K, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Reports. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 60.Kelber JA, et al. Blockade of Cripto binding to cell surface GRP78 inhibits oncogenic Cripto signaling via MAPK/PI3K and Smad2/3 pathways. Oncogene. 2009;28:2324–2336. doi: 10.1038/onc.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasche L, et al. A GRP78-directed monoclonal antibody recaptures response in refractory multiple myeloma with extramedullary involvement. Clin Cancer Res. 2016;22:4341–4349. doi: 10.1158/1078-0432.CCR-15-3111. [DOI] [PubMed] [Google Scholar]
- 62.Dobroff AS, et al. Towards a transcriptome-based theranostic platform for unfavorable breast cancer phenotypes. Proc Natl Acad Sci USA. 2016;113:12780–12785. doi: 10.1073/pnas.1615288113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Wang X, Ferrone CR, Schwab JH, Ferrone S. Intracellular antigens as targets for antibody based immunotherapy of malignant diseases. Mol Oncol. 2015;9:1982–1993. doi: 10.1016/j.molonc.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Ridder GG, Gonzalez-Gronow M, Ray R, Pizzo SV. Autoantibodies against cell surface GRP78 promote tumor growth in a murine model of melanoma. Melanoma Res. 2011;21:35–43. doi: 10.1097/CMR.0b013e3283426805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, et al. Cell membrane gp96 facilitates HER2 dimerization and serves as a novel target in breast cancer. Int J Cancer. 2015;137:512–524. doi: 10.1002/ijc.29405. [DOI] [PubMed] [Google Scholar]
- 66.Kruse PH, Matta J, Ugolini S, Vivier E. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol. 2014;92:221–229. doi: 10.1038/icb.2013.98. [DOI] [PubMed] [Google Scholar]
- 67.Martins M, et al. Listeria monocytogenes triggers the cell surface expression of Gp96 protein and interacts with its N terminus to support cellular infection. J Biol Chem. 2012;287:43083–43093. doi: 10.1074/jbc.M112.422568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shimizu F, et al. Glucose-regulated protein 78 autoantibody associates with blood-brain barrier disruption in neuromyelitis optica. Sci Transl Med. 2017;9:eaai9111. doi: 10.1126/scitranslmed.aai9111. [DOI] [PMC free article] [PubMed] [Google Scholar]