Significance
An LTR retrotransposon, Steamer, was previously identified by virtue of high expression and dramatic amplification in a transmissible cancer in soft-shell clams (Mya arenaria). Here, we investigated genome sequences obtained from both physical collections of bivalves and genome databases and found evidence of horizontal transfer of Steamer-like transposons from one species to another, with jumps between bivalves and even between animals of completely different phyla. Some events were ancient, but some (in particular, those between bivalves) appear to be recent, as the elements are nearly identical in different species. These data show that horizontal transfer of LTR retrotransposons like Steamer has occurred and continues to occur frequently and that the marine environment may be particularly suitable for transfer of transposons.
Keywords: retrotransposons, transposable elements, horizontal transfer, bivalves, cross-phyla horizontal transfer
Abstract
The LTR retrotransposon Steamer is a selfish endogenous element in the soft-shell clam genome that was first detected because of its dramatic amplification in bivalve transmissible neoplasia afflicting the species. We amplified and sequenced related retrotransposons from the genomic DNA of many other bivalve species, finding evidence of horizontal transfer of retrotransposons from the genome of one species to another. First, the phylogenetic tree of the Steamer-like elements from 19 bivalve species is markedly discordant with host phylogeny, suggesting frequent cross-species transfer throughout bivalve evolution. Second, sequences nearly identical to Steamer were identified in the genomes of Atlantic razor clams and Baltic clams, indicating recent transfer. Finally, a search of the National Center for Biotechnology Information sequence database revealed that Steamer-like elements are present in the genomes of completely unrelated organisms, including zebrafish, sea urchin, acorn worms, and coral. Phylogenetic incongruity, a patchy distribution, and a higher similarity than would be expected by vertical inheritance all provide evidence for multiple long-distance cross-phyla horizontal transfer events. These data suggest that over both short- and long-term evolutionary timescales, Steamer-like retrotransposons, much like retroviruses, can move between organisms and integrate new copies into new host genomes.
Transposable elements (TEs) are selfish genetic elements that generate new copies of themselves within the genomes of their host cells and are inherited vertically during replication of their hosts. TEs include both the DNA transposons, which usually replicate through cut-and-paste mechanisms, and the retrotransposons, which replicate through reverse transcription of an RNA transcribed from a DNA copy resident in the host genome (1). These TEs most often increase their copy number by intracellular retrotransposition, leading to new insertions into the genome of the cell they inhabit. In somatic cells, these events can cause mutations and lead to cancers, and, if they occur in germ cells or progenitors of germ cells, can result in increased copy number in the germ-line genome of the host species (2, 3). The gag and pol genes of LTR retrotransposons are related to those of retroviruses (4), and it appears very likely that the vertebrate retrovirus lineage itself arose from an ancestral LTR retrotransposon which acquired an envelope gene (5). Despite this evolutionary relationship with retroviruses, LTR retrotransposons and other TEs are not expected to transmit easily from cell to cell or from individual to individual. It is even harder to understand how these elements can be transmitted from the germ line of one species to another. To do this, the TE must be released from a cell in one individual and then transported into and integrated into the germ line of a different organism. While this is a rare event compared with intracellular transposition, with multiple barriers, there are numerous reports of horizontal transfer of TEs (HTT) from one organism to another (6–10).
We previously identified an LTR retrotransposon, Steamer, in the genomes of soft-shell clams (Mya arenaria) that was highly amplified in the transmissible cancer of that species (2–10 copies per haploid genome in normal individuals vs. 100–300 copies in neoplastic cells) (11, 12). Steamer is a 4,968-bp retrotransposon in the Mag family of the Ty3 lineage of LTR retrotransposons, with a single 1,335-aa gag-pol ORF. As with all other Mag elements, it has no detectible env gene. The dramatically expanded copy number of Steamer in the clonal cancer line may be responsible for the oncogenic phenotype or may contribute to the continued evolution of this contagious cancer lineage. We also identified sequences of retroelements related to Steamer in several bivalves susceptible to bivalve transmissible neoplasia, but those specific retroelements were not amplified in the neoplasias in those species (13). The identification of Steamer did, however, prompt us to look further throughout the genomes of bivalves to understand the diversity of Steamer-like elements (SLEs) and to determine if their phylogenetic relationships suggest vertical inheritance from a bivalve ancestor or more recent HTT between species. We found SLEs in many, but not all, bivalve species and found evidence for multiple, frequent cross-species transfers, including recent transfer of nearly identical elements between soft-shell clams, razor clams, and Baltic clams. We furthermore found evidence for widespread and frequent transfer throughout bivalve evolutionary history and even cross-phyla transfer into many other marine organisms, including vertebrates, sea urchins, and coral.
Results
Multiple Cross-Species Transfers Throughout Bivalve Evolution.
To search for SLEs across the bivalve class, we performed PCR amplification using degenerate primers in conserved positions in the reverse transcriptase-integrase (RT-IN) region of the pol gene in the genomic DNA of 36 bivalve species (and one gastropod) obtained from the Ambrose Monell Cryo Collection (AMCC) of the American Museum of Natural History (AMNH), from collections of multiple independent researchers, and from multiple commercial sources. The integrity of the genomic DNA and the species identities were confirmed by amplification and sequencing of a region of the mitochondrial cytochrome c oxidase subunit I gene (COI). Sequences from this gene could be amplified from only 24 species using reported pan-invertebrate primers (14), but by using other primer variants, mitochondrial COI DNA was amplified and sequenced from all 37 species.
Using degenerate primers in conserved regions of Steamer, SLE sequences were amplified from 19 of the 37 species analyzed (Table S1). The DNAs were cloned from these 18 bivalves and one gastropod, and one to four distinct elements were sequenced from each species. Notably, while these sequences were similar to Steamer, the sequences identified from each species were unique and therefore could not be explained by laboratory contamination with a clonal source of DNA. Additionally, a highly related SLE was identified in the fully sequenced oyster genome (Crassostrea gigas, scaffold 39526) (15). SLE sequences were present in 11 families and nine orders. A previous study of bivalves from the AMCC using highly specific primers (16) identified SLEs nearly identical to Steamer in two bivalves (Baltic clam, Limecola balthica, and Atlantic razor clam, Ensis directus), and we confirmed those findings using the degenerate primers.
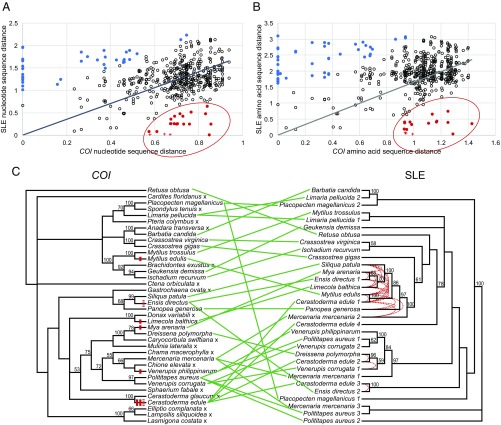
We then compared the SLE phylogeny with the phylogeny of the host organisms. The host phylogenetic tree based on COI sequence agrees with the established taxonomy of bivalve species (Fig. S1) and current molecular phylogenetic analyses (17), but when we aligned the host tree with the SLE tree, there was profound discordance between the COI tree and the SLE tree throughout the resulting tanglegram (Fig. 1). Many SLE sequences are more closely related to each other than would be expected by vertical inheritance, and there is evidence of acquisition of multiple, phylogenetically distinct SLEs in a single species (such as cockles, which host at least four distinct elements). SLEs were not detected in many species, leading to a patchy distribution of SLEs throughout the bivalve lineage, although it is possible that other SLEs are present in some species but were not detected with the primers used here.
Fig. 1.
Comparison of pairwise distances and phylogenetic trees of SLEs and host mitochondrial DNA. (A and B) Nucleotide sequence distances (A) and amino acid sequence distances (B) are plotted for all pairwise comparisons of bivalve SLE sequences (496) and are compared with the distances of the corresponding host COI [circles and crosses; comparisons between sequences reported in Paynter et al. (16) and Steamer from M. arenaria are marked with a cross]. A linear regression was plotted, and pairwise comparisons in which the SLE sequence was higher (blue) or lower (red) than expected are marked, using 1.5 times the IQR as the cutoff. (C, Left) The host phylogenetic tree was made from the alignment of COI nucleotide sequences (Fig. S1) rooted on the gastropod R. obtusa. (Right) The SLE phylogenetic tree was made from the alignment of amino acid sequences from the RT-IN region amplified by conserved primers, rooted on the Polititapes aureus 2/3/Mercenaria mercenaria 3 sequences. The 21 pairwise comparisons in which the SLE distance was lower than expected in either A or B are marked by red dotted lines. The HTT events predicted by both phylogenetic incongruity and pairwise distance comparison are shown on the host COI tree as red marks (the dotted mark represents a case in which pairwise distance comparison data could support either one or two events). Sequences represent amplified sequences from genomic DNA of 36 bivalve species and R. obtusa as well as a sequence from the C. gigas genome assembly (Table S1). PhyML 3.0 was used to generate the trees, and Dendroscope 3.5.9 was used to generate the tanglegram. An “x” marks a species in which no SLE was identified. The species of origin is listed, bootstrap values above 50 are shown, and nodes with bootstrap support below 25 have been condensed.
Additionally, we analyzed each pairwise comparison between bivalve SLEs and compared the distance to the host COI distance (Fig. 1). Many of the comparisons fell along a line expected for vertical inheritance (SLE distance increases as host distance increases). In some comparisons, the SLE distance is low and the host distance is high (marked in red in Fig. 1), providing strong evidence of HTT events. In other comparisons, the SLE distances were much higher than expected (marked in blue in Fig. 1), and this represents comparisons of different lineages of SLE within closely related species (for example, in cockles there are four distinct elements, so there are six pairwise comparisons with a SLE nucleotide distance of 1.1–1.7 and a host distance of zero). Overall, the complete discordance of the host and SLE trees, the patchy distribution of SLEs in the bivalve lineage, and the high similarity of SLEs compared with lower similarity of host sequences strongly argue for many cases of cross-species HTT throughout the bivalve lineage, with vertical maintenance of some elements.
Recent Horizontal Transfer of Steamer Among Soft-Shell Clams, Razor Clams, and Baltic Clams.
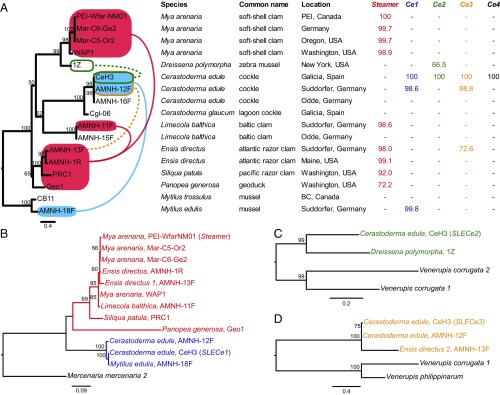
A recent report used highly specific primers to investigate the presence of sequence fragments of Steamer in the AMCC (16). Consistent with this study, we amplified and sequenced a fragment of the pol gene that is nearly identical to the Steamer elements from soft-shell clams (M. arenaria) in multiple Atlantic razor clams (E. directus) and in a single sample of Baltic clam (L. balthica) (Fig. 2). We amplified and cloned two large fragments of retroelements from a sample of E. directus. One clone covers most of the ORF (AMNH-1R-LPM-02, 1,662 bp, 97.5% identical to Steamer) and has an intact RT-IN region but two frameshifts earlier in the element. The other clone was amplified with primers in each LTR, so it covers the entire coding region, and it has multiple frameshifts and stop codons throughout the element (AMNH-1R-L2L2-02, 4,725 bp, 97.2% identical). Atlantic razor clams and soft-shell clams are both bivalves, but they have been genetically isolated from each other for an estimated 300–500 My (18–20), and their COI sequences are only 65% identical. The presence of the nearly identical sequences in these distantly related species strongly argues for recent cross-species horizontal transfer (HT) of the retrotransposon from one species to the other. Additionally, the sequences are too similar to be due to vertical inheritance of the element, but the sequences are unique, and therefore they are not due to contamination with any previously amplified or sequenced DNA.
Fig. 2.
Polymorphism within species and evidence for multiple, recent HTT between bivalve species. (A, Left) A phylogenetic tree of COI sequences from multiple individuals from multiple locations is shown. Proposed HTT events are shown as lines indicating likely routes of transfer of Steamer (red) from E. directus to M. arenaria and L. balthica and of SLECe1 (blue) between M. edulis and populations of C. edule. Dotted lines between sequences similar to SLECe2 and SLECe3 show more distant relationships. The SLECe1 sequence from the individual used as a reference here (CeH3) is 99.2% identical to the previously published sequence from a different individual in the same population (KX018578). (Right) The Steamer-like elements found in each individual sample are shown. Columns correspond to the distinct elements found in M. arenaria and C. edule: Steamer (red), SLECe1 (blue), SLECe2 (green), and SLECe3 (brown). The first member of each group identified is the reference and is listed as 100%. If a member of that group was detected in the sample, the percent identity compared with the group reference is shown (treating gaps as missing data). Samples negative for PCR amplification of each group are marked as “−.” (B) Phylogenetic tree of the SLE clade including Steamer and SLECe1 sequences, rooted with the sequence from M. mercenaria. (C) Phylogenetic tree of SLECe2 and closely related sequences. (D) SLECe3 and closely related sequences. The sequence from S. patula has a single frameshift mutation, the sequence from P. generosa has two closely spaced frameshift mutations that maintain an ORF, and the sequence from D. polymorpha has four frameshift mutations.
To explore the range of distribution of the Atlantic razor clam element in other razor clam species, we investigated genomic DNA from a Pacific razor clam (Siliqua patula), another member of the Pharidae family, and a geoduck (Panopea generosa), another member of the Adapedonta order. A highly similar sequence could be amplified from Pacific razor clams (92.0% nucleotide identity, with one stop codon), and a less related sequence could be amplified from the geoduck (72.2%, with two closely spaced frameshift mutations which combine to maintain an ORF). These results are most consistent with the entry of a retrotransposon into the genome of a common ancestor of the Adapedonta order, followed by vertical inheritance within the Adapedonta order and then more recent cross-species HTT from razor clams into Baltic clams and soft-shell clams (possibly through other intermediate organisms).
Lack of Fixation of SLEs in Some Species Suggests Recent HTT Events.
The finding of a nearly identical element (98.6% both to Steamer from soft-shell clams and to the element from Atlantic razor clams) in one sample of Baltic clam but not in another (Fig. 2) suggests a recent HTT event which has not become fixed in all members of the species. Interestingly, the individual that was positive for Steamer was the one collected in the same location as the Atlantic razor clams, in the Suddorfer Strand in the North Sea of Germany.
In addition to the one SLE identified previously in cockles (13) (previously termed “SLE-Ce” and now “SLECe1”), SLE sequences of three more distinct elements were amplified from cockles (Cerastoderma edule) from Galicia, Spain. Here we tested additional cockle samples from two locations in Germany and found one individual with only two of the four SLEs (>98% identity to that found in cockles from Galicia), while no SLEs were amplified from the individual from the other location. None of the SLE-Ce sequences were detected in the closely related lagoon cockle (Cerastoderma glaucum). Thus, while these SLEs were present in all cockles tested at a single site (Galicia), differences could be observed in geographically separated populations of the same species, showing either that the HTT events were not fixed in the species when the populations diverged or that the elements have been lost from some populations.
Evidence of HTT of SLEs from Cockles.
In addition to evidence of HT of Steamer itself, we found evidence of cross-species HT of other SLEs. An element nearly identical to SLECe1 was amplified from the DNA of a sample of Mytilus edulis collected from the same location as a cockle with the element, strongly suggesting another recent HTT event (Fig. 2). SLECe1 is closely related to Steamer itself (79.3% identical), and since SLECe1 is not found in the closely related cockles (C. glaucum) or mussels (Mytilus trossulus), the data are consistent with the hypothesis that SLECe1 in cockles and mussels derives from an earlier HTT from the Adapedonta lineage or some other source.
Other elements, less closely related to SLEs from cockles, were also found in a zebra mussel (Dreissena polymorpha, 66.2% identical to SLECe2) and an Atlantic razor clam (E. directus, 72.6% identical to SLECe3). These may be older HTT events, but the lack of amplification of the SLECe3-like element in other E. directus individuals suggests that these may have been recent HTT events from unknown sources.
Cross-Phyla Horizontal Transfer of Retroelements.
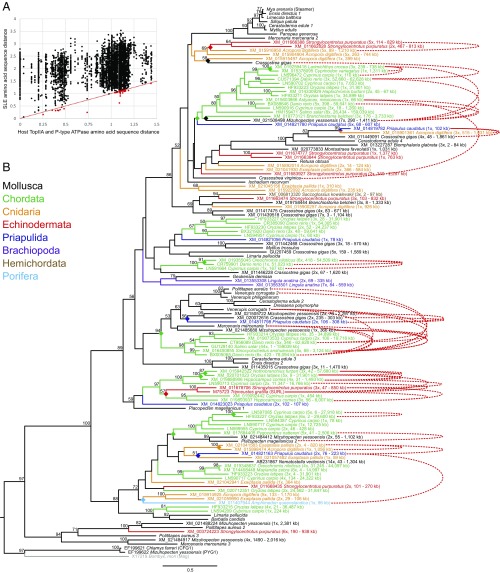
We next looked for SLEs more broadly and systematically by searching the National Center for Biotechnological Information (NCBI) nucleotide database with the same conserved RT-IN region of the pol gene from all known SLEs, including Steamer itself and all SLEs identified here and in our previous studies (32 total). Unexpectedly, SLEs were identified in sequences from organisms of completely different phyla, including Chordata (zebrafish, cichlids, and salmon), Echinodermata (sea urchin), Priapulida (marine priapulid worms), Hemichordata (acorn worms), Cnidaria (acropirid coral), and Porifera (sponges) (Fig. 3). Many of these sequences have been annotated as K02A2.6-like, based on a more distantly related Caenorhabditis elegans retrotransposon (InterPro accession Q09575).
Fig. 3.
Pairwise comparisons and the phylogenetic tree of SLEs provide evidence of cross-phyla HTT. (A) Amino acid sequence distances are plotted for all 7,738 pairwise comparisons of SLEs and are compared with the distances of the corresponding host using combined alignments from TopIIA and P-Type ATPase as host reference genes. A line with a slope of 1 is shown in red. (B) A phylogenetic tree made from the alignment of amino acid sequences from the RT-IN region amplified by conserved primers and from TBLASTN searches of the NCBI nucleotide database. The species of origin is listed, and bootstrap values above 50 are shown. Each hit was used to search the reference genome assembly of the species; the number of contigs with hits and the size range of contigs with hits are in parentheses. The sequences of SURL, CFG1, and PYG1 are included as known references, and Mag is included as an outgroup. Members of different phyla are color coded as shown (the reference, Mag, from the silkworm genome, is gray). Red dotted lines connect tips of the tree for 39 pairwise comparisons in which the SLE distance is lower than the distance of the conserved host genes. Diamonds mark the proposed cross-phyla HTT events that are supported by both phylogenetic evidence and pairwise comparisons.
These TBLASTN hits strongly suggest cross-phyla HT of SLEs, but it is possible that genome assemblies are contaminated with foreign sequences, so we validated each initial hit with a TBLASTN search directly against the species’ current reference genome assembly. We checked the number of contigs in the genome that contain sequences nearly identical to each hit and determined the size of those contigs. Four initial hits were identified only in small contigs (<10 kb) and therefore were excluded from analysis. Each remaining hit (98 total) was found in contigs at least 59 kb in length, with 55 hits found in contigs >1 Mb. This confirms that the SLEs were found in high-confidence genomic sequences and represent bona fide sequences in the genomes of the correct species.
To further validate the finding of cross-phyla HTT, we investigated the hits in the zebrafish genome more closely. Our search identified many sequence fragments with similarity to the RT-IN region and five sequence regions with high identity to the entire Steamer retrotransposon. One sequence (found in CU571394.10) has evidence of a 5-bp target-site duplication (AAGAG) at the predicted ends of the LTRs, and the 5′ and 3′ LTRs are nearly identical (167 of 168 bp), consistent with recent retroelement integration. The other elements have either only one LTR or have LTRs with mismatched sequences immediately adjacent, suggesting recombination between elements as well as insertions and deletions, and none has a complete intact ORF. Genomic DNA from zebrafish embryos from three laboratory lines (AB, TL, and Casper) were assayed for the presence of the three elements that contain two LTRs, using a PCR primer in the SLE and a reverse primer in the flanking genomic sequence. Two of the elements were confirmed to be present in all three lines, and the third was found in the Casper but not in the AB or TL lines (Fig. S2).
While the phylogenetic analysis of the TBLASTN search shows closely related SLEs in very distantly related organisms, which likely represent HTT events (Fig. 3), we additionally analyzed all pairwise comparisons of SLEs and their corresponding hosts. Since mitochondrial mutations occur at variable rates in different phyla, nuclear genes were used as a host comparison. According to the OrthoDB, there are 147 groups of proteins that are present in all of the phyla with SLEs, and there are no proteins that are single copy in 80% or more of the species. We chose two proteins (TopIIA and P-type ATPase) with the lowest number of duplicate genes as host references. Combining the amino acid sequences of these two host reference proteins, we find 39 pairwise comparisons in which the SLE distance is smaller than the distance of host genes. These comparisons mark 12 high-confidence cross-phyla HTT events and six intraphyla HTT events. The host reference genes are highly conserved, so this is a very stringent criterion and likely significantly underestimates HTT.
Interestingly, there is a large clade of SLEs that is only found in fish. This provides evidence either of vertical inheritance through fish evolution or of more restricted fish-specific transfer of this transposon. The pairwise comparisons suggest that there have been at least five cross-species HTT events from one fish genome to another (although we cannot exclude the possibility of an intermediate vector). We also observed pairs of closely related SLE sequences from zebrafish (Danio rerio) and carp (Cyprinus carpio) appearing in four distinct conserved positions in the tree of SLEs. The most likely explanation is that four (or more) distinct elements were transmitted to an ancestor of the two species before their divergence (∼85–125 Mya) (20).
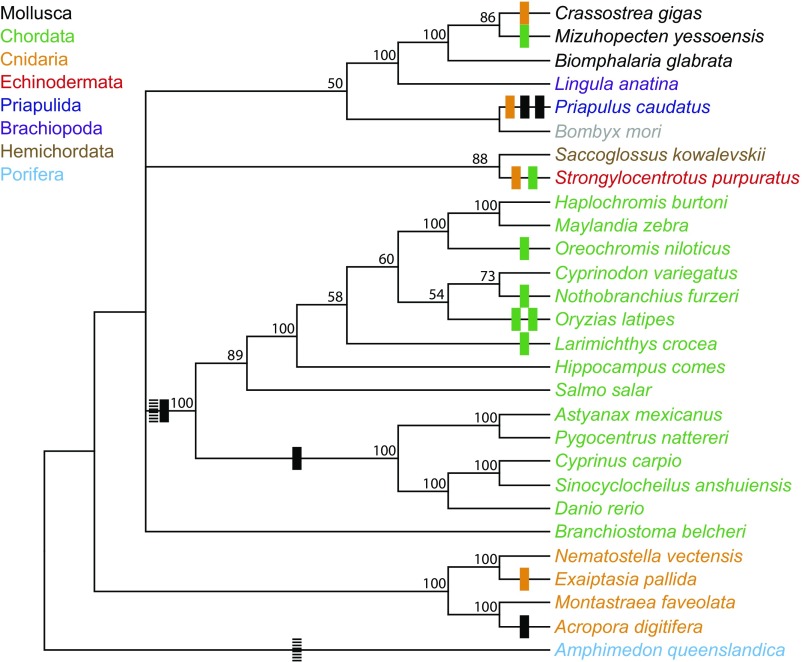
Many groups of closely related SLE sequences are present in completely different organisms, and pairwise analyses of SLE distances provide further evidence of multiple cross-phyla transfer events of closely related elements (Figs. 3 and 4). These high-confidence HTT events include multiple events between bivalves and fish, between fish and echinoderms, and in the SLE-Ce4 clade between species including bivalves, a coral, and a priapulid worm. Notably, from the systematic search of the entire NCBI nonredundant nucleotide database and the wide variety of phyla represented therein, all of the species found to harbor SLEs are aquatic, providing a plausible route of transfer of these elements.
Fig. 4.
Summary of cross-phyla HTT events. A phylogenetic tree was made using the TopIIA and P-type ATPase amino acid sequences of the species identified as harboring SLEs in the TBLASTN search. Bars mark HTT events supported by both phylogenetic analysis and by a lower than expected SLE distance. Bar colors identify the likely SLE donor based on the current evidence; striped bars reflect cases in which the donor could not be predicted.
Discussion
While there have been multiple reports of HT of LTR retrotransposons between species (6–8), the majority of these have been observed within Drosophila (21) and between plant species (22, 23). Additionally, many of the LTR retrotransposon HTT events in Drosophila involve the transfer of Gypsy, a retroelement which has acquired an envelope gene (24), making it functionally a retrovirus despite its falling within the Ty3/gypsy lineage of retrotransposons rather than within the vertebrate retroviruses. Cross-phyla HTT events have been observed with DNA transposons, but as recently as 2010, cross-phyla HT of retrotransposons had been rarely observed (8, 25), and it was suggested that DNA transposons may be more suited to long jumps into highly divergent hosts (7). Recently, however, there have been three reports of ancient “long-distance” HTT between plants and arthropods (26) and between birds and nematodes (27), and there is even a case of Tcn1-like LTR retrotransposons present in both fungi and plants, suggesting an ancient transkingdom HTT event (28). An earlier report has shown that sequences nearly identical to Steamer could be found in two bivalve species in addition to M. arenaria (16), and here we have expanded that study with broad and systematic strategies which show that there has been widespread HTT of SLEs throughout bivalve evolution as well as across phyla throughout the marine environment.
Phylogenetic incongruence and a patchy distribution across phylogeny provide evidence to support claims of HTT, but it has been argued that phylogenetic analysis can be misleading in some cases (7, 29). Additionally, PCR amplification can be negative due to trivial point mutations in primer regions, so a negative result may not necessarily mean that no element is present, and a patchy distribution may not necessarily mean that HTT has occurred. Therefore, we have additionally analyzed pairwise comparisons of SLEs and host sequence to identify cases in which the SLE distances are lower than expected based on host sequences, based on the methods of previous studies of HTT (30, 31). The data from all methods support our conclusions of cross-species and cross-phyla HTT. These pairwise comparisons can be problematic when the host genes are subject to variable mutation rates. For example, mollusks appear to have a higher mitochondrial mutation rate than other phyla, so the COI distance between two bivalves, such as soft-shell clams and mussels, is larger than the distance between soft-shell clams and vertebrates or even sponges, which is clearly at odds with the phylogenetic tree generated by these sequences and with known taxonomy. We therefore did not use COI sequences and instead used nuclear genes in the analysis of cross-phyla HTT. The criteria used to define unexpectedly similar SLEs depend on the host genes selected. The two host genes selected for our analysis of cross-phyla HTT are highly conserved, and they are indeed among a small list of genes conserved across all of the phyla harboring SLEs. Therefore, their use as a measure of host distance means that our analysis is very conservative and likely overlooks many true cases of HTT. Overall, data of all three types (phylogenetic incongruence, patchy distribution, and more similar than expected transposon sequence) support multiple HTT events throughout bivalves and across phyla.
One additional limitation of HTT detection strategies is that they are unlikely to identify HTT between closely related species. This makes the detected number of HTT events a conservative estimate. Assuming that there are greater barriers to HTT into more divergent hosts, then a significant number of HTT events may be occurring within species or within closely related species in an undetectable manner.
Potentially, contamination of either the DNA samples used in PCR or contamination of the genome assemblies themselves could lead to spurious evidence suggesting HTT, but several lines of evidence suggest this is not the case here. Each bivalve SLE sequence and TBLASTN hit was unique, and after the exclusion of four hits from small contigs, the majority of the hits were found in more than one contig, the majority were found in contigs of >1 Mb in length, and all had hits in contigs of at least 59 kb.
While previously reported cross-phyla HTT events were ancient, the identification of three recent HTT events in the current study of bivalves suggests that LTR retrotransposons, at least in the Steamer clade, are active and able to transfer relatively frequently. One previous report of HT of a retrotransposon in the marine environment involved SURL elements, which belong to the same Mag family retrotransposons as the SLEs (32). In this case, investigators found that vertical inheritance predominated within echinoid species but also found evidence of HTT in a few cases between highly related species. While there is evidence for vertical inheritance of many copies of SLEs, we find strong evidence that HTT plays a significant role across multiple phyla, and vertical inheritance can explain only a small number of the bivalve SLE sequences identified.
The timing of HTT events is difficult to determine accurately with the sequence data available here. The lack of fixation of SLEs in some species argues that those HTT events occurred after the most recent species divergence and may have occurred quite recently. The 5′ and 3′ LTRs are identical in the one sequenced full-length endogenous element present in the soft-shell clam genome. This argues for a recent origin, but the short length of the LTR (177 bp) makes an accurate analysis of integration date using a molecular clock difficult. The finding of Steamer in both Pacific and European samples of M. arenaria suggests that the first transmission into the M. arenaria genome occurred at least 800 y ago, as M. arenaria is believed to have been brought to Europe around the 1300s (33). The Steamer sequences from the M. arenaria samples in this study and in 10 more studies published recently (16) are all nearly identical, again suggesting recent entry into the genome or a recent expansion of a single lineage within the species. The SLEs in cockles appear to be recently acquired as well. None of the four cockle SLEs was present in all members of the species, showing that they have not reached fixation, and none was identified in C. glaucum, suggesting that the HTT events all occurred since the divergence of C. edule and C. glaucum.
For the most recent HTT events identified here (Steamer transferring from E. directus to M. arenaria and L. balthica, and SLEs from C. edule transferring to M. edulis and E. directus), the geographic ranges of the hosts clearly overlap. In one case, phylogenetic incongruity suggests HTT between bivalves M. trossulus and L. pellucida; while these samples were taken from very different geographic locations (Pacific and Atlantic coasts, respectively) the host range of M. trossulus is quite large, and it hybridizes with other members of the Mytilus genus, so there would be ample opportunity for close proximity that could allow for HTT. One interesting exception is the predicted HTT between a cockle (C. edule) and a zebra mussel (D. polymorpha). The zebra mussel is the only freshwater mussel in which we identified an SLE, but they are quite invasive in many geographic areas, and the presence of C. edule in estuaries may have allowed for transfer. The SLEs in the two species are only 66% identical to each other, so this is not likely to be a recent HTT event, and may also have occurred in a marine ancestor of the zebra mussel or transferred through an unknown intermediate host.
While many of the species involved in HTT are in close proximity, the exact mechanism of these events remains unknown. Evidence of HTT through host–parasite interactions, such as the ancient HTT between birds and nematodes, suggested that pathogenic interactions might be particularly suitable for HTT (27, 34–36). Notably, we did not observe any sequences corresponding to obvious parasites acting as vehicles for HTT, although this could be due to the limited availability of marine parasite sequence data. The widespread transmissible neoplasias in bivalves may provide a potential vector for the spread of TEs. While most cancers arise within an individual as a result of oncogenic changes within cells of that individual, transmissible clonal cancers that jump from one host to another [first found in Tasmanian devils (37) and dogs (38, 39)] have been found in an increasing number of species. Recently, we found independent lineages of bivalve transmissible neoplasia in at least four marine bivalve species, showing that this is a broad phenomenon across the animal kingdom and that marine invertebrates may be particularly susceptible (12, 13). It has also been shown that the transmissible neoplastic cells of soft-shell clams express high levels of Steamer RNA (11, 40), and reverse transcriptase activity can be detected in hemolymph and cell supernatants (11, 41–43), suggesting that some products of Steamer are being released from neoplastic cells. LTR retrotransposons also generate virus-like particles (VLPs) within the cell, and it is possible that some other virus or envelope-like membrane protein from the cell could transpackage these VLPs. Such VLPs could, in principle, introduce SLE sequences into germ cells and thus into the germ line. Limited searches by EM have not revealed the presence of VLPs in the M. arenaria neoplastic cells.
The consequences of the frequent HT of retrotransposons into new species are unknown. It has been hypothesized that introduction of a new element into a species without a specific control mechanism could be followed by rapid expansion (6), which could lead to pathogenesis. Indeed, the transmissible cancer lineage spreading throughout the soft-shell clam population has suffered a massive amplification of the Steamer retrotransposon. It is unknown whether any of the new integration events in the transmissible cancer lineage are drivers of oncogenesis, but insertional mutagenesis of retroelements can cause oncogenic mutations (3, 44). In the canine transmissible venereal tumor, a canine LINE1 element was found integrated immediately upstream of the c-myc gene (45–47), likely playing a significant role in the evolution of that transmissible cancer. Thus, the introduction of Steamer or SLEs into new species can lead to detrimental mutations and may increase the incidence of both conventional and transmissible cancers, and it may have significant effects on the evolution of these organisms.
The findings here suggest that the marine/aquatic environment may be particularly amenable to HTT due to the ability of particles to spread without the exposure to UV or the dry air of the terrestrial environment. A recent transcriptomic study of the Pacific white shrimp (48) identified TEs of multiple classes which appear to be derived by HTT from other aquatic organisms. Most of the organisms found to harbor SLEs are marine, although several freshwater fish were identified as well. It is unclear whether the transmission of the SLEs in fish occurred in the freshwater environment or in the marine environment of an ancestor of those fish. While it is not possible to directly compare the rates of HTT in the marine environment with that between terrestrial organisms, the results of our broad sampling of different bivalve species combined with a systematic database search (with very stringent criteria) argue that the LTR transposons investigated here have spread widely, but only within the aquatic environment. The cross-species and cross-phyla HTT events reported here also suggest that this is both a recent phenomenon and one that has been occurring throughout long evolutionary timescales. Together, these data provide evidence to suggest that the aquatic environment itself may act as both the vehicle and ecological connection (49) that can allow the spread of TEs from one genomic reservoir into new germ lines.
Materials and Methods
Sample Sources.
Bivalve tissue samples were obtained from several sources including the AMCC of the AMNH, multiple independent research collections and previous studies (11, 13, 50), and commercial sources. Species collected, collection locations, and sources of additional bivalve samples are listed in Table S1. Species names and classifications are used according to the World Register of Marine Species (www.marinespecies.org).
DNA Extraction.
Tissues were frozen or stored in ethanol. Samples from the AMNH collection were extracted using the E.Z.N.A. Mollusc DNA Kit (Omega Bio-tek). DNA extraction of other samples was done using the DNeasy Blood and Tissue Kit (Qiagen) with minor modification, as done previously. Briefly, DNA extraction of tissues included an additional step to reduce the amount of PCR-inhibiting polysaccharides. After tissue lysis, 63 μL of buffer P3 (Qiagen) was added to the lysate and allowed to precipitate for 5 min. The lysate was spun for 10 min at full speed at 4 °C, and the resulting supernatant was mixed with buffer AL (Qiagen) for 10 min at 56 °C and then was mixed with ethanol and added to the column, continuing with the standard protocol.
PCR.
Primers used are listed in Table S2. PfuUltra II Fusion HS DNA Polymerase (Agilent) was used to amplify 10–50 ng of genomic DNA for 35 cycles. PCR products were purified or gel extracted using spin columns (Qiagen), and PCR products were either sequenced directly or cloned using the Zero Blunt TOPO Kit (Invitrogen) and sequenced with primers flanking the cloning site. When multiple, nearly identical clones were sequenced from a single individual, one representative was selected for analysis. The first clone sequenced with an ORF was selected, and the first clone with a frameshift or stop codon was used if no clones had an ORF. An SLE sequence was determined to be a distinct element if it was <80% identical at the DNA sequence level and amino acid level. PCR amplification was considered positive only if an SLE sequence was obtained.
Species Confirmation.
The mitochondrial COI sequence of each species was compared with sequences available on the NCBI database to confirm species’ identities. Of 44 samples from 36 bivalve species and one gastropod, we amplified a COI sequence that was ≥99% identical to the sequence from the same species in the NCBI database in 32 samples (24 species). In eight species, no other COI sequence was available from that species for comparison, and the sequence was 70–85% identical to another member of the same genus. In three samples from the AMCC, Limaria pellucida, Codakia orbiculata, and Retusa obtusa (a gastropod used as an outgroup for the bivalve class), the match to a currently existing NCBI COI sequence from the respective species was 70–90%, with no clear identical sequences in the NCBI database. This suggests high diversity within the species or misclassification at the species level of either the AMCC samples or the previously reported samples in the NCBI database. One sample, Sphaerium fabale, was 99% identical to a different species, Sphaerium striatinum (no S. fabale COI sequence was available), suggesting either that the two species are not distinct or that either the AMCC samples or the previously reported samples had been misclassified at the species level. In each case, the species identification made by the AMCC was used in the analysis.
BLAST Search and Alignment.
A TBLASTN search was conducted on the NCBI nonredundant nucleotide database using the conserved 226-aa region of Steamer pol targeted by primers DHKPL-F1 and PXRPW-R1 (Table S2) and all additional bivalve SLE sequences identified in this study (July 6, 2017). Target sequences were identified with complete coverage across the region that excludes the primer sequences themselves. Sequences with e values above 10−60 were excluded, exact duplicates were excluded, and where multiple similar sequences (>80% identity) were identified within a single species, a single representative was selected. Additionally, bivalve SLE sequences already in the NCBI database, which were a part of the query set, and SURL were excluded, leaving 102 unique sequences.
Each of these 102 hits was used as query for a TBLASTN search of the current genome assembly of the species in which it was found, counting only hits with e values below 10−60, with >80% coverage and >80% identity. The number of hits in each genome assembly, the number of unique contigs containing hits, and the sizes of the contigs were recorded. Four queries in this secondary BLAST search were found in only a single contig <10 kb in length, and therefore they were excluded from analysis, leaving 98 unique sequences, with 407 total hits throughout the genome assemblies.
Phylogenetic Analysis.
Sequences were aligned with MAFFT v 7.311 (51) using the E-INS-i method (as the sequences include a variable region between two conserved regions). Some manual adjustments in DNA alignments were made based on the amino acid alignments. Additional alignments using CLUSTAL yielded similar results. Primer-binding regions were excluded from analysis. Maximum likelihood phylogenetic trees were generated using PhyML 3.0 (52), with nearest neighbor interchange tree improvement and 100 bootstrap replicates, treating gaps in the alignment as missing data. For amino acid trees, frameshifts were also treated as missing data. One sequence contained a “J,” which can stand for a site that is ambiguous for either leucine or isoleucine. We therefore excluded it from analysis by marking it as a missing site. Amino acid trees used the LG model +G+I, and nucleotide trees used GTR +G+I, based on the Akaike Information Criterion analysis of the full SLE amino acid alignment and the full DNA COI alignment. Single trees were visualized using FigTree version 1.4.2 or Dendroscope 3.5.9 (53). The tanglegram was constructed using a Neighbor-Net heuristic, which optimizes branch crossings, implemented in Dendroscope (54).
Pairwise Analysis.
For the within-bivalve analyses, MAFFT-aligned SLE and COI sequences were used (as in phylogenetic analysis). A linear trendline was made with an intercept of zero, and the interquartile range (IQR) of the distance from the trendline was calculated. A cutoff of 1.5 times the IQR was used to determine expected values. For analysis of the TBLASTN hits, OrthoDB (55) was used to identify two host genes likely to be found in all taxa containing SLEs, with minimal duplications. TopIIA (group EOG091G00U2) and P-Type ATPase (group EOG091G022E) genes were selected. BLASTP searches of the relevant genome databases were used to identify the genes in species not included in OrthoDB, and BLASTP was used to identify the top hit in cases of multiple genes and isoforms. In vertebrates, the P-type ATPase is duplicated (atp7a and atp7b); atp7a was selected from each species. Sequences were aligned with MAFFT as above. For bivalves, a sequence was available only for C. gigas and Mizuhopectin yessoensis, so the distance from C. gigas was used as a proxy value for all other bivalves for all cross-phyla comparisons. Strongylocentrotus purpuratus was also used as a proxy for Tripneustes gratilla. Therefore, intrabivalve and intraechinoderm comparisons were excluded from analysis. The F84 model was used for computation of nucleic acid distances with DNADist, and the JTT model was used for computation of amino acid distances with ProtDist, v3.6a2.1 (J. Felsenstein).
Supplementary Material
Acknowledgments
We thank the investigators who collected the samples for the AMCC and the investigators who collected the samples used in previous studies of bivalve transmissible neoplasia, including Antonio Villalba, María J. Carballal, and David Iglesias (Xunta de Galicia, Spain), James Sherry and Carol Reinisch (Environment Canada), and Annette F. Muttray and Susan A. Baldwin (University of British Columbia, Canada); Denise Mayer (New York State Museum, Division of Research and Collections) for the samples of zebra mussels (D. polymorpha) and Elliptio complanata collected from the Hudson River; Josh Barber (Institute of Comparative Medicine and the Zebrafish Core Facility, Columbia University Medical Center) for embryos of zebrafish of multiple strains; Doug Rogers and Camille Speck (Washington State Department of Fish and Wildlife); Carly Strasser and Phillipe St-Onge for additional M. arenaria samples; and Daniel Huson for advice on the use of Dendroscope. The work was supported by the Howard Hughes Medical Institute (S.P.G. and M.J.M.), NIH Training Grant T32 CA009503 (to M.J.M.), and a Research Experiences for Undergraduates Site Grant from the Division of Biological Infrastructure of the National Science Foundation (to A.N.P.). The funding bodies had no part in the design of the study, collection, analysis, interpretation of data, or writing of the paper.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MH012205–MH012241 and MH025768–MH025795).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717227115/-/DCSupplemental.
References
- 1.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Goodier JL. Retrotransposition in tumors and brains. Mob DNA. 2014;5:11. doi: 10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kemp JR, Longworth MS. Crossing the line toward genomic instability: LINE-1 retrotransposition in cancer. Front Chem. 2015;3:68. doi: 10.3389/fchem.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik HS, Henikoff S, Eickbush TH. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 6.Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallau GL, Ortiz MF, Loreto EL. Horizontal transposon transfer in eukarya: Detection, bias, and perspectives. Genome Biol Evol. 2012;4:689–699. doi: 10.1093/gbe/evs055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dotto BR, et al. HTT-DB: Horizontally transferred transposable elements database. Bioinformatics. 2015;31:2915–2917. doi: 10.1093/bioinformatics/btv281. [DOI] [PubMed] [Google Scholar]
- 9.Ivancevic AM, Walsh AM, Kortschak RD, Adelson DL. Jumping the fine LINE between species: Horizontal transfer of transposable elements in animals catalyses genome evolution. BioEssays. 2013;35:1071–1082. doi: 10.1002/bies.201300072. [DOI] [PubMed] [Google Scholar]
- 10.Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci USA. 2013;110:1012–1016. doi: 10.1073/pnas.1205856110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arriagada G, et al. Activation of transcription and retrotransposition of a novel retroelement, Steamer, in neoplastic hemocytes of the mollusk Mya arenaria. Proc Natl Acad Sci USA. 2014;111:14175–14180. doi: 10.1073/pnas.1409945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzger MJ, Reinisch C, Sherry J, Goff SP. Horizontal transmission of clonal cancer cells causes leukemia in soft-shell clams. Cell. 2015;161:255–263. doi: 10.1016/j.cell.2015.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzger MJ, et al. Widespread transmission of independent cancer lineages within multiple bivalve species. Nature. 2016;534:705–709. doi: 10.1038/nature18599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 15.Zhang G, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- 16.Paynter AN, Metzger MJ, Sessa JA, Siddall ME. Evidence of horizontal transmission of the cancer-associated Steamer retrotransposon among ecological cohort bivalve species. Dis Aquat Organ. 2017;124:165–168. doi: 10.3354/dao03113. [DOI] [PubMed] [Google Scholar]
- 17.Combosch DJ, et al. A family-level tree of life for bivalves based on a Sanger-sequencing approach. Mol Phylogenet Evol. 2017;107:191–208. doi: 10.1016/j.ympev.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Plazzi F, Passamonti M. Towards a molecular phylogeny of mollusks: Bivalves’ early evolution as revealed by mitochondrial genes. Mol Phylogenet Evol. 2010;57:641–657. doi: 10.1016/j.ympev.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Kano Y, Kimura S, Kimura T, Warén A. Living monoplacophora: Morphological conservatism or recent diversification? Zool Scr. 2012;41:471–488. [Google Scholar]
- 20.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartolomé C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Baidouri M, et al. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014;24:831–838. doi: 10.1101/gr.164400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roulin A, et al. Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol Biol. 2009;9:58. doi: 10.1186/1471-2148-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 25.Konieczny A, Voytas DF, Cummings MP, Ausubel FM. A superfamily of Arabidopsis thaliana retrotransposons. Genetics. 1991;127:801–809. doi: 10.1093/genetics/127.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Faridi N, Casola C. An ancient transkingdom horizontal transfer of Penelope-like retroelements from arthropods to conifers. Genome Biol Evol. 2016;8:1252–1266. doi: 10.1093/gbe/evw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh A, et al. Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat Commun. 2016;7:11396. doi: 10.1038/ncomms11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novikova O, Smyshlyaev G, Blinov A. Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants. BMC Genomics. 2010;11:231. doi: 10.1186/1471-2164-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capy P, Anxolabéhère D, Langin T. The strange phylogenies of transposable elements: Are horizontal transfers the only explantation? Trends Genet. 1994;10:7–12. doi: 10.1016/0168-9525(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert C, Hernandez SS, Flores-Benabib J, Smith EN, Feschotte C. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol Biol Evol. 2012;29:503–515. doi: 10.1093/molbev/msr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace JK, 2nd, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez P, Lessios HA. Evolution of sea urchin retroviral-like (SURL) elements: Evidence from 40 echinoid species. Mol Biol Evol. 1999;16:938–952. doi: 10.1093/oxfordjournals.molbev.a026183. [DOI] [PubMed] [Google Scholar]
- 33.Strasser M. Mya arenaria—An ancient invader of the North Sea coast. Helgol Meeresunters. 1998;52:309–324. [Google Scholar]
- 34.Panaud O. Horizontal transfers of transposable elements in eukaryotes: The flying genes. C R Biol. 2016;339:296–299. doi: 10.1016/j.crvi.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Wijayawardena BK, Minchella DJ, DeWoody JA. Hosts, parasites, and horizontal gene transfer. Trends Parasitol. 2013;29:329–338. doi: 10.1016/j.pt.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearse AM, Swift K. Allograft theory: Transmission of devil facial-tumour disease. Nature. 2006;439:549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 38.Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Clonal origin and evolution of a transmissible cancer. Cell. 2006;126:477–487. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A. Origins and evolution of a transmissible cancer. Evolution. 2009;63:2340–2349. doi: 10.1111/j.1558-5646.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 40.Siah A, McKenna P, Danger JM, Johnson GR, Berthe FC. Induction of transposase and polyprotein RNA levels in disseminated neoplastic hemocytes of soft-shell clams: Mya arenaria. Dev Comp Immunol. 2011;35:151–154. doi: 10.1016/j.dci.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 41.AboElkhair M, et al. Reverse transcriptase activity associated with haemic neoplasia in the soft-shell clam Mya arenaria. Dis Aquat Organ. 2009;84:57–63. doi: 10.3354/dao02038. [DOI] [PubMed] [Google Scholar]
- 42.AboElkhair M, et al. Reverse transcriptase activity in tissues of the soft shell clam Mya arenaria affected with haemic neoplasia. J Invertebr Pathol. 2009;102:133–140. doi: 10.1016/j.jip.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 43.House ML, Kim CH, Reno PW. Soft shell clams Mya arenaria with disseminated neoplasia demonstrate reverse transcriptase activity. Dis Aquat Organ. 1998;34:187–192. doi: 10.3354/dao034187. [DOI] [PubMed] [Google Scholar]
- 44.Shukla R, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katzir N, Arman E, Cohen D, Givol D, Rechavi G. Common origin of transmissible venereal tumors (TVT) in dogs. Oncogene. 1987;1:445–448. [PubMed] [Google Scholar]
- 46.Katzir N, et al. “Retroposon” insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc Natl Acad Sci USA. 1985;82:1054–1058. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi YK, Kim CJ. Sequence analysis of canine LINE-1 elements and p53 gene in canine transmissible venereal tumor. J Vet Sci. 2002;3:285–292. [PubMed] [Google Scholar]
- 48.Wang X, Liu X. Close ecological relationship among species facilitated horizontal transfer of retrotransposons. BMC Evol Biol. 2016;16:201. doi: 10.1186/s12862-016-0767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venner S, et al. Ecological networks to unravel the routes to horizontal transposon transfers. PLoS Biol. 2017;15:e2001536. doi: 10.1371/journal.pbio.2001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strasser CA, Barber PH. Limited genetic variation and structure in softshell clams (Mya arenaria) across their native and introduced range. Conserv Genet. 2009;10:803–814. [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Huson DH, Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 54.Scornavacca C, Zickmann F, Huson DH. Tanglegrams for rooted phylogenetic trees and networks. Bioinformatics. 2011;27:i248–i256. doi: 10.1093/bioinformatics/btr210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zdobnov EM, et al. OrthoDB v9.1: Cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 2017;45:D744–D749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]