Significance
Cell-surface and secreted glycoproteins are initially synthesized and glycosylated in the endoplasmic reticulum. Glycan structures are trimmed and remodeled as they transit the secretory pathway, resulting in multi-branched complex-type structures. The enzymes that remodel these structures have precise linkage and branch specificities, with the product of one reaction being specifically recognized as the substrate for the following reaction. These reactions include N-acetylglucosaminyltransferase II (MGAT2), an enzyme that initiates complex branch extension by precise recognition of its glycan substrate. The structural basis for MGAT2 substrate recognition is the subject of the present study. Structures of MGAT2-substrate complexes reveal both modular and convergent mechanisms for selective substrate recognition and catalysis and provide a generalized model for template-based synthesis of glycan structures by glycosyltransferases.
Keywords: glycosyltranferase, N-glycan processing, convergent evolution, exosite, substrate recognition
Abstract
Asn-linked oligosaccharides are extensively modified during transit through the secretory pathway, first by trimming of the nascent glycan chains and subsequently by initiating and extending multiple oligosaccharide branches from the trimannosyl glycan core. Trimming and branching pathway steps are highly ordered and hierarchal based on the precise substrate specificities of the individual biosynthetic enzymes. A key committed step in the synthesis of complex-type glycans is catalyzed by N-acetylglucosaminyltransferase II (MGAT2), an enzyme that generates the second GlcNAcβ1,2- branch from the trimannosyl glycan core using UDP-GlcNAc as the sugar donor. We determined the structure of human MGAT2 as a Mn2+-UDP donor analog complex and as a GlcNAcMan3GlcNAc2-Asn acceptor complex to reveal the structural basis for substrate recognition and catalysis. The enzyme exhibits a GT-A Rossmann-like fold that employs conserved divalent cation-dependent substrate interactions with the UDP-GlcNAc donor. MGAT2 interactions with the extended glycan acceptor are distinct from other related glycosyltransferases. These interactions are composed of a catalytic subsite that binds the Man-α1,6- monosaccharide acceptor and a distal exosite pocket that binds the GlcNAc-β1,2Man-α1,3Manβ- substrate “recognition arm.” Recognition arm interactions are similar to the enzyme–substrate interactions for Golgi α-mannosidase II, a glycoside hydrolase that acts just before MGAT2 in the Asn-linked glycan biosynthetic pathway. These data suggest that substrate binding by MGAT2 employs both conserved and convergent catalytic subsite modules to provide substrate selectivity and catalysis. More broadly, the MGAT2 active-site architecture demonstrates how glycosyltransferases create complementary modular templates for regiospecific extension of glycan structures in mammalian cells.
The glycan structures on cell-surface and secreted glycoproteins form a complex interface with the extracellular environment (1, 2). Glycan structures are essential for numerous biological functions including cell signaling, cellular adhesion, and host–pathogen interactions, among many others (1). Cell-surface glycan structures are not template-derived. Instead, they are the products of complex metabolic pathways where biosynthetic enzymes encode the regiospecific branching and extension of mature glycan structures (2, 3). Understanding the details of substrate recognition, catalytic mechanisms, and regulation of these enzymes is key to understanding glycan diversity and their roles in human disease.
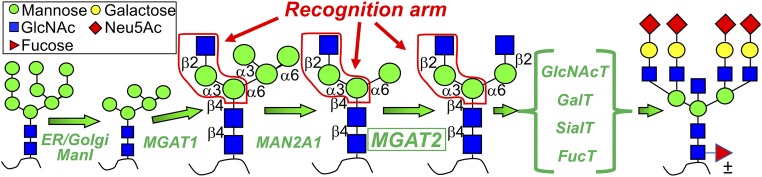
The biosynthesis of Asn-linked glycans (N-glycans) begins with the transfer of a preformed Glc3Man9GlcNAc2 glycan from a lipid-linked precursor to the Asn side chains of polypeptides as they are translocated into the lumen of the endoplasmic reticulum. This glycan structure is cleaved to a Man5GlcNAc2-Asn processing intermediate and further subjected to a complex series of branching, trimming, and extension reactions in the lumen of the Golgi complex to generate the multibranched, complex-type glycans found on cell-surface and secreted glycoproteins (2) (Fig. 1). The enzymatic steps required for the synthesis of these complex-type structures follow a discrete hierarchy based on the substrate specificities of the respective enzymes. However, little is known regarding the structural basis for this substrate specificity. The first step in the glycan branching pathway is the addition of a β1,2GlcNAc to the core α1,3Man residue by N-acetylglucosaminyltransferase I (MGAT1) to produce the GlcNAcMan5GlcNAc2-Asn intermediate (4). This addition is a prerequisite for several subsequent reactions (SI Appendix, Fig. S1), including the cleavage of terminal α1,3Man and α1,6Man residues by Golgi α-mannosidase II (MAN2A1), followed by the addition of the second β1,2GlcNAc branch on the core α1,6Man residue by N-acetylglucosaminyltransferase II (MGAT2). Other enzymes that are dependent upon MGAT1 action include the GlcNAc branching enzymes MGAT3, MGAT4, and MGAT5, B4GALT1 (for addition of a β1,4Gal residue) and FUT8 (adds a core α1,6Fuc) (Fig. 1 and SI Appendix, Fig. S1) (4). Several of these enzymes compete for the same GlcNAcMan5GlcNAc2-Asn substrate (SI Appendix, Fig. S1), and often the product of one reaction will preclude the action of other enzymes (4).
Fig. 1.
Processing of N-glycans from Man9GlcNAc2 to complex-type structures includes GlcNAc addition by MGAT1, Man trimming by MAN2A1, and GlcNAc addition by MGAT2. The recognition arm bound by MAN2A1 and MGAT2 is shown by the red outline (SI Appendix, Fig. S1).
Previous studies suggest that the GlcNAc-β1,2Man-α1,3Manβ arm functions as a common binding site (“recognition arm”) for MAN2A1, MGAT2, MGAT3, MGAT4, and FUT8, even though none of these enzymes except MGAT4 act directly on this arm (4). The crystal structure of Drosophila Golgi α-mannosidase II (MAN2A1) in complex with its GlcNAcMan5GlcNAc substrate provided the first and only structural evidence showing the role of the recognition arm in substrate specificity (5). MAN2A1 uses two distinct substrate-binding subsites: a catalytic subsite where α-Man bond cleavage occurs and an adjoining recognition arm-binding exosite that anchors the substrate. However, nothing is known about how the recognition arm provides substrate specificity for the glycosyltransferases (GTs).
MGAT2 represents the committed step in the synthesis of complex-type N-glycan structures (Fig. 1). This enzyme extends a GlcNAc-β1,2- linkage on the Man-α1,6Manβ- arm of the trimannosyl N-glycan core. MGAT2 has been characterized from mammalian, plant, and insect sources (6) and employs a UDP-GlcNAc donor in a Mn2+-dependent inverting catalytic mechanism (7). Detailed kinetic analysis and inhibitor studies have been performed (7), including an extensive mapping of glycan determinants on the acceptor structure important for substrate specificity (8). These data confirm that both the Man-α1,6Manβ- acceptor arm and an unmodified GlcNAc-β1,2Man-α1,3Manβ- recognition arm are required for MGAT2 action, but the structural basis for this restricted substrate specificity remains unknown.
Here we have determined the structural basis for substrate recognition by MGAT2. Using a recently developed platform for recombinant expression of human glycosylation enzymes in mammalian cells (3), we generated a secreted form of the human MGAT2 catalytic domain. The structures of a bound donor analog, Mn2+-UDP, and a MGAT2:GlcNAcMan3GlcNAc2-Asn acceptor complex revealed the structural basis of substrate specificity, including the convergent evolution of an exosite pocket that binds the acceptor arm similarly to the exosite observed in MAN2A1. These data provide a framework for understanding the restricted substrate specificity of MGAT2 and related GTs and highlight the use of modular template subsites for sugar donor and acceptor interactions in combination with adjacent catalytic subsites to produce glycosidic linkages. It is the combination of analogous modular subsites in GTs that defines the regiospecific synthesis of glycan structures in mammalian cells.
Results
Protein Production and Crystallization of MGAT2.
An MGAT2 catalytic domain expression construct (residues 29–447) was generated by replacement of the NH2-terminal membrane anchor with a fusion peptide cassette to target the secretion of the recombinant fusion protein product in mammalian cells (3). Expression in transiently transfected HEK293S (GnTI-) cells was followed by Ni2+-NTA chromatography, concurrent cleavage of fusion tags, and trimming of glycan structures to a single GlcNAc residue and further purification (SI Appendix, Fig. S2 B and C). The resulting enzyme preparation had kinetic constants for GlcNAc transfer that were similar to those of the intact fusion protein (SI Appendix, Table S1).
Structural Features of Human MGAT2.
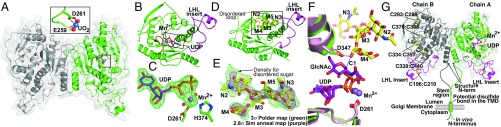
A UO2 derivative of the catalytic domain of MGAT2 (MGAT2:UO2) was crystallized and solved at 2.0 Å resolution using single-wavelength anomalous diffraction (SI Appendix, Table S2). The structure revealed two molecules in the asymmetric unit, with the NH2 terminus of each chain (residues 29–83) and a loop (residues 376–387 in chain A and residues 376–391 in chain B) being disordered. There are extensive interactions between the two peptide chains (Fig. 2A), but size exclusion-multiangle light scattering (SEC-MALS) revealed a monomeric protein in solution (SI Appendix, Fig. S2C). Chains A and B superimpose 351 Cα atoms with an rmsd of 0.73 Å, with the largest conformational difference between the chains being a 7.3° rotation of residues 181–224 in a loop-helix-loop segment (LHL181–224), which makes unique crystal contacts in each chain (SI Appendix, Fig. S3C). There is good electron density for the predicted N89 glycan site in chain A (SI Appendix, Fig. S3A), but the corresponding residue in chain B is exposed to solvent and is disordered.
Fig. 2.
The MGAT2 structure, substrate binding, proposed mechanism, and proposed transmembrane structure. (A) The MGAT2:UO2 asymmetric unit (monomers green and gray). (Inset) The interactions between DxD motif residues (green sticks) and the bound UO2 (U and O atoms are blue and red spheres, respectively). (B) MGAT2:UDP structure (Mn2+, purple sphere; UDP, purple sticks) with LHL insert (magenta) and residues ordered following Mn2+ binding (Loop 375–387, orange). Box indicates the region for the difference map in C. (C) A 1.6-Å difference density map (Fo − Fc) of the donor analog contoured at 3σ before modeling UDP and Mn2+. (D) The MGAT2:Acc complex with acceptor bound (yellow sticks) and the LHL181–224 loop (magenta) indicated. Box indicates the region for the difference map shown in E. (E) A 2.8-Å difference density map (Fo − Fc) contoured at 3σ (purple) was calculated after omitting acceptor (yellow sticks) and subjecting the model to simulated annealing. An unbiased Polder map (24) contoured at 3σ (green) illustrates weak density that would otherwise be obscured by bulk solvent (SI Appendix, SI Materials and Methods). (F) UDP-GlcNAc modeled in MGAT2:UDP (green) using the MGAT1:UDP-GlcNAc complex (pink, Protein Data Bank ID code 1FOA) (12). Ligands are colored as in B–E. Proposed mechanism involves the catalytic base D347 deprotonating the O2 hydroxyl of M4, which attacks the C1 of the UDP-GlcNAc donor (blue dashed lines). (G) The proposed membrane-bound form of the MGAT2:UDP dimer is shown. UDP (purple sticks), Mn2+ (slate spheres), disulfide bonds (yellow spheres), LHL181–224 loop (magenta), and diagrammatic representation of the “stem region” (green and gray lines) and NH2-terminal transmembrane anchor for the full-length enzyme found in vivo.
The overall fold of human MGAT2 consists of an eight-stranded twisted β-sheet with 12 α-helical segments resembling a GT-A Rossmann-like fold (9) (Fig. 2 A and B). Many GT-A fold enzymes require a divalent cation (usually Mn2+) coordinated by a DxD motif to interact with the phosphodiester of the sugar nucleotide donor (9). In MGAT2:UO2, the DxD motif is composed of residues EED259–261 with E259 and D261 coordinated with the UO2 molecule (Fig. 2A).
The Crystal Structure of UDP Bound MGAT2.
We solved the crystal structure of a Mn2+:UDP complex (MGAT2:UDP) to a resolution of 1.6 Å, revealing two molecules in the asymmetric unit (SI Appendix, Table S2). The Mn2+-UDP complex is bound in the active site of chain A, but only Mn2+ is bound in chain B (Fig. 2G). Due to differences in the crystal packing of residues 294–320 in chain B, E316 is positioned to prevent the UDP from binding and coordinating with the metal ion (SI Appendix, Fig. S3B). Therefore, all analyses with respect to the UDP in the MGAT2 active site were confined to chain A.
The MGAT2:UDP and MGAT:UO2 structures superimpose 339 Cα atoms with an rmsd of 0.7 Å. The largest difference is a 12.5° rotation of the LHL181–224 segment due to the unique crystal contacts made by this segment in the two chains (SI Appendix, Fig. S3C). The other difference between the two structures involves the ordering of Loop375–387 in both chains of MGAT2:UDP (Fig. 2B), which reveals an additional disulfide bridge (Cys378:Cys386, SI Appendix, Fig. S3C). Loop375–387 contributes H374 to the octahedral coordination sphere of the Mn2+, which is also coordinated by the oxygen atoms of the α- and β-phosphates of the UDP, Oδ2 of D261 in the DxD motif, and two water molecules (Fig. 2C). Ordering of active-site loop regions upon binding of a sugar nucleotide donor has been observed in other GT structures (9, 10). However, Loop375–387 is also ordered in chain B, which does not contain a bound UDP. While it is possible that the coordination of the Mn2+ ion by H374 facilitates the folding of the Loop375–387, the loop in chain B is also in a crystal contact, which will favor an ordered conformation.
The Crystal Structure of Acceptor Bound MGAT2.
We solved the structure of MGAT2 in complex with the glycan acceptor GlcNAcMan5GlcNAc2-Asn (MGAT2:Acc) at 2.8 Å resolution (Fig. 2 D and E and SI Appendix, Table S2). There is clear electron density for the terminal GlcNAc-β1,2- residue on the recognition arm and weaker, but interpretable, density for four additional residues of the glycan acceptor in both chains of the MGAT2:Acc complex (Fig. 2E). The remaining GlcNAc at the reducing end of the acceptor is solvent-exposed and disordered. The terminal Man-α1,6- residue of the acceptor extends into the active site where the nucleophilic O2 hydroxyl donates a hydrogen bond to D347 (Fig. 2F and SI Appendix, Fig. S3E). The remainder of the acceptor adopts an extended conformation across the surface of the enzyme, which positions the terminal GlcNAc-β1,2- residue of the recognition arm in an exosite pocket enclosed by LHL181–224 (Fig. 2D). F219, F227, and Y344 bracket the sides of the exosite to form a complementary surface that packs against the GlcNAc residue (SI Appendix, Fig. S3E). Hydrogen bonds are formed between the terminal GlcNAc residue and N428, H221, D217, G430, R198, and E224. The core β-Man forms H-bonds with Y344 and N345 (SI Appendix, Fig. S3E). A comparison of the MGAT2:UDP and MGAT2:Acc (chain B) structures shows that acceptor binding induces an 11.9° rotation of LHL181–224 to form the exosite pocket (SI Appendix, Fig. S3C). Kinetic analysis of mutants in the exosite residues ranged from minor reductions in kcat/Km for the donor and acceptor (26- to 30-fold for R198A), to intermediate effects (2,030- to 3,860-fold for Y344A), to profound reduction in catalysis (4,480- to 105-fold for D217A) (SI Appendix, Fig. S4B and Table S1).
The MGAT2:UO2, MGAT2:UDP, and MGAT2:Acc crystal structures are uniquely packed into different space groups (SI Appendix, Table S2), but all reveal the same dimer. In MGAT2:UO2, the dimer represents the asymmetric unit (Fig. 2A), while both MGAT2:UDP and MGAT2:Acc dimers are formed following the application of the appropriate crystallographic symmetry operators (Fig. 2G). The SEC-MALS analysis (SI Appendix, Fig. S2C) shows a monomer in solution, which suggests that the dimer observed in the crystal structure is weakly associated, and was stabilized by the highly crowded conditions encountered during crystallization. In fact, bimolecular fluorescence complementation studies have previously indicated that full-length, transmembrane-tethered MGAT2 forms a homodimer in vivo (11). It is not unreasonable to assume that MGAT2 may dimerize in the crowded environment of the cell, especially when tethered to the Golgi membrane, which will reduce the entropic cost of dimerization (Fig. 2G).
Modeling of the Donor Sugar in MGAT2.
The structure of an intact UDP-GlcNAc donor complex could not be obtained for MGAT2, presumably because of hydrolysis of the sugar donor. In contrast, intact UDP-GlcNAc donor and donor analog complexes were solved for MGAT1 (12). MGAT2 and MGAT1 superimpose 737 equivalent atoms with an rmsd of 1.5 Å (Fig. 2F). We used this superposition to model the UDP-GlcNAc donor in MGAT2. Without any additional optimization, the resulting model shows that GlcNAc C1 is reasonably positioned for an in-line nucleophilic substitution by the O2 hydroxyl of the Man-α1,6- acceptor residue. D347 is positioned to act as the catalytic base for deprotonating the nucleophilic hydroxyl, consistent with the predicted inverting catalytic mechanism (Fig. 2F). The model also suggests that the position of the GlcNAc residue in the active site is stabilized by H-bonding interactions with the side chains of W346 and E259 analogous to similar interactions in MGAT1. Additional H-bond interactions are predicted between N318 and Y294 and the GlcNAc acetyl group of the donor.
The resulting model is consistent with kinetic analysis of alanine mutants for substrate interacting residues (SI Appendix, Fig. S4B and Table S1). First, mutation of the proposed catalytic base, D347A, resulted in a complete loss of enzyme activity. The residues predicted to form H-bonds with O3 and O4 of the donor (E259A and W346A) also resulted in a profound reduction in donor kcat/Km. Finally, the Y294A and N318A substitutions are expected to disrupt interactions with the N-acetyl group and resulted in relatively modest reductions (11- to 37-fold) in kcat/Km.
Convergent Acceptor Exosites for MGAT2 and MAN2A1.
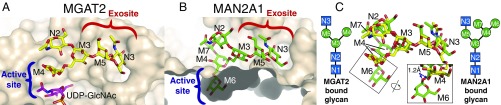
The interactions and conformation of the GlcNAc-β1,2Man-α1,3Manβ- recognition arm within the MGAT2 exosite (Fig. 3A) are reminiscent of those observed in MAN2A1 (Fig. 3B). This similarity is striking since these two proteins have different structural folds and catalyze distinct reactions. Both enzymes bind the recognition arm using a broad exosite surface that extends away from the active site and forms a pocket specific for the terminal GlcNAc residue (Fig. 3 A and B). Both exosites reveal a similar contact surface for binding the recognition arms in almost the same conformations. In addition to the surface complementarity, the binding specificity for the recognition arms in both enzymes is accomplished using similarly positioned side chains to satisfy the H-bonding requirements (MGAT2 Y344, F219, and E224 are analogous to MAN2A1 Y267, W299, and Q64). The exosite binding of the recognition arms also results in a similar positioning of the substrates with respect to the catalytic residues in both enzymes, despite the fact that the active sites and the reactions catalyzed are entirely distinct. Specifically, a superposition of the recognition arms positions the O2 hydroxyl of the Man-α1,6- acceptor in the MGAT2:Acc structure ∼1.2 Å from the equivalent glycosidic oxygen in the Man-α1,6-Man- linkage (the linkage cleaved by MAN2A1) in the MAN2A1:substrate complex (Fig. 3C, Insets). These observations are strong evidence that MGAT2 and MAN2A1 have converged upon similar specificity mechanisms based on exosite surface complementarity to the recognition arm and explain the importance of the terminal GlcNAc residue in the maturation of complex glycan structures.
Fig. 3.
Comparison of bound substrates for MGAT2 and MAN2A1. The structures of (A) MGAT2:GlcNAcMan3GlcNAc (acceptor in yellow sticks with purple sticks for modeled UDP-GlcNAc) and (B) MAN2A1:Man5GlcNAc (substrate in green sticks) are shown in tan surface representation. Exosite (red bracket) and active-site (blue bracket) positions are indicated in each structure. (C) An overlay of the two substrate structures is shown along with cartoon representations of the respective bound glycans with coloring as shown in Fig. 1. Alignment was based on the M5 residue in each respective structure. The boxes indicate similar regions of the aligned substrates differing by a rotation of view. The O2 of the M4 residue in MGAT2:Acc (the position of GlcNAc transfer) and the glycosidic oxygen of the M4-M6 linkage (the linkage cleaved by MAN2A1) for the MAN2A1:substrate complex are separated by only ∼1.2 Å.
Structural Comparison with Other GTs.
In most metazoans, MGAT2 is the sole member of CAZy family GT16 (13). However, a query of the Dali protein structure comparison server (14) revealed strong similarities to numerous other GT-A fold inverting and retaining GTs, including GT2, GT13, GT27, GT62, and GT78 enzymes from mammals, fungi, and bacteria, each displaying varied sugar donor and/or acceptor specificities (SI Appendix, Table S3). The most similar enzymes are the GT13 GlcNAc transferases, MGAT1 and POMGNT1, which synthesize the same GlcNAc-β1,2Man- linkages as MGAT2 using distinct acceptor substrates; MGAT1 employs a Man-α1,3Manβ- acceptor and POMGNT1 transfers to a Man-αThr/Ser-peptide acceptor. The most significant differences in structure between POMGNT1 and MGAT1 versus MGAT2 involve insertions and extensions to the core GT-A fold as well as differences in loop sequences that link the GT-A fold elements. MGAT2 has a large insertion (LHL181–224) that is not conserved in the two GT13 enzymes (Fig. 2 B and D and SI Appendix, Fig. S5 A and D). The COOH-terminal extensions for the enzymes are also quite distinct. MGAT2 residues 377–447 form extended interactions that wrap around the circumference of the globular catalytic domain (SI Appendix, Fig. S5A). In contrast, the COOH-terminal extensions in POMGNT1 (residues 503–647) and MGAT1 (residues 316–447) form an antiparallel β-sheet domain structure with additional α-helical segments that are positioned on the opposite side of the active site relative to the LHL181–224 insertion in MGAT2 (SI Appendix, Fig. S5A).
Modular Architecture for GT Substrate Interactions.
The different donor and acceptor specificities among the GT-A fold enzymes suggests that modular active-site architecture has been employed to evolve expanded catalytic diversity among the GTs (Fig. 4D). The donor template module binds the sugar donor using side chains associated with the GT-A fold. Despite low sequence identities with MGAT2 (18 and 17% for rabbit MGAT1 and human POMGNT1, respectively; SI Appendix, Fig. S5D), the GT-A fold in all three enzymes is highly conserved (rmsds of 1.5 Å for 737 Cα atoms in MGAT1 and 1.3 Å for 793 Cα atoms for POMGNT1 versus MGAT2, respectively; SI Appendix, Fig. S5A) and employs identical or similar amino acids for interacting with the UDP portion of the donor (SI Appendix, Fig. S3D). All three enzymes employ an EED sequence for the DxD motif, a conserved Arg residue forming a salt bridge with the α-phosphate of the sugar nucleotide and a His, Glu, and peptide bond carbonyl that forms H-bonds with the nucleotide and ribose. MGAT2 forms additional H-bond interactions with the uracil substituents (SI Appendix, Fig. S3D). The uracil C5 position in MGAT2 also faces toward the solvent in the binding site, explaining why the enzyme can be purified using an immobilized 5-HgUDP-GlcNAc resin (6). A comparison of the MGAT2:Acc complex with a related α1,6-mannosyltransferase, Mnn9 (15), a retaining GT-A fold enzyme that employs GDP-Man as donor (SI Appendix, Table S3), indicates a similar positioning of the nucleotide relative to the GT-A fold core, but entirely different interactions with the nucleotide diphosphate (SI Appendix, Fig. S5C). These data demonstrate that interacting residues within the donor template module confer unique donor-binding specificities independent of the context of the underlying conserved GT-A scaffold (Fig. 4D and SI Appendix, Table S3).
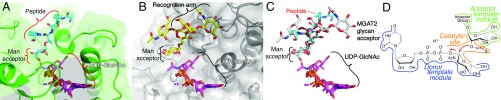
Fig. 4.
Comparison of the MGAT2 and POMGNT1 acceptor complex structures. (A) The POMGNTI:UDP:glycopeptide acceptor complex (Protein Data Bank ID code 5GGI) (25), depicted as a green surface with the glycopeptide as cyan sticks and the (B) MGAT2:acceptor and MGAT2:UDP complexes (gray surface, acceptor as yellow sticks) were aligned. Peptide and Man acceptor residue (POMGNT1), recognition arm and Man acceptor residue (MGAT2), bound UDP and modeled UDP-GlcNAc (purple sticks, Mn2+ as small magenta spheres) are indicated by brackets. (C) Overlay of the acceptor and donor structures from A and B. (D) Generalized model for GT active-site modules, where the “Donor template module” facilitates selective sugar-nucleotide donor binding, the “Acceptor template module” selectively binds the acceptor to appropriately position the hydroxyl nucleophile, and the “Catalytic site” facilitates base-catalyzed nucleophilic attack by the acceptor to the C1 of the sugar donor, in this case using an inverting catalytic mechanism.
The structurally distinct acceptor template modules for MGAT2, MGAT1, and POMGNT1 reflect the diversity of linkages synthesized by the enzymes. There are no overlaps in positions of acceptor-binding residues between the three enzymes and no similarities in acceptor interactions between the MGAT2 and POMGNT1 acceptor complexes (Fig. 4 A and B) other than the position of the Man residue that acts as the nucleophile in the GT reaction. Much of the acceptor template module is built from insertions and extensions to the core GT-A fold. For example, the exosite in MGAT2 is formed partly by a large insertion (LHL181–224) that is not conserved with the other two GT13 enzymes (SI Appendix, Fig. S5A). In contrast, POMGNT1 binds a Man-αThr-peptide acceptor in a cleft in the active site largely through H-bonding with the Man residue in the Man-αThr-peptide (D476, R480, and R605 with Man O2, O3, and O3, respectively; Fig. 4B). The R605 residue is contributed from the COOH-terminal globular extension that is unique to POMGNT1 and MGAT1, but distinct from MGAT2. The equivalent cleft position in MGAT2 is blocked by the COOH-terminal peptide extension (residues 424–429) that wraps around the globular catalytic domain (SI Appendix, Fig. S5B). This COOH-terminal extension forms the base of the exosite surface in MGAT2. Significant differences in proposed acceptor-binding site structures also exist between MGAT2 and MGAT1, but details of acceptor interactions for the latter enzyme have not yet been determined. Thus, each enzyme has evolved independent approaches for presentation of the appropriate acceptor hydroxyl group for catalysis based on elements inserted into the GT-A fold.
The donor and acceptor modules of MGAT2 position the C1 of the donor sugar for nucleophilic attack by the deprotonated acceptor hydroxyl in the catalytic site (Fig. 2F). Conserved interactions are predicted for positioning the GlcNAc donor in MGAT1, MGAT2, and POMGNT1, including identically positioned Glu (E259 in MGAT2) and Trp (W346 in MGAT2) residues interacting with the GlcNAc O3 and O4 hydroxyls, respectively. The catalytic base (D347 in MGAT2) is also identically positioned relative to the Man O2 hydroxyl acceptor in all three enzymes (Fig. 2F). It is striking that the acceptor O2 hydroxyl is identically positioned for nucleophilic attack in both the MGAT2:Acc and POMGNT2:Acc complexes, despite the fact that all other interactions with the respective acceptors are distinct. Thus, the catalytic site for MGAT2 specifies the inverting catalytic mechanism by positioning of the donor GlcNAc C1 relative to the Man O2 nucleophile and the Asp catalytic base, with minimal conservation in how the positioning of the remainder of the acceptor is achieved.
Disease-Causing Mutations in MGAT2.
A human deficiency in MGAT2 leads to carbohydrate-deficient glycoprotein syndrome type IIa (CDG IIa) characterized by facial dysmorphy, ventricular septal defects, and severely retarded psychomotor development (16). To date, five MGAT2 mutations have been identified in CDG IIa patients, all residing within the catalytic domain (H262R, S290F, N318D, C339ter, and K237N) (17–19). Four of the mutations (H262R, S290F, N318D, C339ter) exhibit significantly reduced (compound heterozygote N318D:C339ter) (17) or a complete absence of enzyme activity (H262R, S290F) (16, 19). Three of the residues are nonconserved (H262, N318, and K237), and N318 and K237 H-bond directly or indirectly with the UDP-GlcNAc donor (SI Appendix, Fig. S6 C and D). The S290 is a conserved residue that stabilizes the core of the GT-A fold through H-bonds to peptide-bond backbone residues (SI Appendix, Fig. S6B). The H262R, S290F, and K237N mutations would break these critical hydrogen bonds and introduce steric clashes to destabilize the protein (SI Appendix, Fig. S6 A, B, and D, respectively). The isosteric N318D mutation (SI Appendix, Fig. S6C) resulted in reduced activity (17) similar to the 30- to 37-fold reduction in kcat/Km for the N318A mutation (SI Appendix, Fig. S4B and Table S1), presumably through charge repulsion and destabilization of its H-bonding to the proposed catalytic base, D347. The C339ter mutation deletes a major portion of the GT-A fold, including H374 that coordinates the divalent cation, and would likely lead to a severely destabilized protein.
Discussion
N-glycan maturation is catalyzed by a collection of enzymes that have exceptionally restricted substrate specificities for glycan trimming, branching, and extension. Seminal studies, predominantly in Harry Schachter’s laboratory, initially characterized MGAT1 and MGAT2 (4, 6, 7), including the detailed kinetic analysis that probed determinants and restrictions for sugar donor and acceptor structures (8, 20). Those studies established many of the enzymatic “rules” for ordered N-glycan modifications (4, 21) and created a framework for understanding the hierarchal steps in glycan maturation within the secretory pathway. These studies demonstrated that MGAT1 modification of the Man5GlcNAc2-Asn intermediate produced the key GlcNAc-β1,2Man-α1,3Manβ- recognition arm required for substrate binding by numerous subsequent glycan modification enzymes (SI Appendix, Fig. S1).
Convergent Acceptor Interactions.
The role of the GlcNAc-β1,2Man-α1,3Manβ- recognition arm in substrate binding and catalysis was first observed in the Drosophila MAN2A1:substrate complex (5). That structure showed how the enzyme achieved its precise substrate specificity by using an exosite to select for glycans containing the recognition arm. Here, we have shown that MGAT2 evolution has converged upon an analogous exosite to achieve recognition arm specificity. In both enzymes, the terminal GlcNAc residue added by MGAT1 sits at the base of an exosite pocket, which provides a significant portion of the enzyme:substrate-binding energy. The similarities in exosite interactions with the recognition arm in both enzyme:substrate complexes are striking because the two enzymes have completely distinct protein folds and catalytic mechanisms (GT versus glycoside hydrolase). Thus, the MGAT2 and MAN2A1 exosite pockets are formed using structural elements inserted into the unrelated folds to produce surfaces that are complementary to the same recognition arm. The convergence to a shared exosite topology also places the terminal sugar substrate in a similar position in the respective active sites, despite the distinct catalytic mechanisms. These data explain the critical role that the recognition arm plays in substrate selection and suggest that other N-glycan–processing enzymes may utilize this glycan structure to tether similar substrates in their active sites.
Structural Basis for Restricted Substrate Specificity.
While details of the acceptor and donor requirements for MGAT2 were established several decades ago (7, 8), the structural basis for those requirements can now be explained within the framework of the MGAT2 structure (SI Appendix, Fig. S4A). MGAT2 acts on the GlcNAcMan3GlcNAc2-Asn substrate only after MGAT1 and MAN2A1 action largely because these latter enzymes complete the recognition arm and unmodified acceptor arm that are critical determinants for interactions with the MGAT2 active site. The exosite pocket encloses the terminal GlcNAc residue of the recognition arm, and additional tight interactions with the remainder of the recognition arm explain the inability of the enzyme to act on substrates extended by either B4GALT1 or MGAT3 (SI Appendix, Fig. S4A). Fine details of acceptor requirements based on deoxy-sugar or chemically extended glycan structures (8) are also in close agreement with the steric restrictions or solvent exposure of the bound acceptor complex. Similar observations are also found for details of the bound donor sugar nucleotide (7). In addition, kinetic studies of active-site mutants confirm the model for donor and acceptor interactions and provide a structural explanation for restricted substrate specificity (SI Appendix, Fig. S4B).
Modular Assembly of GT Active Sites.
Glycan structures are not directly derived from genomic templates; instead, they are encoded by the template geometries and activities of the respective GT active-site structures. The regiospecificities of the GT reactions are achieved by a combination of three modular structural features: (i) a donor template module with selectivity for the type of sugar donor; (ii) the catalytic site to provide anomeric specificity for sugar transfer based on an inverting or retaining catalytic mechanism; and (iii) an acceptor template module that positions the acceptor sugar containing the nucleophilic hydroxyl within the active site (Fig. 4D). For MGAT2, the conserved GT-A fold scaffold creates a Mn2+-dependent UDP-GlcNAc binding site as the donor template module. The catalytic site supports the inverting catalytic mechanism to produce an exclusive β1,2GlcNAc anomer product that is conserved among several other CAZy GT families. The key difference between members of the closely related CAZy GT families concerns the specificity for binding distinct acceptor substrates. In MGAT2, acceptor substrate specificity is dictated by an acceptor template module that uses an exosite to bind the glycan recognition arm and appropriately position the acceptor arm for sugar transfer. The similarity of this acceptor template module to the exosite observed in MAN2A1 suggests that evolution has converged upon a common solution for acceptor recognition. The modular structure that we describe also provides a framework for understanding how the specificity of other GTs can be reshaped through the intrinsic and extrinsic selective pressures of evolution (22). Both the donor template module and the catalytic site are built from side chains originating from the conserved GT-A fold. In this context, donor and reaction specificity can be achieved through simple mutations within a common scaffold. In contrast, the acceptor template modules are formed from structural elements inserted into the GT-A fold. This suggests a mechanism for how the broad diversity of glycan structures can evolve. Protein evolution is subject to both folding and functional constraints (23). Because the acceptor template module is formed from elements inserted into the stable GT-A scaffold, the evolution of acceptor specificity is decoupled from the folding constraint and can evolve more rapidly. This model suggests that it is the independent changes within the donor and acceptor template modules and catalytic site that have led to the expanded diversification of glycan structures found on cell-surface and secreted glycoproteins and glycolipids in mammalian cells.
Materials and Methods
SI Appendix, SI Materials and Methods, provides a detailed discussion of the materials and methods used in this study.
Protein Expression and Purification.
Wild-type or site-directed mutants of human MGAT2 were expressed in HEK293 cells and purified as described in SI Appendix, SI Materials and Methods.
Generation of Mutants.
Site-directed mutagenesis was performed using the Q5 site-directed mutagenesis kit in the pGEn2-MGAT2 expression vector as described in SI Appendix, SI Materials and Methods.
Substrate/Ligand Preparation.
The GlcNAcMan3GlcNAc2-Asn substrate analog was generated by enzymatic modification of purified egg yolk sialoglycopeptide and purified as described in SI Appendix, SI Materials and Methods.
Crystallization and X-Ray Diffraction.
Crystals of MGAT2 were generated from purified enzyme preparations and premixed with either Mn2+:UDP or GlcNAcMan3GlcNAc2-Asn substrate analogs as described in SI Appendix, SI Materials and Methods. Crystal growth using hanging-drop vapor diffusion, diffraction data collection and processing, and structure solution by single-wavelength anomalous diffraction or molecular replacement are described in SI Appendix, SI Materials and Methods.
Enzyme Assays and Time Course of Glycan Digestion.
Enzyme assays were performed using GlcNAcMan3GlcNAc2 as substrate, and enzymatic products were detected using the UDP-Glo assay as described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Bobby Ng and Hudson Freeze for providing information on CDG IIa patient mutations. This research was supported by NIH Grants P41GM103390 and P01GM107012 (to K.W.M., G.-J.B., and Z.A.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.C. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5VCM, 5VCR, and 5VCS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716988115/-/DCSupplemental.
References
- 1.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moremen KW, et al. Expression system for structural and functional studies of human glycosylation enzymes. Nat Chem Biol. 2018;14:156–162. doi: 10.1038/nchembio.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schachter H. The ‘yellow brick road’ to branched complex N-glycans. Glycobiology. 1991;1:453–461. doi: 10.1093/glycob/1.5.453. [DOI] [PubMed] [Google Scholar]
- 5.Shah N, Kuntz DA, Rose DR. Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site. Proc Natl Acad Sci USA. 2008;105:9570–9575. doi: 10.1073/pnas.0802206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendiak B. Mannosyl (α-1,6-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase (MGAT2) In: Taniguchi N, et al., editors. Handbook of Glycosyltransferases and Related Genes. 2nd Ed Springer; Tokyo: 2014. [Google Scholar]
- 7.Bendiak B, Schachter H. Control of glycoprotein synthesis. Kinetic mechanism, substrate specificity, and inhibition characteristics of UDP-N-acetylglucosamine:α-D-mannoside β 1-2 N-acetylglucosaminyltransferase II from rat liver. J Biol Chem. 1987;262:5784–5790. [PubMed] [Google Scholar]
- 8.Reck F, et al. Synthetic substrate analogues for UDP-GlcNAc: Man α 1-6R β(1-2)-N-acetylglucosaminyltransferase II. Substrate specificity and inhibitors for the enzyme. Glycoconj J. 1994;11:210–216. doi: 10.1007/BF00731220. [DOI] [PubMed] [Google Scholar]
- 9.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 10.Ramakrishnan B, Qasba PK. Crystal structure of lactose synthase reveals a large conformational change in its catalytic component, the β1,4-galactosyltransferase-I. J Mol Biol. 2001;310:205–218. doi: 10.1006/jmbi.2001.4757. [DOI] [PubMed] [Google Scholar]
- 11.Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J Biol Chem. 2010;285:17771–17777. doi: 10.1074/jbc.M110.103184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unligil UM, et al. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: Catalytic mechanism and a new protein superfamily. EMBO J. 2000;19:5269–5280. doi: 10.1093/emboj/19.20.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 14.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Striebeck A, Robinson DA, Schüttelkopf AW, van Aalten DM. Yeast Mnn9 is both a priming glycosyltransferase and an allosteric activator of mannan biosynthesis. Open Biol. 2013;3:130022. doi: 10.1098/rsob.130022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schachter H, Jaeken J. Carbohydrate-deficient glycoprotein syndrome type II. Biochim Biophys Acta. 1999;1455:179–192. doi: 10.1016/s0925-4439(99)00054-x. [DOI] [PubMed] [Google Scholar]
- 17.Alazami AM, et al. Congenital disorder of glycosylation IIa: The trouble with diagnosing a dysmorphic inborn error of metabolism. Am J Med Genet A. 2012;158A:245–246. doi: 10.1002/ajmg.a.34347. [DOI] [PubMed] [Google Scholar]
- 18.Cormier-Daire V, et al. Congenital disorders of glycosylation IIa cause growth retardation, mental retardation, and facial dysmorphism. J Med Genet. 2000;37:875–877. doi: 10.1136/jmg.37.11.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan J, Dunn J, Jaeken J, Schachter H. Mutations in the MGAT2 gene controlling complex N-glycan synthesis cause carbohydrate-deficient glycoprotein syndrome type II, an autosomal recessive disease with defective brain development. Am J Hum Genet. 1996;59:810–817. [PMC free article] [PubMed] [Google Scholar]
- 20.Reck F, et al. Synthetic substrate analogues for UDP-GlcNAc: Man α 1-3R β 1-2-N-acetylglucosaminyltransferase I. Substrate specificity and inhibitors for the enzyme. Glycoconj J. 1995;12:747–754. doi: 10.1007/BF00731234. [DOI] [PubMed] [Google Scholar]
- 21.Brockhausen I, Narasimhan S, Schachter H. The biosynthesis of highly branched N-glycans: Studies on the sequential pathway and functional role of N-acetylglucosaminyltransferases I, II, III, IV, V and VI. Biochimie. 1988;70:1521–1533. doi: 10.1016/0300-9084(88)90289-1. [DOI] [PubMed] [Google Scholar]
- 22.Varki A. Evolutionary forces shaping the Golgi glycosylation machinery: Why cell surface glycans are universal to living cells. Cold Spring Harb Perspect Biol. 2011;3:a005462. doi: 10.1101/cshperspect.a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebschner D, et al. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr D Struct Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuwabara N, et al. Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of α-dystroglycan. Proc Natl Acad Sci USA. 2016;113:9280–9285. doi: 10.1073/pnas.1525545113. [DOI] [PMC free article] [PubMed] [Google Scholar]