Significance
We present here that adenovirus type 52 (HAdV-52) attaches to target cells through a mechanism not previously observed in other human pathogenic viruses. The interaction involves unusual, transient, electrostatic interactions between the short fiber capsid protein and polysialic acid (polySia)-containing receptors on target cells. Knowledge about the binding interactions between polySia and its natural ligands is relatively limited, and our results therefore provide additional insight not only into adenovirus biology but also into the structural basis of polySia function. Since polySia can be found in high expression levels in brain and lung cancers where its presence is associated with poor prognosis, we suggest that this polySia-binding adenovirus could be useful for design of vectors for gene therapy of these cancers.
Keywords: human adenovirus, short fiber, polysialic acid, glycan receptor, glycan microarray
Abstract
Human adenovirus 52 (HAdV-52) is one of only three known HAdVs equipped with both a long and a short fiber protein. While the long fiber binds to the coxsackie and adenovirus receptor, the function of the short fiber in the virus life cycle is poorly understood. Here, we show, by glycan microarray analysis and cellular studies, that the short fiber knob (SFK) of HAdV-52 recognizes long chains of α-2,8-linked polysialic acid (polySia), a large posttranslational modification of selected carrier proteins, and that HAdV-52 can use polySia as a receptor on target cells. X-ray crystallography, NMR, molecular dynamics simulation, and structure-guided mutagenesis of the SFK reveal that the nonreducing, terminal sialic acid of polySia engages the protein with direct contacts, and that specificity for polySia is achieved through subtle, transient electrostatic interactions with additional sialic acid residues. In this study, we present a previously unrecognized role for polySia as a cellular receptor for a human viral pathogen. Our detailed analysis of the determinants of specificity for this interaction has general implications for protein–carbohydrate interactions, particularly concerning highly charged glycan structures, and provides interesting dimensions on the biology and evolution of members of Human mastadenovirus G.
Human adenoviruses (HAdVs) are common human pathogens associated with gastrointestinal, ocular, and respiratory infections. To date, 84 different HAdV types have been identified, and they are grouped into seven species (Human mastadenovirus A to G) (1). HAdVs are nonenveloped viruses whose icosahedral capsid is composed of three major proteins, the fiber, the penton base, and the hexon, all of which are known to mediate binding to host cells. The fiber protein, with a terminal knob domain, binds to cellular receptors such as the coxsackie and adenovirus receptor (CAR) (2–4), desmoglein-2 (5), CD46 (6–8), or sialic acid (Sia)-containing glycans (9–11). The penton base interacts with cellular integrins, thereby facilitating endocytosis (12, 13) and endosomal release (14, 15). The hexon protein is the main component of the viral capsid and binds with high affinity to coagulation factors IX and X, resulting in liver tropism through indirect binding to heparan sulfate on hepatocytes (16–18), and shields the virion from neutralizing antibodies and complement-mediated destruction (19).
HAdV-52 was isolated in 2003 from a small outbreak of gastroenteritis (20). The virus diverged from other HAdVs and was classified into the new species Human mastadenovirus G (HAdV-G), which otherwise exclusively contains Old World monkey AdVs. HAdVs are normally equipped with only one fiber protein, but HAdV-52, along with species HAdV-F types HAdV-40 and -41, differ from all other known HAdVs by having two different fiber proteins, one short (coded by gene fiber-1) and one long (fiber-2) (20–22). We showed recently that the knob domain of HAdV-52 long fiber (52LFK) binds to CAR and that the knob domain of the short fiber (52SFK) binds to Sia-containing glycoproteins on target cells (23). However, the identity and structure of the cellular Sia-containing glycans have remained unknown.
Sia-containing glycans serve as receptors for a large number of viral pathogens, including influenza A virus, coronavirus, rotavirus, polyomavirus, and many others (24). Variations in Sia specificity determine host and tissue tropism, pathogenicity, and transmission of multiple viruses. Here, we show by glycan microarray analysis that the 52SFK recognizes long chains of sialic acid residues, known as polysialic acid (polySia), with higher affinity than any other tested glycan. PolySia is a rare posttranslational modification of only nine identified carrier proteins to our knowledge; among them are the cell adhesion molecules NCAM (25) and SynCAM-1 (26) as well as Neuropilin-2 (27) and the dendritic cell chemokine receptor CCR7 (28). Polysialylation is best known as a modulator of developmental plasticity in the nervous system, but more recently, additional roles in the development of a number of organs, such as the liver, kidney, heart, and testes, have been unraveled (reviewed in ref. 29). In the adult brain, polySia expression is markedly down-regulated and only retained in few areas that maintain plasticity such as the hippocampus, olfactory bulb, and hypothalamus (reviewed in refs. 30–32). However, polySia is not exclusively associated with the brain. Recent studies demonstrate additional regulatory roles in innate immune responses (28, 33–36), and in regenerative or antiinflammatory processes (37–42). Furthermore, polySia is found at high expression levels on several types of cancer including glioma (43–45), neuroblastoma (46, 47), and lung cancer (48, 49). By means of X-ray crystallography, NMR, molecular dynamics (MD) simulation, and cellular analyses, we reveal here a function for polySia as a cellular receptor for HAdV-52. The 52SFK possesses a unique polySia-binding mode featuring transient polar interactions and electrostatic contributions that extend beyond a fixed anchoring epitope engaging the nonreducing end of the polySia chain. We further provide an evolutionary analysis of the newly found polySia binding pocket within Human mastadenovirus G.
Results
HAdV-52 Short Fiber Knob Binds to PolySia.
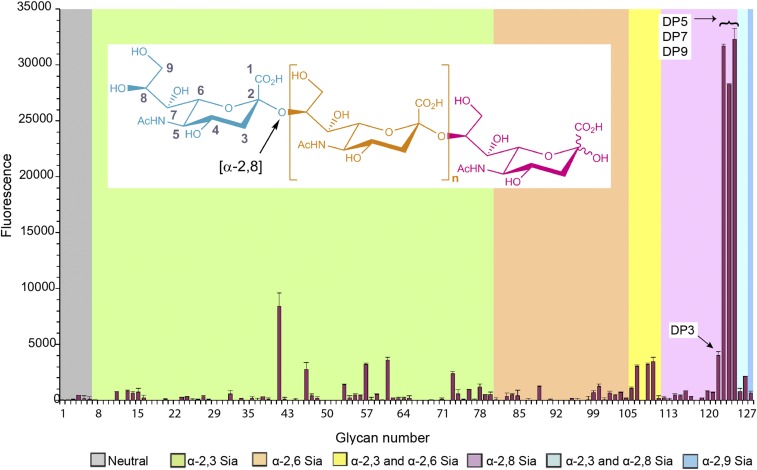
We showed previously that the binding of HAdV-52 to human epithelial cells is sialic acid dependent and occurs via the SFK (23). To date, the precise compositions and structures of glycans that can be optimally engaged by 52SFK remain unknown. We performed glycan microarray analysis of 52SFK with 128 different sialylated glycans, in an attempt to characterize the glycan receptor of HAdV-52. Very strong binding signals were observed with 52SFK for a group of linear α-2,8-linked oligoSia that represent fragments of naturally occurring polySia (Fig. 1 and Table S1). The maximal response was observed at a degree of polymerization (DP) greater than 3 (DP5 to -9). This binding was much greater than for α-2,3- and α-2,6-linked sialic acids in the array. The relatively weak binding detected with the probe oligoSia DP3 is likely to be due to the ring-opened status of the core monosaccharide as a consequence of the reductive amination procedure used for preparing the neoglycolipid probe (50). This suggests that the high-affinity interaction with 52SFK requires at least three intact α-2,8-linked sialic acid residues. To confirm the ability of 52SFK to interact with polySia and to evaluate the specificity of this interaction, we developed an ELISA with immobilized, Escherichia coli-derived polySia (colominic acid; DP ∼ 80–100) and analyzed the binding of recombinant knob domains from HAdV-52 short fiber, the Sia-binding HAdV-37 fiber (37FK), and the CAR-binding HAdV-5 (5FK) and HAdV-52 long fiber (52LFK). The 52SFK bound efficiently to polySia, while the two CAR-binding FKs did not show any binding to this compound (Fig. 2A). 37FK, which binds with relatively high affinity to the branched, disialylated GD1a glycan using a different binding site (11, 23), bound less strongly to polySia than 52SFK. We therefore conclude that HAdV-52 is able to interact preferentially with polySia via the knob domain of its short fiber while having low affinities for a number of monosialylated glycans.
Fig. 1.
Glycan array analysis of HAdV-52 SFK interactions with sialylated glycans. The microarray consists of lipid-linked oligosaccharide probes; the sequences are listed in Table S1. The probes are arranged according to terminal sialic acid linkage, oligosaccharide backbone chain length, and sequence. The various types of terminal sialic acid linkages are indicated by the colored panels as defined at the Bottom of the figure. Numerical scores for the binding intensity are shown as means of fluorescence intensities of duplicate spots at 5 fmol/spot. Error bars represent one-half of the difference between the two values. The three probes that are most strongly bound are DP5–DP9 = α-2,8-linked sialic acids with a degree of polymerization (DP) between 5 and 9 (from Left to Right, in steps of 2). Inlay: structure of polySia, depicted are up to n = ∼100 sialic acid residues that are linearly connected via an α-2,8-linkage (orange). Blue, nonreducing end; pink, reducing end.
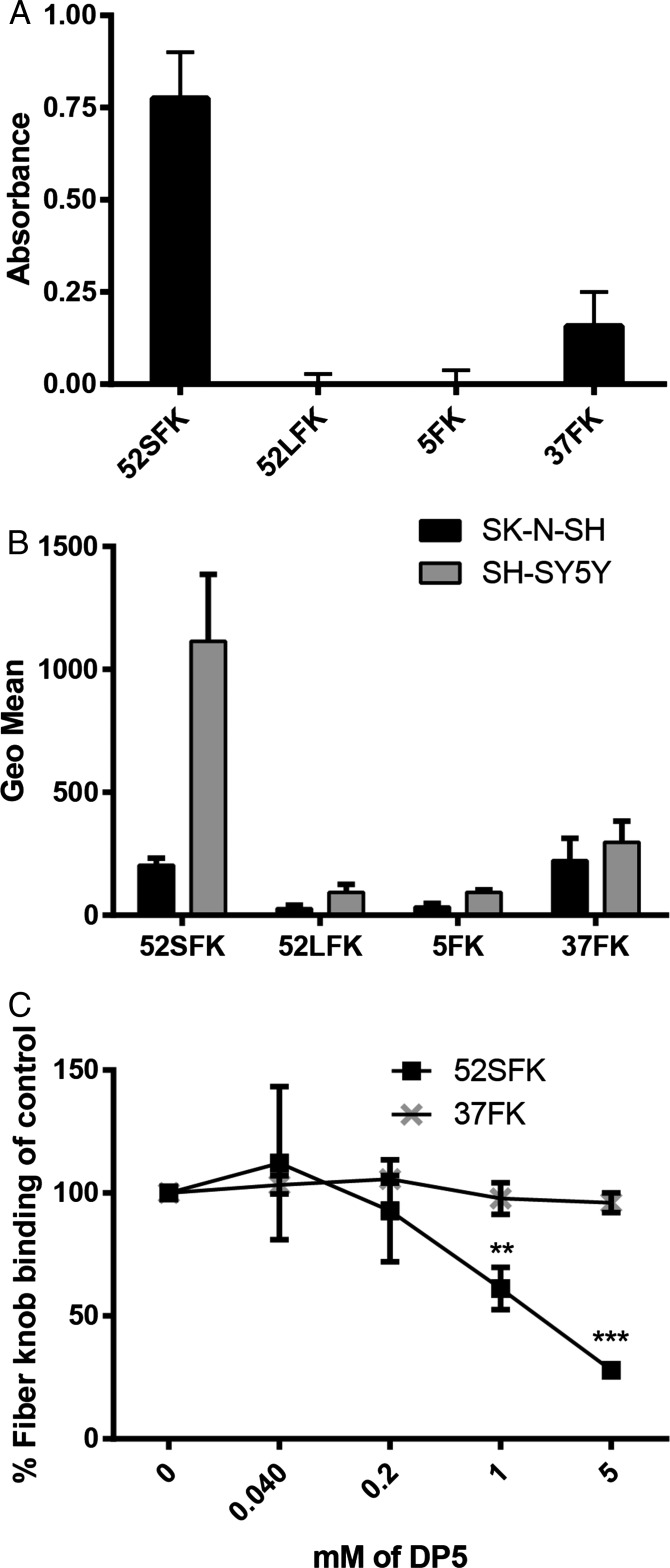
Fig. 2.
HAdV-52 SFK binds to polySia. (A) HAdV FK binding to immobilized E. coli-derived polySia (colominic acid, DP ∼ 80–100). Relative absorbance is shown. (B) Flow cytometry-based quantification of HAdV FK binding to human neuroblastoma cells expressing (SH-SY5Y) or lacking (SK-N-SH) polySia. (C) Flow cytometry-based quantification of 52SFK and 37FK binding to SH-SY5Y cells after FK preincubation with increasing concentrations of pentasialic acid (DP5). FK, fiber knob; LFK, long fiber knob; SFK, short fiber knob. All experiments were performed three times with duplicate samples in each experiment. Error bars represent mean ± SD. **P < 0.01; ***P < 0.001.
HAdV-52 Binds to PolySia on Human PolySia-Expressing Cells.
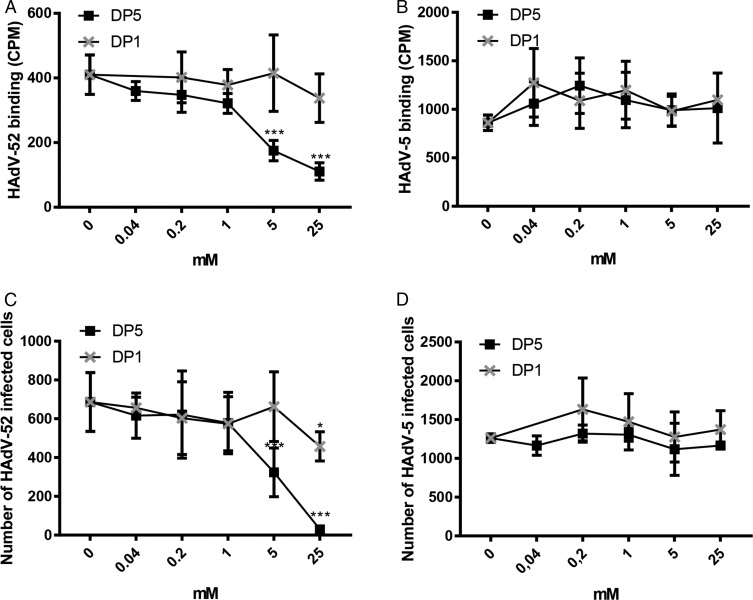
To test the relevance of polySia recognition by HAdV-52 in a cellular context, we used the human polySia-expressing neuroblastoma cell line SH-SY5Y and its polySia-lacking parental cell line SK-N-SH as models for virus binding and infection (51). The levels of polySia on these cells were confirmed by flow cytometry using the anti-polySia antibody mAb735 (Fig. S1). 52SFK gave five times higher binding signals with polySia-expressing SH-SY5Y cells compared with the control cell line, whereas none of the control knobs, including 37FK, showed a comparable interaction pattern (Fig. 2B). Next, we used monosialic acid-binding lectins to evaluate the relative levels of glycans with terminal sialic acids on the two cell lines to exclude the possibility that the higher 52SFK binding to SH-SY5Y was due to a higher level of glycans with terminal monosialic acids on these cells rather than preferential binding to polySia. All three lectins tested, Maackia amurensis I and II (MAL I and II; binds to α-2,3-linked Sia), Sambucus nigra lectin (SNA; binds to α-2,6-linked Sia), and wheat germ agglutinin (WGA; binds to terminal sialic acid as well as to N-acetyl-d-glucosamine) bound stronger to SK-N-SH cells than to SH-SY5Y cells (Fig. S1), indicating that the parental, polySia-negative SK-N-SH cells have a higher total density of terminal sialic acids. Furthermore, preincubation of 52SFK with soluble oligoSia (DP5) reduced 52SFK binding to SH-SY5Y cells up to 75%, while no effect was observed on 37FK binding (Fig. 2C). Preincubating the whole HAdV-52 virions with oligoSia (DP5) also efficiently reduced binding to and infection of SH-SY5Y cells, whereas sialic acid monosaccharide (DP1) did not have as much of an effect (Fig. 3 A and C). Neither of the two glycans tested reduced HAdV-5 binding to or infection of SH-SY5Y cells (Fig. 3 B and D). Based on these results, we conclude that HAdV-52 virions show a clear preference for polySia-expressing cells over cells lacking polySia, that this feature is not shared by monosialic acid- or CAR-binding HAdVs, and that the interactions with polySia are mediated by the 52SFK.
Fig. 3.
OligoSia efficiently reduces HAdV-52 virion binding to and infection of SH-SY5Y cells. Binding of (A) 35S-labeled HAdV-52 and (B) 35S-labeled HAdV-5 virions to SH-SY5Y cells after preincubation with soluble monosialic acid (DP1) or pentasialic acid (DP5). Infection of SH-SY5Y with (C) HAdV-52 and (D) HAdV-5 after preincubation with DP1 or DP5. The experiments were performed three times with duplicate samples in each experiment. Error bars represent mean ± SD. *P < 0.05; ***P < 0.001.
PolySia Is Engaged at the Nonreducing End, Similarly to Monosialylated and Disialylated Glycans.
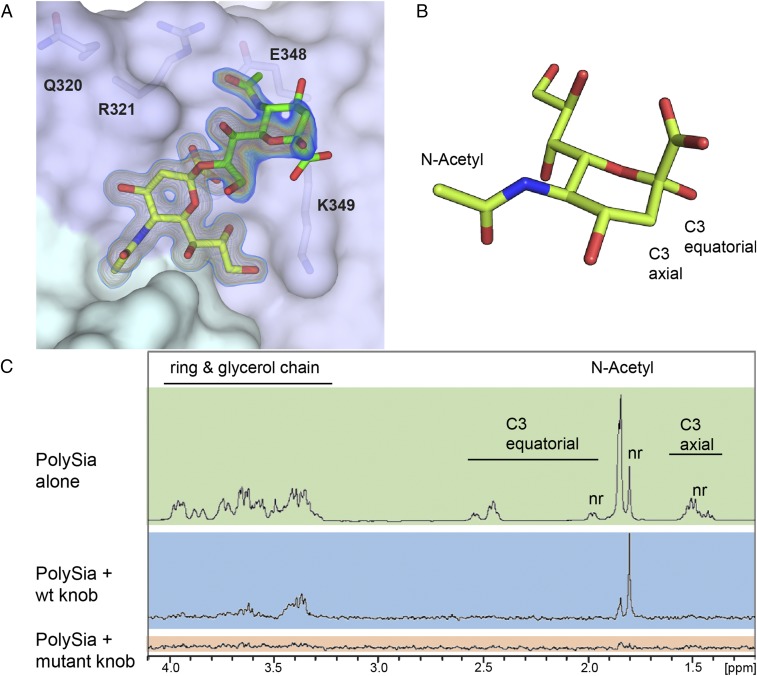
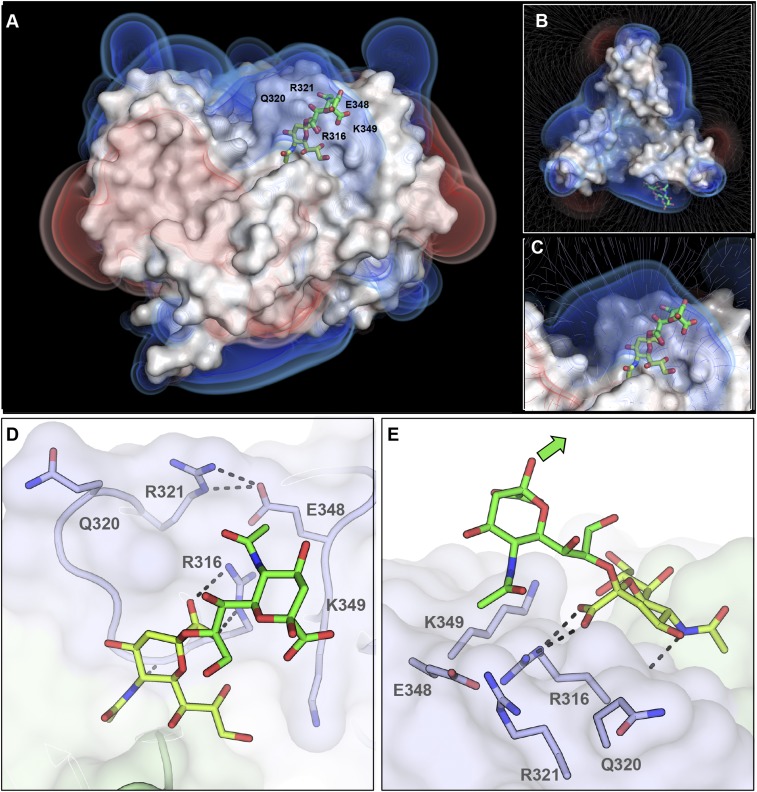
Using 2-O-methyl-sialic acid as a ligand, we previously identified a sialic acid-binding site on the lateral side of 52SFK (23). This binding site includes a stretch of three adjacent residues that together form a prominent RGN motif (R316–G317–N318). This site is located on a different part of the knob from the binding site of 37FK, which engages sialic acid near its threefold axis. The features responsible for the increased affinity for polySia are unknown, and it seems plausible that additional contacts or an additional epitope that went undetected in earlier studies are formed between 52SFK and polySia. Consequently, we solved the complex crystal structures of 52SFK with three oligoSia glycans (DP3, -4, or -5) as well as the GD3 glycan (Neu5NAcα2,8Neu5NAcα2,3Galβ1,4Glc, representing a disialic acid motif). All complex structures produced similar results, as shown exemplary for DP3 in Fig. 4. Surprisingly, well-defined electron density was found only for a single sialic acid moiety in the canonical binding pocket in all cases. The electron density around O8 and its direction relative to the protein clearly indicate that it is the nonreducing end of the glycan chain that is engaged, and the observed binding mode is identical to the one observed for monosialic acid. In all cases except for GD3, we observed additional electron density for a second sialic acid moiety projecting from the pocket toward the solvent. The overall density for this moiety is weaker, deteriorating from the glycerol group to the pyranose ring and indicating increased flexibility. All structures showed similar angles for the α-2,8-glycosidic linkage (Fig. S2). Interestingly, the second sialic acid moiety does not seem to contribute any direct contacts in the overall interaction, except for a van der Waals contact between its N-acetyl group and E328. This contact seems to cause a local decrease of electron density and a slight rotation of the N-acetyl group. The third (and all following) sialic acids could not be unambiguously traced in any of the structures. To verify our observations in solution, we performed saturation transfer difference (STD)–NMR spectroscopy to screen for glycan protons of DP3 and DP5 that are consistently placed within 5–6 Å of the protein (shown exemplary for DP3 in Fig. 4 B and C). The spectrum of the glycans alone compared well with the literature (52, 53). Since all of the sialic acid repeats were in a highly similar chemical environment in solution, the respective peaks overlap—with the exception of the nonreducing end, which experiences an upfield shift. The experiment showed saturation transfer occurring almost exclusively at the nonreducing end, while the other moieties received only a very moderate spin saturation occurring exclusively in the N-acetyl group region, which is consistent with the contacts observed in the crystal structures. In the case of the R316A mutant, which disrupts the canonical RGN motif and prevents 52SFK attachment to sialic acid on A549 cells (23), saturation transfer was completely abrogated. Together, these results demonstrate that 52SFK engages polySia exclusively via its canonical sialic acid binding site, without any additional binding sites on the knob domain.
Fig. 4.
α-2,8-Linked oligoSias are engaged in the canonical binding pocket of HAdV-52 SFK via their nonreducing end. (A) Complex structure of 52SFK and trisialic acid (DP3). Shown is a 2Fo − Fc map calculated at 1 σ (blue) and 1.5 σ (orange) after refinement. The nonreducing sialic acid moiety is colored in yellow, and the adjacent moiety in green. The third sialic acid moiety could not be resolved. (B) Schematic representation of sialic acid in the α-conformation. The positions of distinctive protons for NMR are indicated. (C) STD-NMR of 52SFK and DP3. Green box, DP3 alone; blue box, STD spectrum of the 52SFK:DP3 complex; red box, STD spectrum of the R316A-52SFK:DP3 complex; nr, nonreducing end.
Transient Hydrogen Bonds and Electrostatic Effects Are Major Determinants of 52SFK:PolySia Interactions.
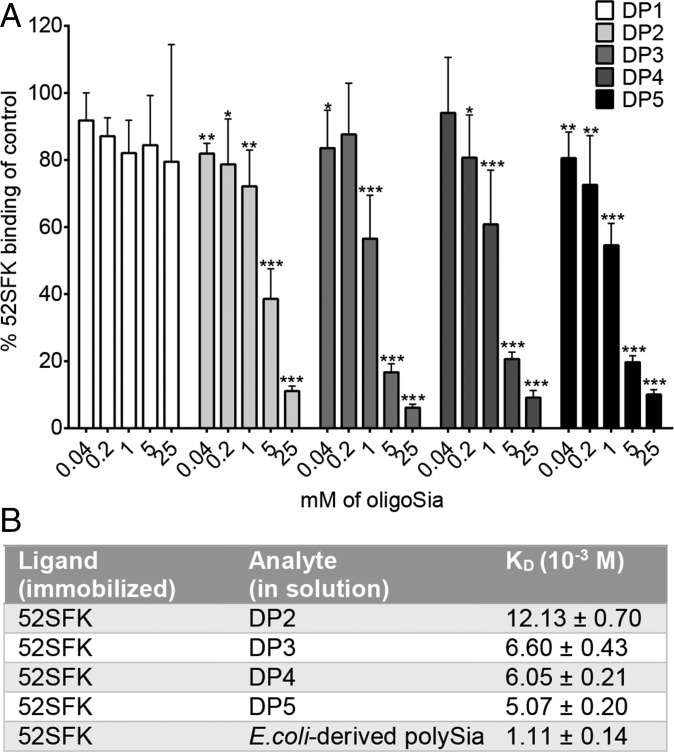
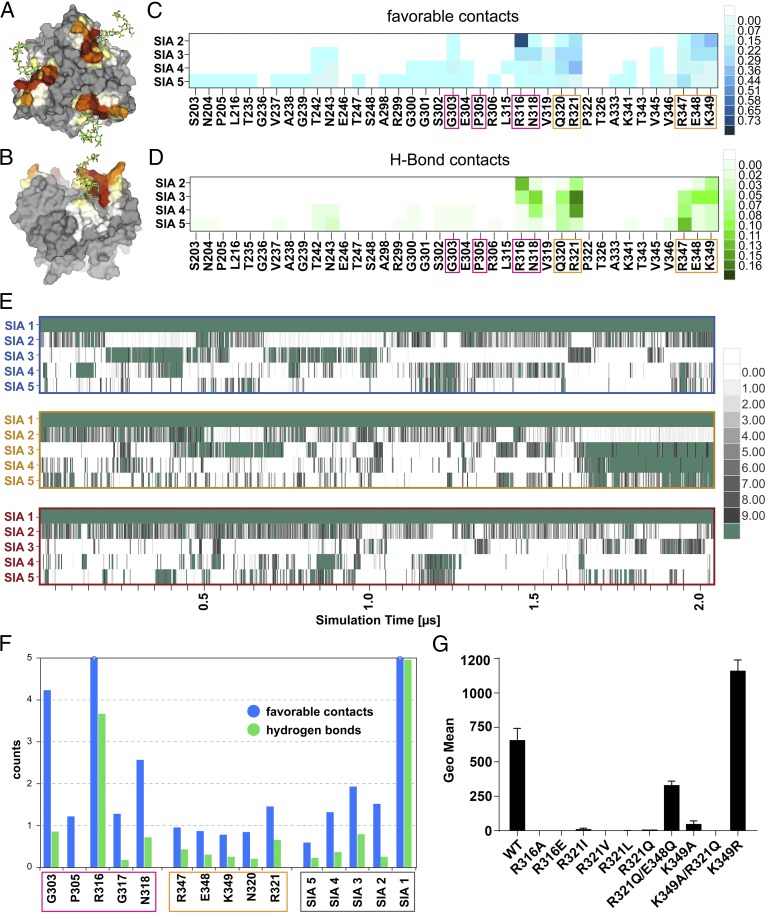
A length of more than three sialic acid residues is required for a strong interaction with 52SFK, as seen in our glycan array data (Fig. 1). According to a cell attachment inhibition experiment, which does not underlie the steric constraints of chip-bound probes, a DP of 3 was sufficient to substantially decrease 52SFK binding at low concentration in solution. A decrease was also observed with DP2, but only at higher concentrations (Fig. 5A). Similar results were acquired from surface plasmon resonance experiments with immobilized FKs and oligoSia in solution, where the biggest increase in affinity was shown between DP2 and DP3 (Fig. 5B). In combination with the structural data, these findings suggest that effects other than classical directed short-range contacts account for the increased binding affinity of higher-order polySia compounds of DP3 or more. Given the polyanionic character of polySia, we hypothesized that these effects might be caused by electrostatic interactions, which are nondirected and can occur over longer distances than direct interactions such as hydrogen bonds or van der Waals contacts. Indeed, an inspection of the electrostatic potential of the 52SFK revealed a positively charged rim located around the sialic acid binding site, which we termed the “steering rim.” The rim is mainly formed by residues Q320, R321, R316, and K349 (Fig. 6 A–D). According to in-solution NMR studies, the polyanionic polySia seems to at least transiently adopt a left-handed helical conformation (54). However, polySia is expected to be rather flexible in solution due to its linear, nonbranched structure and the conformationally less restricted α-2,8-glycosidic linkage (42). In the DP3 complex structure, the second sialic acid moiety is positioned above the ε-amino group of K349. We reasoned that if the polySia glycan roughly followed the left-handed helical arrangement proposed in the literature with energy-minimal glycosidic torsion angles similar to those observed between the first two moieties (Fig. S2), the carbohydrate chain would protrude away from the protein surface into the bulk solvent (indicated in Fig. 6E). Since such an arrangement is unlikely to enhance the affinity for polySia, we performed an MD simulation of the complex between 52SFK and DP5 on the microsecond timescale in explicit solvent. Throughout this simulation, DP5 shows a flexible structure with dynamic partial helical features (Movie S1). Consistent with the results from our STD-NMR experiments, only the nonreducing end is stably associated with the protein (Figs. 6E and 7 A and B). However, the simulation shows that the other sialic acid residues transiently approach the protein surface and form favorable contacts with a variety of amino acids, most of which are located in the steering rim and the closely adjacent R347 (Fig. 7 A–D). While the sialic acid moieties adjacent to the nonreducing end mainly interact with a subset of residues located in the canonical pocket and steering rim, the moieties toward the reducing end show a much more variable interaction pattern with low occupancies for individual contacts. In total, however, the large majority of contacts are being formed with the canonical pocket or steering rim, respectively. The dimensions of DP5 are similar to the combined radius of the binding pocket and steering rim (Fig. 7 A and B). In particular, the fifth sialic acid engages in a large number of low-intensity interactions with residues outside the rim according to our simulations (Fig. 7 C and D), which might explain why the enhancing effect of additional Sia moieties is fading beyond DP5 (Fig. 1) and why colominic acid is only moderately more potent than DP5 given the size difference (Fig. 5B). Over the time course of the simulation, the sialic acids display an alternating pattern of transient interactions, and there are most of the time at least two pyranoses that directly interact with the protein (Fig. 7E). This avidity effect is only possible if there are at least three Sia moieties. The average number of favorable interactions found in the canonical pocket and the steering rim per residue are shown in Fig. 7F. Overall, the data agree remarkably well with the other experiments and strongly suggest that transient contact interactions of the third to fifth sialic acid moiety are responsible for the increased binding affinity, while the nonreducing sialic acid is a necessary feature and engaged in a shape-complementary binding site. The involvement of additional noncontact electrostatic interactions could further contribute to binding affinity (Fig. S3). Despite the excellent agreement between the experiments and the results derived from MD simulations, it should be noted that even sampling of conformational space on the microsecond timescale might not be long enough to sample all possible interactions between the HAdV-52 SFK and oligoSia. Therefore, the results from MD simulations should be taken with some caution.
Fig. 5.
A DP of 3 (or more) strengthens the interactions with 52SFK. (A) Flow cytometry-based quantification of 52SFK binding to SH-SY5Y cells after FK preincubation with increasing concentrations of oligoSia. The experiment was performed three times with duplicate samples in each experiment. Error bars represent mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001. (B) Surface plasmon resonance analysis of 52SFK binding to disialic acid (DP2), trisialic acid (DP3), tetrasialic acid (DP4), pentasialic acid (DP5), and E. coli-derived polySia (DP ∼ 80–100).
Fig. 6.
Representation of the HAdV-52 SFK steering rim. Poisson–Boltzmann electrostatic potential isosurfaces and field lines for the protein were calculated at ±1, ±0.75, and ±0.5 kT/e. The positively charged rim can be seen in blue. Bound trisialic acid (DP3) is shown as green sticks. (A) Side view. (B) Top view including field lines. (C) Detailed view of the binding pocket including field lines. (D) Detailed view of the binding pocket showing the relative placement of glycan and steering rim residues. Residues of the steering rim are highlighted as sticks. R321 and E348 are forming a salt bridge, as do R316 and the carboxyl group at the nonreducing end of DP3. The orientation is the same as in A. (E) Side view of the interaction site. The second sialic acid moiety is projecting away from the protein surface. The green arrow indicates the expected direction of the adjacent sialic acid moieties. (D and E) The nonreducing sialic acid moiety is colored in yellow, and the adjacent moiety in green.
Fig. 7.
MD simulation of the interactions between 52SFK and DP5. Three pentasialic acid (DP5) molecules interacting with the three identical binding pockets of 52SFK were simulated over a time of 2 μs. (A and B) The interaction profile of DP5 with the protein is mapped onto 52SFK in a “heat map” style. Noninteracting residues are colored in gray, and interacting residues are scored from white (few interactions) to brown (strongly interacting). (A) All three pockets are shown from a top view. (B) One of the simulated binding pockets is shown from a side view. (C and D) Detailed interactions contributed by the additional sialic acid moieties in polySia. Amino acids of the canonical binding site are boxed in pink, and residues of the steering rim in orange. (C) Residue–residue interaction matrix showing the average number of favorable atom contacts between individual amino acids and sialic acids (SIA 2–5, counted from the nonreducing end) over the whole simulation. (D) Analogous plot showing the average number of hydrogen bonds. (E) Time-resolved trajectory plot of the number of atom contacts per sialic acid residue (numbered from the nonreducing end) in the three binding sites (individual rows) averaged over 2.5-ns increments. Atom contacts are counted as favorable if one of the following conditions are satisfied: H-bond donor/acceptor atom distance <3.2 Å or C–C atom distance of <4.2 Å. The average number of interactions is depicted according to the color legends on the Right for each panel. (F) Summary of the interactions of polySia with the 52SFK canonical pocket and steering rim. The number of favorable atom contacts and hydrogen bonds per residue is averaged over the three binding sites. Boxing of the amino acid residues is analogous to C and D; sialic acids are boxed in gray. (G) Flow cytometry-based analysis of HAdV-52 SFK mutant binding to polySia-expressing SH-SY5Y cells. The experiment was performed three times with duplicate samples in each experiment. Error bars represent mean ± SD.
To provide additional experimental support for our hypothesis, we produced FKs with mutations in the steering rim and analyzed knob binding to polySia-expressing SH-SY5Y cells. The K349A mutant almost completely lost its cell binding capacity, and similar effects were observed for the R321Q and analogous mutants (Fig. 7G). When mutated, the residue R321 can no longer counterbalance the charge of the proximal side chain of E348, which then likely repels the polyanionic polySia and might thus contribute to an unexpectedly strong loss in binding. Indeed, if E348 is also mutated to a noncharged residue, the effect of the R321Q mutation is largely reversed (Fig. 7G). This implies that R321 interacts more weakly with polySia than R316 and K349 do, which fits well with the assumption of a flexible “pseudohelical” arrangement.
The PolySia Binding Site and the Steering Rim Are Conserved in Closely Related Simian Adenoviruses.
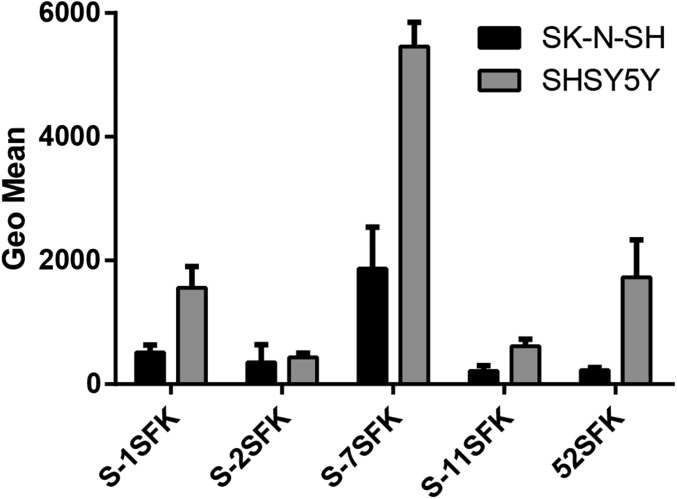
The polySia-binding RGN motif is conserved in the short fibers of other closely related members of species HAdV-G: simian adenovirus (SAdV)-1, -2, -7, and -11, as well as SAdV-19 (SAdV-C, which acquired its short fiber from an unknown type/species) (55), but it is not found in any other known nonhuman or human AdV, including the SFKs of HAdV-40 and -41 (HAdV-F) (Fig. S4A). Interestingly, the three positively charged residues forming the steering rim are also functionally conserved in these SAdV types, but in different permutations (RRK, RKK, RRR) (Fig. S4A). Another functionally important residue is Q320, which aids in the production of an electropositive field in the steering rim and is functionally conserved in all of the SAdV types of HAdV-G (but not in SAdV-19). No other HAdV FK with known structure exhibits a comparable steering rim (Fig. S4B). In fact, the lateral part of the knob is typically used for protein interfaces, for example, for CAR or CD46 (56). However, since the two fibers of HAdV-52 display a clear division of labor, the 52SFK likely serves as a purely Sia-binding FK and thus can accommodate Sia-containing glycans at a more prominently exposed lateral binding site than for example on HAdV-37. In the HAdV-41 SFK, which is the only other structurally characterized SFK, the disordered G strand is thought to obstruct the electropositive patch on the side (57), making a Sia interaction unlikely. HAdV-5 possesses an electropositive patch but lacks a shape-complementary Sia-binding site and has not been reported to use sialic acid as attachment receptor. Instead, it has been used as a negative control in many experiments (Figs. 2 and 3). This further supports our hypothesis that polySia binding is a specific ability limited to a small subset of AdVs. We assayed the polySia specificity and binding capacity of FKs belonging to this subset in a cell attachment assay with cells expressing or lacking polySia. All of the examined short knobs except that of SAdV-2 SFK bound better to polySia-expressing cells than to the control cell line (Fig. 8). One possible explanation for the inability of SAdV-2 SFK to bind polySia, despite a conserved steering rim, could be that this knob harbors a sequence more distantly related to the other knobs (Fig. S4C), which might result in a different overall arrangement of the residues. Nonetheless, 52SFK displayed the strongest discrepancy between SH-SY5Y and SK-N-SH cells, indicating a more specific interaction of 52SFK with polySia rather than a general high binding to both cell lines as seen for SAdV-7SFK (Fig. 8).
Fig. 8.
AdV SFK binding to polySia-expressing/lacking cells. Flow cytometry-based quantification of simian (S) and human AdV SFK binding to human neuroblastoma cells expressing (SH-SY5Y) or lacking (SK-N-SH) polySia. The experiment was performed three times with duplicate samples in each experiment. SFK, short fiber knob. Error bars represent mean ± SD.
Discussion
We show here that HAdV-52 specifically engages cell surface-expressed polySia via its SFK, employing long-lived direct protein–carbohydrate contacts as well as transient longer-range electrostatic steering forces. With this study, we have identified an additional role of polySia, as a cellular receptor for a human pathogenic virus. Although a growing number of polySia-binding proteins have been identified (58–64), there are relatively few in-depth structural analyses on the determinants of specificity for polySia, and to date no other polySia-binding protein has been reported to use the unusual binding mode presented here. We therefore believe that our analysis provides a useful framework for a better understanding of general aspects of the interactions of polySia with its binding partners, and it remains to be seen whether polySia reacts with other binding partners in a manner similar to that predicted for HAdV-52.
PolySia was identified as a potential receptor for 52SFK by glycan microarray screening (Fig. 1). In that same array, 52SFK showed weaker binding to a number of glycans with single capping sialic acids, mainly α-2,3-linked, as we described in our previous study (23). In a cellular context, however, blocking or removing α-2,3-linked sialic acids from the cell surface had only a minor effect on HAdV-52 attachment (23). Thus, in comparison, polySia is a more effective ligand. The topology of the polySia binding site of HAdV-52 allows it to maintain a large pool of glycan ligands while developing increased affinity for a specific subset of surface molecules using just a single binding site. 52SFK can engage differently linked sialylated glycans, which bind with their terminal, nonreducing sialic acid moieties to the same epitope using identical direct contacts. The strong preference for α-2,8-linked polySia compounds is generated through a multitude of transient contacts between residues surrounding the binding site and sialic acid residues that are distal to the nonreducing end of the polySia chain. These transient contacts ensure that most of the time at least two Sia moieties are simultaneously associated with the protein, providing an avidity effect. In this manner, monosialylated and disialylated glycans are still able to interact with the knob with lower affinities, but long-range electrostatic and transient polar interactions enable higher-affinity binding of oligosialic acids with a higher DP (DP ≥ 3). In a physiological context, this might reflect the ability of HAdV-52 to adapt to different surface glycan landscapes presented by different cells, hosts, or even commensal bacteria. In humans, it is unknown in which contexts HAdV-52 might encounter polySia for cell attachment. The two most efficient attachment factors of HAdV-52, polySia and CAR, have different expression profiles in the human body and are recognized by the two separate HAdV-52 fiber proteins. In light of its limited genome size, the virus likely draws an evolutionary advantage from being able to interact with two attachment factors. The close evolutionary relationship between HAdV-52 and simian AdVs, and the polySia-binding capacity of these AdVs (Fig. 8), also indicate that polySia might play a role as a cellular attachment factor for viruses that infect other mammals. Sialic acid-containing glycans are known to serve as attachment factors for a number of animal AdVs such as turkey and canine AdV (65, 66). The observed interaction between HAdV-52 and polySia therefore provides an interesting angle to the known rules that govern virus:glycan receptor interactions, which may be translated to other glycan-binding pathogens.
PolySia has been detected in a number of cancer tissues by immunohistochemical staining, and its expression is frequently associated with high tumor aggressiveness and invasiveness, resulting in poor clinical prognosis (43, 67, 68). Cancers expressing polySia are also often recurrent and nonresponsive to conventional treatments (43), and therefore attention has been drawn to novel therapeutic approaches, including AdV vectors for gene delivery and the use of modified oncolytic AdVs. In a recent approach, the FK of HAdV-5 was substituted with endosialidase NF, a tail spike protein from the bacteriophage K1F to generate an efficient polySia-targeting oncolytic vector (69). HAdV-52, a naturally occurring, unmodified HAdV that already binds polySia, could form the basis for a viable alternative strategy for developing oncolytic vectors, especially in the light of its low seroprevalence rates and reduced liver tropism (23, 70). The specificity for polySia can also be increased further by mutating K349 to an arginine (Fig. 7G). With this in mind, we suggest that HAdV-52–based vectors could have a potential for treatment of cancers characterized by elevated polySia expression.
Materials and Methods
Please see SI Materials and Methods for information regarding cells, viruses, and glycans used in the study, and for detailed descriptions of production of fiber knobs, glycan microarray, ELISA, flow cytometry, virus binding and infection experiments, STD-NMR, crystallization, surface plasmon resonance, MD simulations, and statistical analysis.
Supplementary Material
Acknowledgments
The neoglycolipid-based glycan microarrays contain several saccharides provided by collaborators whom we thank as well as members of the Glycosciences Laboratory for their collaboration in the establishment of the microarray system. We gratefully acknowledge support from the Swiss Light Source staff during X-ray data collection. This investigation was supported by Swedish Cancer Society Grant CAN 2015/695, Cancer Research Foundation in Northern Sweden Grant AMP 16-794, Knut and Alice Wallenberg Foundation Grant KAW 2013.0019, German Research Foundation Grant DFG-SFB685, ViroCarb Grant FOR 2327, Marie Curie Initial Training Network “ADVance” Grant FP7-290002, the Glycobiology/Glycobiotechnology Program of the Baden-Württemberg Foundation, and Welcome Trust Grants WT093378MA/Z/10/Z/ and WT099197/Z/12/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallographic dataset of the relevant protein/glycan complex structure has been deposited in the RCSB Protein Data Bank (PDB), www.rcsb.org/pdb/ (6G47).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716900115/-/DCSupplemental.
References
- 1.Kaján GL, Kajon AE, Pinto AC, Bartha D, Arnberg N. The complete genome sequence of human adenovirus 84, a highly recombinant new Human mastadenovirus D type with a unique fiber gene. Virus Res. 2017;242:79–84. doi: 10.1016/j.virusres.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson JM, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 3.Tomko RP, Xu R, Philipson L. HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roelvink PW, et al. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat Med. 2011;17:96–104. doi: 10.1038/nm.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat Med. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 7.Segerman A, et al. Adenovirus type 11 uses CD46 as a cellular receptor. J Virol. 2003;77:9183–9191. doi: 10.1128/JVI.77.17.9183-9191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marttila M, et al. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J Virol. 2005;79:14429–14436. doi: 10.1128/JVI.79.22.14429-14436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnberg N, Edlund K, Kidd AH, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 10.Arnberg N, Kidd AH, Edlund K, Olfat F, Wadell G. Initial interactions of subgenus D adenoviruses with A549 cellular receptors: Sialic acid versus alpha(v) integrins. J Virol. 2000;74:7691–7693. doi: 10.1128/jvi.74.16.7691-7693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson EC, et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat Med. 2011;17:105–109. doi: 10.1038/nm.2267. [DOI] [PubMed] [Google Scholar]
- 12.Belin MT, Boulanger P. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1993;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 13.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 14.Wickham TJ, Filardo EJ, Cheresh DA, Nemerow GR. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Guan T, Cheresh DA, Nemerow GR. Regulation of adenovirus membrane penetration by the cytoplasmic tail of integrin beta5. J Virol. 2000;74:2731–2739. doi: 10.1128/jvi.74.6.2731-2739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker AL, et al. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood. 2006;108:2554–2561. doi: 10.1182/blood-2006-04-008532. [DOI] [PubMed] [Google Scholar]
- 17.Waddington SN, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Lenman A, et al. Coagulation factor IX mediates serotype-specific binding of species A adenoviruses to host cells. J Virol. 2011;85:13420–13431. doi: 10.1128/JVI.06088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, et al. Coagulation factor X shields adenovirus type 5 from attack by natural antibodies and complement. Nat Med. 2013;19:452–457. doi: 10.1038/nm.3107. [DOI] [PubMed] [Google Scholar]
- 20.Jones MS, 2nd, et al. New adenovirus species found in a patient presenting with gastroenteritis. J Virol. 2007;81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd AH, Chroboczek J, Cusack S, Ruigrok RW. Adenovirus type 40 virions contain two distinct fibers. Virology. 1993;192:73–84. doi: 10.1006/viro.1993.1009. [DOI] [PubMed] [Google Scholar]
- 22.Yeh HY, Pieniazek N, Pieniazek D, Gelderblom H, Luftig RB. Human adenovirus type 41 contains two fibers. Virus Res. 1994;33:179–198. doi: 10.1016/0168-1702(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 23.Lenman A, et al. Human adenovirus 52 uses sialic acid-containing glycoproteins and the coxsackie and adenovirus receptor for binding to target cells. PLoS Pathog. 2015;11:e1004657. doi: 10.1371/journal.ppat.1004657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matrosovich M, Herrler G, Klenk HD. Sialic acid receptors of viruses. Top Curr Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finne J, Finne U, Deagostini-Bazin H, Goridis C. Occurrence of alpha 2-8 linked polysialosyl units in a neural cell adhesion molecule. Biochem Biophys Res Commun. 1983;112:482–487. doi: 10.1016/0006-291x(83)91490-0. [DOI] [PubMed] [Google Scholar]
- 26.Galuska SP, et al. Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc Natl Acad Sci USA. 2010;107:10250–10255. doi: 10.1073/pnas.0912103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werneburg S, Mühlenhoff M, Stangel M, Hildebrandt H. Polysialic acid on SynCAM 1 in NG2 cells and on neuropilin-2 in microglia is confined to intracellular pools that are rapidly depleted upon stimulation. Glia. 2015;63:1240–1255. doi: 10.1002/glia.22815. [DOI] [PubMed] [Google Scholar]
- 28.Kiermaier E, et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science. 2016;351:186–190. doi: 10.1126/science.aad0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galuska CE, Lütteke T, Galuska SP. Is polysialylated NCAM not only a regulator during brain development but also during the formation of other organs? Biology (Basel) 2017;6:E27. doi: 10.3390/biology6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnaar RL, Gerardy-Schahn R, Hildebrandt H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev. 2014;94:461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colley KJ, Kitajima K, Sato C. Polysialic acid: Biosynthesis, novel functions and applications. Crit Rev Biochem Mol Biol. 2014;49:498–532. doi: 10.3109/10409238.2014.976606. [DOI] [PubMed] [Google Scholar]
- 32.Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- 33.Rey-Gallardo A, et al. Polysialylated neuropilin-2 enhances human dendritic cell migration through the basic C-terminal region of CCL21. Glycobiology. 2010;20:1139–1146. doi: 10.1093/glycob/cwq078. [DOI] [PubMed] [Google Scholar]
- 34.Rey-Gallardo A, Delgado-Martín C, Gerardy-Schahn R, Rodríguez-Fernández JL, Vega MA. Polysialic acid is required for neuropilin-2a/b-mediated control of CCL21-driven chemotaxis of mature dendritic cells and for their migration in vivo. Glycobiology. 2011;21:655–662. doi: 10.1093/glycob/cwq216. [DOI] [PubMed] [Google Scholar]
- 35.Curreli S, Arany Z, Gerardy-Schahn R, Mann D, Stamatos NM. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem. 2007;282:30346–30356. doi: 10.1074/jbc.M702965200. [DOI] [PubMed] [Google Scholar]
- 36.Stamatos NM, et al. Changes in polysialic acid expression on myeloid cells during differentiation and recruitment to sites of inflammation: Role in phagocytosis. Glycobiology. 2014;24:864–879. doi: 10.1093/glycob/cwu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchiya A, et al. Polysialic acid/neural cell adhesion molecule modulates the formation of ductular reactions in liver injury. Hepatology. 2014;60:1727–1740. doi: 10.1002/hep.27099. [DOI] [PubMed] [Google Scholar]
- 38.El Maarouf A, Petridis AK, Rutishauser U. Use of polysialic acid in repair of the central nervous system. Proc Natl Acad Sci USA. 2006;103:16989–16994. doi: 10.1073/pnas.0608036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Induced expression of polysialic acid in the spinal cord promotes regeneration of sensory axons. Mol Cell Neurosci. 2007;35:109–119. doi: 10.1016/j.mcn.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Lentiviral-mediated expression of polysialic acid in spinal cord and conditioning lesion promote regeneration of sensory axons into spinal cord. Mol Ther. 2007;15:1796–1804. doi: 10.1038/sj.mt.6300220. [DOI] [PubMed] [Google Scholar]
- 41.Werneburg S, et al. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia. 2016;64:1314–1330. doi: 10.1002/glia.23004. [DOI] [PubMed] [Google Scholar]
- 42.Ulm C, et al. Soluble polysialylated NCAM: A novel player of the innate immune system in the lung. Cell Mol Life Sci. 2013;70:3695–3708. doi: 10.1007/s00018-013-1342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki M, et al. Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology. 2005;15:887–894. doi: 10.1093/glycob/cwi071. [DOI] [PubMed] [Google Scholar]
- 44.Petridis AK, Wedderkopp H, Hugo HH, Maximilian Mehdorn H. Polysialic acid overexpression in malignant astrocytomas. Acta Neurochir (Wien) 2009;151:601–603 discussion 603–604. doi: 10.1007/s00701-009-0324-3. [DOI] [PubMed] [Google Scholar]
- 45.Amoureux MC, et al. Polysialic acid neural cell adhesion molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC Cancer. 2010;10:91. doi: 10.1186/1471-2407-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Figarella-Branger DF, Durbec PL, Rougon GN. Differential spectrum of expression of neural cell adhesion molecule isoforms and L1 adhesion molecules on human neuroectodermal tumors. Cancer Res. 1990;50:6364–6370. [PubMed] [Google Scholar]
- 47.Glüer S, Schelp C, Gerardy-Schahn R, von Schweinitz D. Polysialylated neural cell adhesion molecule as a marker for differential diagnosis in pediatric tumors. J Pediatr Surg. 1998;33:1516–1520. doi: 10.1016/s0022-3468(98)90488-2. [DOI] [PubMed] [Google Scholar]
- 48.Lantuejoul S, Moro D, Michalides RJ, Brambilla C, Brambilla E. Neural cell adhesion molecules (NCAM) and NCAM-PSA expression in neuroendocrine lung tumors. Am J Surg Pathol. 1998;22:1267–1276. doi: 10.1097/00000478-199810000-00012. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka F, et al. Expression of polysialic acid and STX, a human polysialyltransferase, is correlated with tumor progression in non-small cell lung cancer. Cancer Res. 2000;60:3072–3080. [PubMed] [Google Scholar]
- 50.Chai W, Stoll MS, Galustian C, Lawson AM, Feizi T. Neoglycolipid technology: Deciphering information content of glycome. Methods Enzymol. 2003;362:160–195. doi: 10.1016/S0076-6879(03)01012-7. [DOI] [PubMed] [Google Scholar]
- 51.Valentiner U, Mühlenhoff M, Lehmann U, Hildebrandt H, Schumacher U. Expression of the neural cell adhesion molecule and polysialic acid in human neuroblastoma cell lines. Int J Oncol. 2011;39:417–424. doi: 10.3892/ijo.2011.1038. [DOI] [PubMed] [Google Scholar]
- 52.Brisson JR, Baumann H, Imberty A, Pérez S, Jennings HJ. Helical epitope of the group B meningococcal alpha(2-8)-linked sialic acid polysaccharide. Biochemistry. 1992;31:4996–5004. doi: 10.1021/bi00136a012. [DOI] [PubMed] [Google Scholar]
- 53.Ray GJ, et al. Complete structural elucidation of an oxidized polysialic acid drug intermediate by nuclear magnetic resonance spectroscopy. Bioconjug Chem. 2014;25:665–676. doi: 10.1021/bc400456g. [DOI] [PubMed] [Google Scholar]
- 54.Battistel MD, Shangold M, Trinh L, Shiloach J, Freedberg DI. Evidence for helical structure in a tetramer of α2-8 sialic acid: Unveiling a structural antigen. J Am Chem Soc. 2012;134:10717–10720. doi: 10.1021/ja300624j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Podgorski II, Pantó L, Papp T, Harrach B, Benkö M. Genome analysis of four Old World monkey adenoviruses supports the proposed species classification of primate adenoviruses and reveals signs of possible homologous recombination. J Gen Virol. 2016;97:1604–1614. doi: 10.1099/jgv.0.000465. [DOI] [PubMed] [Google Scholar]
- 56.Cupelli K, Stehle T. Viral attachment strategies: The many faces of adenoviruses. Curr Opin Virol. 2011;1:84–91. doi: 10.1016/j.coviro.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 57.Seiradake E, Cusack S. Crystal structure of enteric adenovirus serotype 41 short fiber head. J Virol. 2005;79:14088–14094. doi: 10.1128/JVI.79.22.14088-14094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanato Y, Kitajima K, Sato C. Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology. 2008;18:1044–1053. doi: 10.1093/glycob/cwn084. [DOI] [PubMed] [Google Scholar]
- 59.Sato C, Yamakawa N, Kitajima K. Measurement of glycan-based interactions by frontal affinity chromatography and surface plasmon resonance. Methods Enzymol. 2010;478:219–232. doi: 10.1016/S0076-6879(10)78010-1. [DOI] [PubMed] [Google Scholar]
- 60.Ono S, Hane M, Kitajima K, Sato C. Novel regulation of fibroblast growth factor 2 (FGF2)-mediated cell growth by polysialic acid. J Biol Chem. 2012;287:3710–3722. doi: 10.1074/jbc.M111.276618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isomura R, Kitajima K, Sato C. Structural and functional impairments of polysialic acid by a mutated polysialyltransferase found in schizophrenia. J Biol Chem. 2011;286:21535–21545. doi: 10.1074/jbc.M111.221143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishra B, et al. Functional role of the interaction between polysialic acid and extracellular histone H1. J Neurosci. 2010;30:12400–12413. doi: 10.1523/JNEUROSCI.6407-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagae M, et al. Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: Insight into the binding preference for polysialic acid. J Biol Chem. 2013;288:33784–33796. doi: 10.1074/jbc.M113.496224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haselhorst T, et al. Endosialidase NF appears to bind polySia DP5 in a helical conformation. ChemBioChem. 2006;7:1875–1877. doi: 10.1002/cbic.200600252. [DOI] [PubMed] [Google Scholar]
- 65.Seiradake E, et al. The cell adhesion molecule “CAR” and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLoS Pathog. 2009;5:e1000277. doi: 10.1371/journal.ppat.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh AK, et al. Structure and sialyllactose binding of the carboxy-terminal head domain of the fibre from a siadenovirus, turkey adenovirus 3. PLoS One. 2015;10:e0139339. doi: 10.1371/journal.pone.0139339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka F, et al. Prognostic significance of polysialic acid expression in resected non-small cell lung cancer. Cancer Res. 2001;61:1666–1670. [PubMed] [Google Scholar]
- 68.Falconer RA, Errington RJ, Shnyder SD, Smith PJ, Patterson LH. Polysialyltransferase: A new target in metastatic cancer. Curr Cancer Drug Targets. 2012;12:925–939. doi: 10.2174/156800912803251225. [DOI] [PubMed] [Google Scholar]
- 69.Martin NT, et al. Targeting polysialic acid-abundant cancers using oncolytic adenoviruses with fibers fused to active bacteriophage borne endosialidase. Biomaterials. 2018;158:86–94. doi: 10.1016/j.biomaterials.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 70.Bányai K, et al. Searching for HAdV-52, the putative gastroenteritis-associated human adenovirus serotype in Southern Hungary. New Microbiol. 2009;32:185–188. [PubMed] [Google Scholar]