Significance
Drought remains a critical obstacle to meeting the food demands of the coming century. Understanding the interplay between drought stress, plant development, and the plant microbiome is central to meeting this challenge. Here, we demonstrate that drought causes enrichment of a distinct set of microbes in roots, composed almost entirely of monoderms, which lack outer membranes and have thick cell walls. We demonstrate that under drought, roots increase the production of many metabolites, and that monoderms inhabiting the drought-treated rhizosphere exhibit increased activity of transporters connected with some of these same compounds. The discovery of this drought-induced enrichment and associated shifts in metabolite exchange between plant and microbe reveal a potential blueprint for manipulating plant microbiomes for improved crop fitness.
Keywords: sorghum, root, microbiome, drought, metatranscriptome
Abstract
Drought stress is a major obstacle to crop productivity, and the severity and frequency of drought are expected to increase in the coming century. Certain root-associated bacteria have been shown to mitigate the negative effects of drought stress on plant growth, and manipulation of the crop microbiome is an emerging strategy for overcoming drought stress in agricultural systems, yet the effect of drought on the development of the root microbiome is poorly understood. Through 16S rRNA amplicon and metatranscriptome sequencing, as well as root metabolomics, we demonstrate that drought delays the development of the early sorghum root microbiome and causes increased abundance and activity of monoderm bacteria, which lack an outer cell membrane and contain thick cell walls. Our data suggest that altered plant metabolism and increased activity of bacterial ATP-binding cassette (ABC) transporter genes are correlated with these shifts in community composition. Finally, inoculation experiments with monoderm isolates indicate that increased colonization of the root during drought can positively impact plant growth. Collectively, these results demonstrate the role that drought plays in restructuring the root microbiome and highlight the importance of temporal sampling when studying plant-associated microbiomes.
Plant-associated microbiomes play a role in determining plant fitness (1, 2), and it is well established that the composition of the plant microbiome is influenced by many host-associated and environmental factors (3–12). Recent research has demonstrated that changes in soil moisture availability have significant effects on plant-associated microbiomes (13, 14). As drought represents one of the most significant obstacles to agricultural productivity (15) and is expected to increase in severity and frequency in the coming century (16), efforts to ensure global food security are focusing on improving drought response in the world’s crops. Compared with traditional breeding and genetic engineering-based approaches, manipulation of the crop microbiome (17, 18) is relatively fast to develop and bring to market, and has the potential to confer benefits to a wide range of hosts (19, 20). However, to maximize the benefits of microbially mediated tools aimed at improving drought tolerance, a better understanding of the causes and timing of recently documented drought-induced shifts in the plant microbiome (13, 14) is needed.
It is well established that the timing of drought onset and duration of drought affect the extent to which crop productivity (21) is negatively impacted, and that some plant genotypes are more susceptible to drought stress early or late in development. As plant root microbiomes are largely recruited from the surrounding soil early in plant development (22), we hypothesize that plant microbiomes will also be differentially affected by the timing of drought events, with early microbiome development being the most susceptible to disruption. Previous research from our laboratory suggests that the timing of drought may indeed affect the extent of changes in plant microbiome composition, but limited temporal sampling in this study prevented us from fully exploring this hypothesis (13). More generally, while the temporal dynamics of the human microbiome have been thoroughly investigated (23, 24), few high-resolution experiments have been conducted in plant systems, even under normal watering regimes (25), despite evidence that plant age is an important determinant of microbiome composition (26).
In this study, we demonstrate that drought significantly delays the early development of the root microbiome of Sorghum bicolor, an important food staple in the developing world and a proposed model for studying plant drought response (27). Furthermore, we show that rewatering early drought-stressed plants leads to a rapid return to a normal but delayed progression of microbiome development. We observe that drought leads to reduced microbial diversity within the root-associated microbiome and increases the relative abundance of nearly all monoderm bacteria, which differ from diderm bacteria in that they lack an outer membrane and typically have thicker cell walls (28). We present evidence that in the rhizosphere, drought is correlated with increased transcription of many monoderm transporter genes associated with specific amino acids and carbohydrates. Conspicuously, we also observe increases in the production of many of these same compounds within the plant root, suggesting that drought-induced shifts in root metabolism are connected with shifts in root-associated microbiome composition. Finally, we performed in vivo colonization experiments that indicate that drought causes increased root colonization of monoderm strains, and that this colonization is correlated with increased root biomass, specifically under drought stress. Thus, we show that drought significantly impacts root bacterial composition and activity in a time-dependent manner with potential implications for crop fitness.
Results
Sorghum Microbiome Development Is Delayed by Drought Stress.
To explore the effects of sorghum development on root-associated bacterial communities, and to determine how drought influences microbiome recruitment, we conducted a large-scale field experiment in Kearney, located in California’s Central Valley, in which sorghum plants were sampled weekly across 17 time points (TP1–TP17) for a variety of plant- and microbiome-related traits. To account for potential interactions between cultivar-specific differences in host tolerance to drought and microbiome-related phenotypes, we chose to use two varieties of sorghum (BTx642 and RTx430) that are characterized by different drought tolerance phenotypes (SI Appendix, Fig. S1). To explore our hypothesis that drought has greater impact on the development of the early root microbiome compared with the late root microbiome, we subjected plants to one of three irrigation treatments (SI Appendix, Figs. S2 and S3): (i) preflowering drought, with no water applied between planting and the onset of flowering (TP0–TP8); (ii) postflowering drought, with no water applied after flowering (TP10–TP17); and (iii) control, with normal irrigation applied throughout the experiment (TP3–TP17). For all three treatments, plants were sown into prewatered fields (TP0) and left unirrigated for 2 wk (TP1–TP2) to allow seedlings to establish stable root systems.
To determine the level of drought stress achieved by our three treatments, we quantified the percent depletion of available soil moisture (ASM), which is correlated with the amount of water available to the plant root system. We observed that the depletion in ASM was significantly higher during both drought treatments but recovered rapidly following rewatering at TP9 in the preflowering treatment (SI Appendix, Fig. S4). To directly measure the effect of the drought treatments on plant performance, measurements of the crop water stress index, which serves as an approximation for reductions in levels of active leaf transpiration (29, 30), were taken at a subset of time points throughout the experiment. These data demonstrate that both preflowering and postflowering drought treatments lead to corresponding increases in plant stress (SI Appendix, Fig. S5). In accordance with this, we observed that plant height was reduced in both cultivars under preflowering drought treatment compared with control and failed to fully recover until the end of the experiment (SI Appendix, Fig. S6). Despite measurable changes in plant height and transpiration, no discernible differences in grain productivity were observed for either drought treatment compared with the control in both genotypes (SI Appendix, Fig. S7). Finally, qPCR using primers designed to amplify transcripts from two dehydrin genes (DS1 and DS6), which have been demonstrated to be molecular markers of drought stress in plants generally (31) and in sorghum specifically (32–34), demonstrated significant overexpression of these two genes (DS1: average log2-fold change = 12.7; P < 0.001 and DS6: average log2-fold change = 10.5; P < 0.001) in drought-stressed roots compared with control-treated roots at the peak of the preflowering drought treatment (TP8) (SI Appendix, Table S1). Similar, although smaller, increases in expression for both genes were observed at the peak of the postflowering drought treatment (TP17) (DS1: average log2-fold change = 5.2; P < 0.05 and DS6: average log2-fold change = 5.1; P < 0.05) (SI Appendix, Table S1). Collectively, these data suggest that both of the applied drought treatments significantly reduced soil moisture and induced drought stress in the plant host.
To determine drought’s influence on plant-associated microbial communities, samples of bulk soil, the rhizosphere, and the root endosphere were collected each week (SI Appendix, Fig. S2) starting 7 d after seedling emergence (TP1) and continuing until senescence (TP17) (SI Appendix, Material and Methods). Bacterial community composition across all time points (SI Appendix, Fig. S3) was investigated for each sample type (soil, rhizosphere, and root) using Illumina MiSeq sequencing of the V3–V4 region of the 16S rRNA gene. During initial plant development (TP0–TP2), when all three treatments were identical, microbial diversity in the rhizosphere and root samples exhibited a rapid increase following seedling emergence (TP1–TP2), followed by an equally rapid but smaller decrease 1 wk later (TP3) (Fig. 1 A–C). This is consistent with shifts in phylogenetic diversity observed for microbial communities undergoing endogenous heterotrophic succession (35) during which diversity peaks at a midpoint in community development when there is coalescence of an intermediate supply of limiting resources. Following this initial period of colonization, a comparison of drought and control treatments revealed that both preflowering and postflowering drought treatments lead to a significantly decreased level of Shannon’s diversity within the root (∼20%) and rhizosphere (∼15%) relative to the control (Fig. 1 B and C); by contrast, levels of Shannon’s diversity in the soil remained largely unchanged (Fig. 1A).
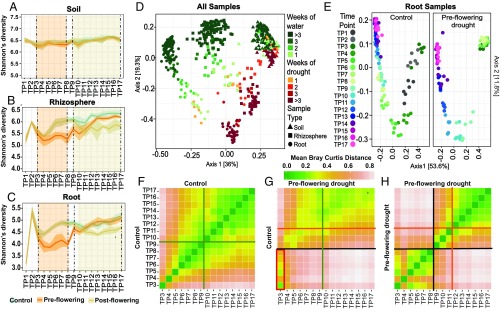
Fig. 1.
Drought impacts root microbiome development. Mean Shannon’s diversity across the soil (A), rhizosphere (B), and roots (C) at each time point under preflowering drought (orange lines), postflowering drought (yellow lines), and control (blue lines) treatments. The shaded areas above and below each line represent standard deviation from the mean. The orange- and yellow-shaded regions demarcated by vertical dashed lines indicate the periods in which preflowering drought and postflowering drought were applied, respectively. (D) PCoA of Bray Curtis distances for all control and preflowering drought samples. Soils (▲), rhizospheres (■), and root samples (●) are indicated. The color of each shape indicates the number of weeks of applied watering (shades of green) or drought treatment (shades of orange and red). (E) PCoA of Bray Curtis distances for all control and preflowering drought root samples colored by time point. Individual time points (TP1–TP17) are represented by distinct colors, with initial time points (TP1–TP2) shown as dark gray (control plot only), early time points (TP3–TP8) shown as shades of green, and late time points (TP9–TP17) shown as shades of blue and purple. (F–H) Heat maps of the mean pairwise Bray Curtis dissimilarity between all root sample replicates within the specified pairs of treatments and time points. A comparison of control samples versus control samples (F), a comparison of preflowering drought versus control samples (G), and a comparison of preflowering drought versus preflowering drought samples (H) are shown. Shades of green and pink represent low and high Bray Curtis distances, respectively. The orange and green lines indicate the mean flowering times in drought and control treatments, respectively, while the black lines represent the rewatering event at the end of drought treatment. (G, Lower Left) Red rectangle highlights the strong similarity between drought-treated samples at TP3–TP8 and the control treated samples belonging to TP3.
To explore the relative impacts of preflowering drought treatment, plant genotype, and plant age on bacterial community composition compared with other well-characterized factors, such as sample type, unconstrained principal coordinate analyses (PCoAs) of Bray Curtis distances were performed. These analyses conducted with preflowering drought and control samples demonstrate that samples belonging to roots, the rhizosphere, and soils clustered in distinct groups (Fig. 1D). Additionally, samples showed separation according to how long they had been under weekly water (Fig. 1D, shades of green) or drought (Fig. 1D, shades of orange and red) treatment. A permutational multivariate analysis of variance (PERMANOVA) analysis was performed using the Bray Curtis distances for all control and preflowering drought samples. These analyses indicate that bacterial communities were influenced most strongly by sample type, time point, and treatment (SI Appendix, Fig. S8), and that genotype and replicate were not found to significantly influence community composition in global analyses (SI Appendix, Table S2) or at any individual time point (SI Appendix, Table S3). As genotype did not significantly impact community composition, all remaining 16S rRNA analyses were conducted on aggregated data treating the two genotypes as additional replicates. Individual PCoAs performed separately for each sample type revealed that root (Fig. 1E) and rhizosphere (SI Appendix, Fig. S9A) communities exhibit larger and more dynamic compositional changes than do soil communities (SI Appendix, Fig. S9B), as indicated by a greater separation between time points and greater combined variation explained by the first two axes in the PCoA plots (roots = 65.4%, rhizosphere = 54.2%) compared with soil samples (soils = 29.5%). The “arch”-like shape of the plots for root and rhizosphere samples is characteristic of microbial environments in which there is a high degree of turnover along an environmental or temporal gradient (36), and suggests that the plant microbiome exhibits strong niche differentiation during development and may be subject to dramatic shifts in biochemistry.
These analyses also revealed that root and rhizosphere communities exhibited a stalled progression of bacterial community development under drought treatment relative to control samples (Fig. 1E and SI Appendix, Fig. S9A), as indicated by the distinct relative positions of drought and control samples at each time point (denoted by unique colors). However, despite these differences in the timing of microbial community development, both sets of samples follow a common trajectory. This delay is also evident in a heat map of the relative Bray Curtis dissimilarities between drought- and control-treated root samples across all time points (Fig. 1G), which demonstrates that throughout the preflowering drought treatment (TP3–TP8), the compositional profiles of drought-stressed plant roots most closely resemble root microbiomes of younger control plants from TP3 (Fig. 1G, red rectangle). The delayed transition in the root microbiome could potentially be explained by a drought-induced slowing of plant development, as we observed that preflowering drought also resulted in later flowering times and decreased plant growth rates (SI Appendix, Fig. S6). However, a side-by-side comparison of between-time-point community dissimilarity within replicates of control (Fig. 1F), and a similar comparison within replicates of preflowering treatments (Fig. 1H), revealed a much greater distinction between roots collected before and after watering in drought-stressed plants than between roots collected before and after flowering in the control treatment, suggesting that the effect of drought on the root microbiome exceeds the effect imposed by plant development.
Early Root Endosphere Communities Characterized by Dynamic Shifts in Dominant Taxa.
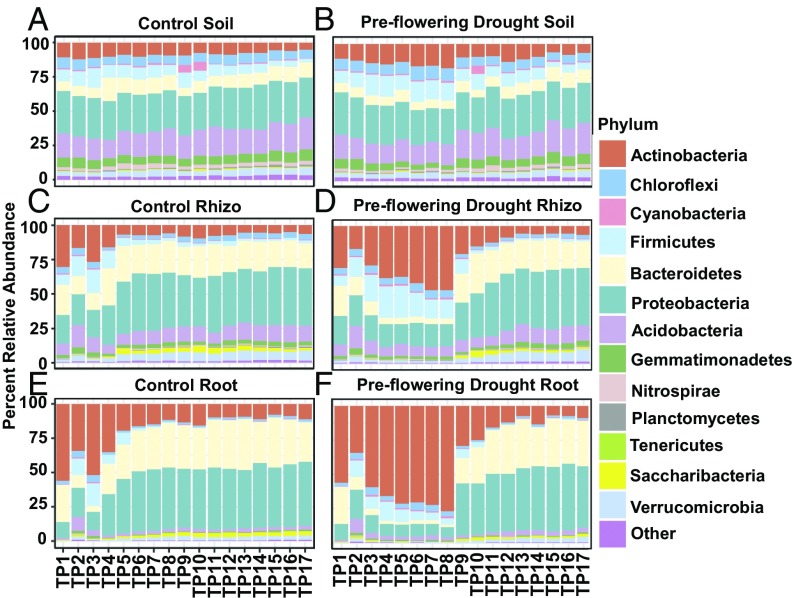
To better understand the delayed progression that drought induces in the root microbiome, we first sought to understand the temporal dynamics of the root microbiome under normal irrigation. Phylum-level relative abundance plots revealed that the root and rhizosphere communities exhibit an initial period of dynamic recruitment (TP1–TP6), followed by a later period of relative stability (Fig. 2 C and E); similar early shifts are not observed in the surrounding soil communities (Fig. 2A), indicating that this is not simply a result of changes in the surrounding source microbiome from which roots recruit their endophytes. In accordance with this, root and rhizosphere microbiomes exhibit relatively large compositional dissimilarity between adjacent time points in the initial six time points (SI Appendix, Fig. S10) compared with later in the experiment and with the microbial communities of the surrounding soil. This early compositional volatility of the root microbiome is driven by significant shifts in the relative abundance of a large number of dominant phyla, including Actinobacteria, Firmicutes, and Proteobacteria (Fig. 2E and SI Appendix, Fig. S11); by comparison, the most significant phylum-level shifts in abundance later in development (TP7–TP17) are in less abundant lineages (SI Appendix, Fig. S11). While the above results suggest that later development of the root microbiome is relatively stable, evidence of temporal restructuring of the sorghum root and rhizosphere microbiome throughout plant development is observable for some microbial lineages when finer taxonomic resolution is used, as demonstrated by family-level changes in both the Proteobacteria and Actinobacteria (SI Appendix, Fig. S12) that continued throughout the experiment.
Fig. 2.
Relative abundance for the most abundant bacterial phyla. Percent relative abundance of the top 13 most abundant phyla for control (A, C, and E) and drought (B, D, and F) treatments across soils (A and B), rhizospheres (C and D), and roots (E and F). All time points (TP1–TP17) are arranged in order along the x axis in each panel.
To identify bacterial lineages enriched within the root relative to the soil during early development in control conditions, we used a negative binomial model in the R package DESeq (37) to model operational taxonomic unit (OTU)-level root endosphere and soil community abundances across time points (TP3–TP8). Of the 366 genera containing OTUs with significant differences between soil and roots 35.5% showed strong enrichment within roots, and a phylogenetic reconstruction of differentially abundant lineages (SI Appendix, Fig. S13A) indicates a nonrandom distribution to this enrichment. Of the top eight most abundant phyla, the three that exhibited the highest overall preference for the root environment (Proteobacteria, Bacteroidetes, and Verrucomicrobia), as demonstrated by the percentage of genera enriched within roots compared with soils (SI Appendix, Fig. S13B), are typically considered diderm lineages. Among the three monoderm phyla (28) (Actinobacteria, Firmicutes, and Chloroflexi), two had the lowest relative percentages of genera enriched in roots; only Actinobacteria showed moderate levels of root enrichment relative to the other diderm phyla in our analysis. Taken together, these results suggest that early development of the root microbiome is dynamic, with large shifts in many dominant taxa, and that the root microbiome is preferentially colonized by diderm bacteria when plants are grown under normal watering conditions.
Drought induces enrichment of monoderm lineages in early root endosphere communities.
To determine how drought affects the observed developmental dynamics of the root microbiome, we next compared the phylum-level compositional profile of preflowering drought- and control-treated samples. In roots grown under preflowering drought treatment, we observed an enrichment for Actinobacteria and Firmicutes that is consistent with recently published reports (13, 14, 38) and a likely contributing factor for the observed decrease in microbial diversity within root-associated samples in this study (Fig. 2). This enrichment progressed over the course of drought treatment (6 wk) until watering resumed (Fig. 2). Strikingly, within 1 wk following rewatering, the root microbiome of previously drought-treated plants rapidly returned to a pattern of community progression observed in younger control roots (Figs. 1 E and G and 2). This rapid rewatering-induced shift in root endosphere composition is driven by a more than 200% increase in the relative abundance of the diderm lineages Proteobacteria and Bacteroidetes (P < 0.001) and a similarly large decrease in the monoderm phyla Actinobacteria (P = 0.012) (Fig. 2). As the relative enrichment in Actinobacteria could be the result of an absolute increase in their abundance, or an absolute decrease in other dominant taxa, we measured the absolute abundance of several lineages in drought and control root samples through qPCR using lineage-specific primers. These results demonstrate that in this field experiment, drought treatment leads to an overall decrease in total bacterial abundance, but that Actinobacteria and Firmicutes show significantly greater resistance to these shifts compared with Proteobacteria (SI Appendix, Fig. S14). Additionally, we observed that the decreases were greater for all community members at the peak of drought (TP8; SI Appendix, Fig. S14A) compared with earlier in the drought treatment (TP4; SI Appendix, Fig. S14B), which suggests that the absolute decrease in abundance is correlated with the length of drought treatment. Taken together, these results suggest that the root microbiome composition of field-grown sorghum is sensitive to drought perturbation early in plant development, and that this perturbation results in decreased abundance of the total bacterial community and a phylum-level relative enrichment of select monoderm bacterial lineages.
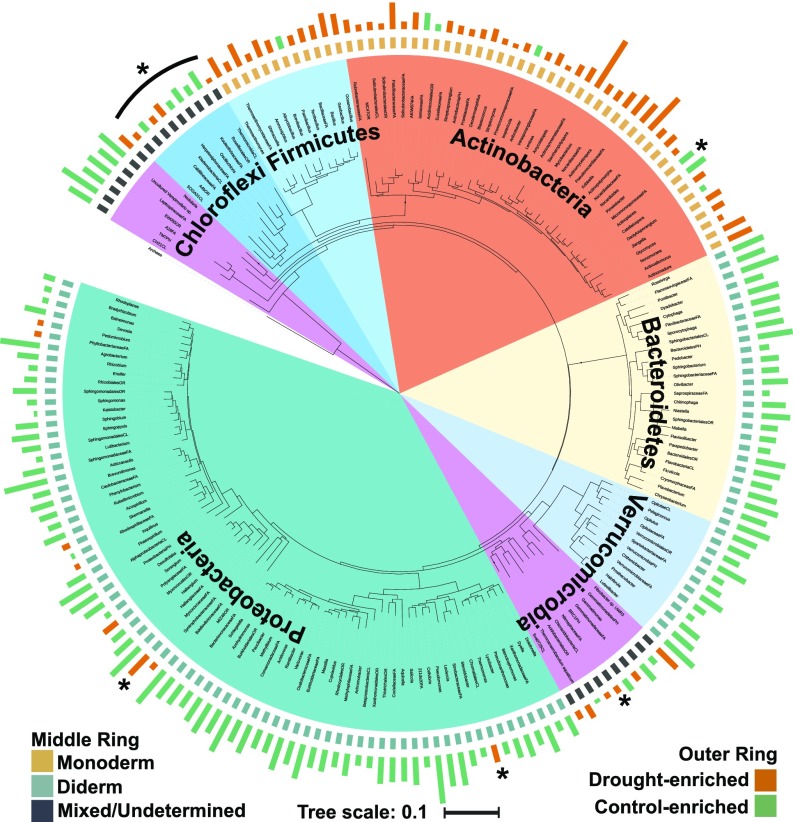
To identify at higher taxonomic resolution specific bacteria that exhibit relative abundance patterns that differ between control and preflowering drought, we used a negative binomial model as in the previous analysis to select OTUs with patterns of enrichment or depletion in roots between drought and control treatments. A total of 1,029 OTUs belonging to 195 of the 390 genera observed within the root endosphere during the preflowering time period (TP3–TP8) were identified as having significantly altered abundance between control and preflowering drought treatments, including 66 drought-enriched genera and 129 drought-depleted genera. A phylogenetic tree (Fig. 3) constructed from a representative 16S rRNA OTU sequence from each of these differentially abundant root genera offers further evidence that drought-enriched taxa are highly phylogenetically clustered, and demonstrates that drought-enriched lineages belong almost exclusively to monoderm phyla (SI Appendix, Fig. S15). Several notable exceptions to this pattern (Fig. 3, asterisks) are elaborated upon in detail in Discussion. Comparative analyses of genera containing OTUs with significant changes under drought in roots, rhizospheres, and soils yielded similar phylogenetic patterns of enrichment and depletion (SI Appendix, Fig. S16A); only 18 of 254 genera had opposing patterns of enrichment and depletion across any of the three sample types (SI Appendix, Fig. S16B). However, among the remaining 236 OTUs with enrichment patterns that were not inconsistent across the three sample types, the total number of lineages with significant changes and the magnitude of these shifts were consistently lower in soils than in either root-associated compartment (SI Appendix, Fig. S16).
Fig. 3.
Phylogenetic tree of all drought-enriched and -depleted root genera. The phylogenetic tree at the center of the figure was constructed from one representative OTU sequence from all genera that contained preflowering drought-enriched or -depleted OTUs in root samples. The inner colored ring represents the phylum each genus belongs to (legend as in Fig. 2). The middle ring indicates the expected status of each genus as belonging to phyla commonly considered monoderms (tan), to phyla commonly considered diderms (light blue), or to phyla for which monoderm status remains uncharacterized or is mixed (dark blue). The outer ring of colored bars represents the relative log2-fold enrichment (red) or depletion (blue) of each genus within drought-treated roots compared with control roots. The asterisks indicate select lineages that are elaborated upon in Discussion.
As the microbial community analysis conducted above was performed on samples taken in a single field experiment, we next sought to test whether the patterns of monoderm enrichment under drought were observable in other environments. To this end, we performed an analysis of a dataset derived from a similar but smaller experiment conducted at a field station in Berkeley, California, in which sorghum plants of the same two genotypes grown under both preflowering drought and control treatments were sampled weekly for 16S rRNA microbiome profiling across 12 time points. In accordance with data from the main field trial, conducted in Kearney, California, we observed a strong phylogenetic clustering of drought-enriched taxa and enrichment of monoderms (SI Appendix, Fig. S17 A and B) within roots in the Berkeley field trial. Similarly, relative abundance of Actinobacteria was observed to increase in roots exposed to drought (TP5–TP12) (SI Appendix, Fig. S17B), although at levels lower than those found in the Kearney field experiment. The decreased enrichment may be due to the timing of drought treatment, which was initiated later in development (week 5), or to the less severe levels of soil moisture depletion in the Berkeley experiment (SI Appendix, Fig. S18) that resulted from lower mean temperatures and incidence of evening fog at this location. However, these results suggest that in broad terms, the temporal patterns of monoderm enrichment we observe under drought are both common to multiple environments and potentially related to the level and timing of stress that is imposed.
Drought causes greater disruption to early compared with late root communities.
We hypothesized that the root microbiome of older plants is less sensitive to drought stress due to prior establishment of endophyte communities during early development. To test this, we conducted an analysis of samples collected weekly from plants grown during the same field season in Kearney but subjected to drought only after plants had flowered (postflowering drought). As noted previously, comparison of the postflowering drought and control-treated communities revealed that, as with preflowering drought, median Shannon’s diversity decreased under drought in roots and rhizospheres, but not in the surrounding soil (Fig. 1 A–C). However, an exploration of phylum-level relative abundance revealed that the overall root-associated bacterial community composition during the postflowering period was relatively stable and similar to that of the control (SI Appendix, Fig. S19). PCoA of Bray Curtis distances in postflowering- and control-treated samples demonstrates a relative lack of clustering by treatment, especially for root and rhizosphere samples, compared with previous analysis of preflowering drought and control samples (SI Appendix, Fig. S20). Similarly, PERMANOVA reveals that the amount of variation explained by postflowering drought treatment is less than 40% of that explained by preflowering drought treatment (SI Appendix, Fig. S8 and Tables S2–S5). Finally, an analysis of mean Bray Curtis differences between drought- and control-treated roots revealed that after the first week of drought treatment, preflowering drought treatment had greater differences compared with control than did postflowering drought treatment at every subsequent time point (SI Appendix, Fig. S21). These results demonstrate that while drought occurring late in plant development is accompanied by a reduction in microbial diversity similar to that observed in early plant development, it has a significantly smaller impact on root microbial community composition.
To explore the degree to which OTUs exhibiting shifts in abundance during preflowering and postflowering drought overlapped, we identified OTUs with significant compositional changes between postflowering and control treatments across TP10–TP17 (n = 1,059) through negative binomial modeling. A comparison of this list of postflowering drought-altered OTUs with the list of preflowering drought-altered OTUs (TP3–TP8, n = 1,029) generated previously revealed that while overall phylum-level distribution patterns and total counts of OTUs per phylum appeared similar (SI Appendix, Fig. S22A), more than half of the OTUs in both sets were significantly altered in only one of the two treatments (SI Appendix, Fig. S22B). Notably, OTUs with shifts only in preflowering drought (SI Appendix, Fig. S22C, Right) accounted for threefold more read counts across all root samples than those with shifts unique to postflowering drought (preflowering = 20.7%, postflowering = 6.4%). This result may explain why OTUs with differential abundance in postflowering drought, although large in number, have relatively little impact on community composition. The data also demonstrate that the OTUs with significant shifts in both treatments (n = 491; SI Appendix, Fig. S22C) account for 67.6% of all read counts in root samples. Furthermore, a comparison of the respective fold enrichments of these common OTUs under preflowering and postflowering drought revealed that 98% (n = 41 of 42) of the OTUs enriched under drought in both treatments belonged to monoderm phyla, while 94% (n = 131 of 139) of OTUs depleted in both treatments were diderms. Collectively, these results suggest that while preflowering and postflowering drought treatments of roots exhibit conserved phylogenetic patterns of monoderm enrichment, their responses differ in magnitude and the specific OTUs exhibiting changes in abundance.
Actinobacterial Transcription Increases in Drought-Treated Rhizospheres.
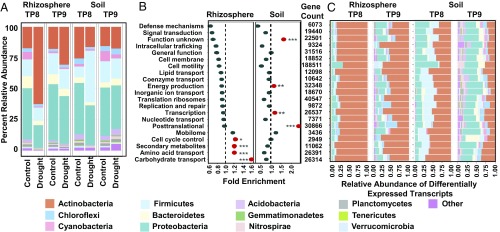
To determine if the shifts in root-associated microbiome composition during preflowering drought are correlated with changes in microbiome function, we performed metatranscriptome sequencing of the rhizosphere and soil communities at the peak of preflowering drought stress (TP8) and following rewatering (TP9). As with the amplicon data, samples from the two genotypes showed a high degree of correlation, and in all subsequent analyses, we treat the two genotypes as additional replicates (SI Appendix, Table S6). Of 556,826 genes in our dataset, we identified 13,300 drought-enriched genes (DEGs) in the rhizosphere (61% of total rhizosphere DEGs; SI Appendix, Table S7); by comparison, only 4,685 genes showed enrichment under drought in soils (12% of total soil DEGs; SI Appendix, Table S8). Unexpectedly, only 1,457 of all enriched genes were enriched under drought in both the soil and the rhizosphere (SI Appendix, Fig. S23).
In accordance with the results from our amplicon analysis, in a global analysis of all transcripts, we observed a significant increase in the expression of actinobacterial transcripts during drought in the rhizosphere (Fig. 4A) that mirrors their observed increase in relative abundance analysis of 16S rRNA data (Fig. 2 C and D). These results suggest that increases in the abundance of Actinobacteria are unlikely to be accounted for by sporulation, which would lead to largely dormant cells with reduced transcriptional activity (39). In contrast to the rhizosphere, similar analyses in soils exhibited no increase in actinobacterial transcripts but did see increased expression of transcripts belonging to Firmicutes, another monoderm lineage (Fig. 4A). As taxonomic analyses based on global metatranscriptome datasets can be susceptible to biases resulting from the fact that not all genes are equally represented in databases and are equally good phylogenetic markers, we performed a similar taxonomic analysis using only a subset of nine core housekeeping genes (gyrA, recA, rpoB, rpoA, gyrB, gap, rho, ftsZ, and secA) (40) (SI Appendix, Fig. S24). Within rhizospheres, the relative abundance profiles demonstrate largely similar patterns, including a large enrichment for actinobacterial core gene transcripts under drought treatment that is diminished upon rewatering. However, this analysis did reveal some differences compared with the global analysis; for instance, the increase in Firmicutes transcripts in drought-treated soils observed previously was not reproduced in the core gene analysis. As the relative abundance of Firmicutes transcripts in this core gene-based analysis was drastically reduced at all time points and in all treatments compared with the global analysis, we expect that the discrepancy may be accounted for by a relative lack of Firmicutes representation for the nine core genes in our taxonomic database. Taken together, these results suggest that while rhizosphere- and soil-associated microbial communities may respond similarly to drought stress in terms of taxonomic profile, they respond differently in terms of the quantity and types of genes exhibiting differential expression. This result is similar to that observed in the previous comparison of differential abundance in the 16S rRNA datasets (SI Appendix, Fig. S16), in which drought-treated root and soils yielded similar phylogenetic patterns of OTU enrichment and depletion but differences in terms of the quantity and size of these changes.
Fig. 4.
Drought impacts root microbiome transcription. (A) Percent relative abundance across the top 13 phyla for all transcripts in the metatranscriptome data for which taxonomies could be assigned from rhizospheres (Left) and soils (Right) for all control and drought-treated samples at TP8 and TP9. (B) GO enrichment analysis for all genes showing enrichment under drought for both rhizospheres (Left) and soils (Right) at TP8. The values on the x axis indicate the fold enrichment ratio of the relative percentages of genes up-regulated under drought in each category relative to the total relative percentage of genes in the corresponding category within the entire dataset. Categories for which there were fewer than five differentially expressed genes were omitted. The red circles indicate categories for which the enrichment had a P value of <0.05 in a hypergeometric test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). (C) Relative abundance across the top 13 phyla for all transcripts for which taxonomies could be assigned and which showed differential expression by treatment from rhizospheres (Left) and soils (Right) at TP8 and TP9, separated according to GO categories (y axis). Categories for which there were fewer than five differentially expressed genes were omitted. The legend for colors used for each phylum is as in Fig. 2.
Drought Increases Actinobacterial Transcription Related to Metabolite Transport.
We next sought to establish if the drought-induced shifts in our rhizosphere metatranscriptomes were correlated with changes in the expression of specific bacterial functions. Within the rhizosphere, we observed a significant increase at the peak of drought (TP8) in transcripts associated with carbohydrate transport and metabolism, amino acid transport and metabolism, and secondary metabolite biosynthesis (Fig. 4B); by contrast, soils exhibited a relative decrease in all three of these categories and an enrichment for a distinct set of functions, including energy production, transcription, and posttranslational modification (Fig. 4B). Additionally, we observed that gene categories exhibiting enrichment under drought changed after recovery (TP9) for both the rhizosphere and soil (SI Appendix, Fig. S25A). A finer resolution analysis of functional subcategories influenced by drought in the rhizosphere at TP8 revealed that a significant number of the most enriched gene subfunctions were related to resource transport, including those for both amino acids (10 of 33; Fisher’s exact test: P < 0.039; SI Appendix, Table S9) and carbohydrates (11 of 50; Fisher’s exact test: P < 0.0005; SI Appendix, Table S10), although it is worth noting that ATP-binding cassette (ABC)–type transporters are three-component systems, which may be artificially inflating this observed enrichment. In accordance with our taxonomic analysis of the metatranscriptome data, these data also revealed that the majority of DEGs in the rhizosphere (∼90%) in all functional categories belong to Actinobacteria (Fig. 4C), compared with ∼50% in the soil community (Fig. 4C). Finally, to determine if the enrichment in carbohydrate and amino acid transport and metabolism gene ontology (GO) categories is merely a consequence of the increased relative abundance of Actinobacteria or, alternatively, a shift in Actinobacterial function, we performed an analysis of the GO functional category assignments of all actinobacterial transcripts (SI Appendix, Fig. S25B). This analysis revealed that the drought-induced shifts in rhizosphere function are driven by significant changes in gene expression within the actinobacterial lineage in almost all GO functional categories, by carbohydrate and amino acid transport and metabolism, and by increased expression of ABC transporters (SI Appendix, Fig. S25C). Taken together, these data suggest that drought has a significant effect on the transcriptional activity of the root-associated microbiome, that rhizosphere genes associated with carbohydrate and amino acid metabolism and transport show increased expression under drought, and that the altered transcriptional activity in the rhizosphere microbiome during drought is largely due to shifts in actinobacterial activity and function.
Drought-Induced Shifts in Root Metabolism Correlate with Altered Rhizosphere Transcriptional Activity.
To investigate whether drought-induced shifts in the rhizosphere microbiome transcriptional activity, specifically the increased expression of transporters of carbohydrates and amino acids, are correlated with shifts in sorghum root metabolism, we performed untargeted metabolomics on sorghum roots using gas chromatography–mass spectrometry (GC-MS) at the peak of preflowering drought (TP8) and after rewatering (TP9). Through comparative analyses across treatments, we identified a large number of identifiable drought-enriched root metabolites (n = 114), including a variety of carbohydrates and amino acids (SI Appendix, Table S11). The most significantly enriched metabolite is glycerol-3-phosphate (G3P), which is 4.34 log10-fold more abundant in drought-treated than control roots (SI Appendix, Table S11). Interestingly, among the significantly drought-enriched carbohydrate gene subcategories in the rhizosphere metatranscriptomes (SI Appendix, Table S10), we observed a strong enrichment of ABC-type transporters of G3P, which had the largest number of up-regulated genes in the dataset (n = 191; P = 0.0001328).
As the observed increase in G3P within roots could potentially be produced by either the plant or the microbes in the system, we performed a high-performance liquid chromatography (HPLC) analysis of G3P levels in gnotobiotically grown sorghum seedling roots following drought treatment. These results indicate that in the absence of microbes, G3P is produced at levels twofold and 100-fold higher in roots exposed to 2 and 4 wk of drought, respectively, compared with controls (SI Appendix, Fig. S26). To further demonstrate that the G3P enrichment is host-generated, we performed qPCR with primers designed to quantify expression of several genes in the G3P transport and catabolism pathway on field-grown root samples collected during the peak of drought (TP8). These results revealed significant increases in genes involved in G3P transport across the plant cell plasma membrane (G3PP; SI Appendix, Table S12) and decreases in two genes [cytosolic glycerol-3-phosphate dehydrogenase (cGPDH) and glycerol-3-phosphate acyltransferase 6 (GPAT6)] responsible for converting G3P to other products [dihydroxyacetone phosphate (DHAP) and precursors of cutin biosynthesis]. These observations are consistent with a model in which G3P accumulates under drought within plant root tissues and is subsequently transported into the apoplast, perhaps to help reduce oxidative stress faced by the cell (SI Appendix, Fig. S27). Furthermore, the metabolomics data demonstrate that drought leads to the accumulation of a variety of other carbohydrates and amino acids within the root (SI Appendix, Table S11), and that many of these have potentially related gene categories with significant enrichment in the rhizosphere, including ribose, asparagine, proline, maltose, glucose, and threonine (17) (SI Appendix, Tables S9 and S10). Notably, many fewer metabolites were found to be differentially enriched between drought and control (n = 7) at the peak of postflowering drought treatment (SI Appendix, Table S13), and G3P is not among them. We also observed that the relative enrichment of Actinobacteria in postflowering drought-treated roots at TP17 is roughly threefold lower than their enrichment in the preflowering-treated roots at TP8. Taken together, these results suggest that a variety of root metabolites that are enriched under drought stress may be imported and used by root-associated Actinobacteria, and that the relatively large drought-induced shifts in community structure in early compared with late development are correlated with correspondingly larger shifts in the plant metabolome.
Isolates of Actinobacteria Are Enriched in Roots Under Drought.
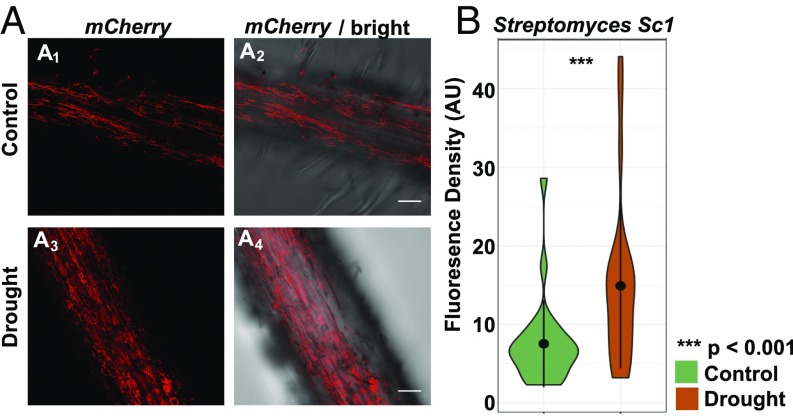
Evidence from our field trials suggests that the increased abundance of Actinobacteria during drought is likely due, in part, to the greater absolute depletion of most other bacterial lineages, but does not rule out some form of direct positive selection by the plant through an as yet undiscovered mechanism. To better distinguish between these potential causes of enrichment and to determine if actinobacterial enrichment under drought contributes to changes in host phenotype, we performed controlled inoculation experiments in plants grown in sterile soils. In these experiments, individual bacterial isolates cultivated from field-grown, drought-stressed sorghum roots were applied individually to presterilized soil in a gnotobiotic growth system containing 2-d-old sterilized sorghum seedlings. Three isolates, including two actinobacterial strains (Streptomyces coelicolor Sc1 and Streptomyces ambofaciens Sc2) as representatives of drought-enriched lineages and one proteobacterial strain (Pseudomonas syringae Ps1) as a representative of drought-depleted lineages, were chosen for this experiment. To quantify levels of microbial colonization of the root following 2 wk of imposed drought stress, we performed qPCR with primers specific to either Actinobacteria or Proteobacteria. In contrast to the qPCR results obtained from field-grown samples, we observed that both actinobacterial strains showed approximately twofold increases in abundance within drought-treated roots compared with control roots (SI Appendix, Fig. S28). By comparison, the Proteobacteria exhibited an approximately threefold reduction in abundance. To confirm the increased root colonization of Actinobacteria during drought, we tagged both of the Streptomyces isolates Sc1 and Sc2 with an mCherry fluorescence gene and used confocal microscopy to image colonization of root tissue during drought and control treatments in inoculation experiments performed as described above. We observed that both of the tagged strains showed roughly twofold higher levels of colonization following drought compared with control, as demonstrated by increased fluorescence density (Fig. 5 and SI Appendix, Fig. S29). Similarly, treatment of plants with PEG 6000, an alternative cause of osmotic stress often used to simulate drought in controlled experiments (41), also showed significantly increased colonization compared with control (SI Appendix, Fig. S30). These results demonstrate that individual Actinobacteria can have an absolute enrichment under drought in the absence of competition from other microbes, indicating that niche expansion alone is unlikely to fully account for the observed phenomenon. Unexpectedly, both strains also induced modest but significant increases in root growth following inoculation, and this growth promotion was only observable following exposure to drought treatment (SI Appendix, Fig. S31). Taken together, these data suggest that in a controlled laboratory setting and in the absence of other microbes, the observed increase in actinobacterial abundance during drought is accompanied by increased colonization, and that this increase is potentially beneficial to plant fitness.
Fig. 5.
Streptomyces exhibit increased colonization of sorghum roots under drought. Confocal fluorescence (A1 and A3) and bright-field (A2 and A4) imaging of colonization of mCherry-tagged Streptomyces strain Sc1 on control (Top) and drought-treated (Bottom) roots of genotype RTx430. (Scale bars: 50 μm.) Violin plots of the fluorescence intensity are measured using confocal fluorescence microscopy across 24 control (green) and 24 drought-treated (orange) root samples. AU, arbitrary unit.
Discussion
Our study provides a high-resolution characterization of the effects of plant development and drought on the sorghum root microbiome. We identified a high degree of microbial dynamism within the sorghum root during early development and demonstrate that early root microbiome development leads to relatively greater enrichment in diderm taxa compared with monoderms, with strong preferential enrichment for Proteobacteria and Bacteroidetes, as has been observed elsewhere (4). However, microbial coexistence depends, by necessity, on trade-offs, with distinct lineages having fitness advantages under different sets of conditions determined by the surrounding biotic and abiotic environments (42). We propose that within the root and rhizosphere, diderm lineages with relatively superior colonization ability under well-watered conditions are less suited to survive a set of selective pressures caused by drought. Similar examples of temporally variable selection of community composition via environmental fluctuation have been observed in other micro- and macroecosystems (43, 44).
In contrast to prior published results (13), root samples from our field experiment exhibited an absolute decrease in actinobacterial abundance following drought treatment that grew more pronounced as drought progressed. However, drought treatment following inoculation with individual Actinobacteria in gnotobiotically grown sorghum was shown to lead to an absolute increase in abundance of Actinobacteria. Similar discrepancies have been observed in the study of drought’s impact on the abundance of total bacterial abundance in soils (45). In general, total biomass has been observed to decrease under drought (46, 47), but some studies have observed bacterial abundance to remain constant (48) or even to increase (49). A variety of confounding factors could be responsible for these inconsistencies, including the duration and intensity of the drought treatment, temperature, edaphic factors such as soil pH and physicochemistry, nutrient availability, and methods of measurement. Despite the observed differences in absolute abundance in this study, broad patterns of relative abundance remain consistent between field and laboratory experiments. In particular, the large relative decrease in the absolute abundance of Proteobacteria and increase in Actinobacteria in both the field and gnotobiotic systems are consistent with the observed changes in relative abundance observed for these lineages in our amplicon dataset.
One unexpected result from this study is the degree of resilience exhibited by the root microbiome, which undergoes, within 1 wk of rewatering, a dramatic shift from monoderm back to diderm dominance. A study of resilience in other microbial communities found that less than half of all studied microbiomes (including soil, marine, and host-associated microbiomes) subjected to either short- or long-term disturbance events were capable of compositional recovery (50). A possible explanation for the observed resilience in our study could be the relatively faster growth rates of some diderms compared with monoderms (51), which could allow even a few surviving diderm individuals to recover to predisturbance population levels upon release from the selective pressures that lead to their reduced relative abundance under drought (50).
While the cause of the monoderm enrichment under drought currently remains unknown, unexpected enrichment and depletion patterns for several lineages (Fig. 3, asterisks) suggest that this phenomenon may be associated with the structure and thickness of the peptidoglycan cell wall layer. Actinoplanes, an Actinobacteria that is a notable exception to the general drought enrichment observed for this phylum, has recently been shown to have a novel cell wall type with considerable alterations in peptidoglycan structure compared with the majority of other actinobacterial lineages (52, 53). Within the Proteobacteria, a largely diderm phylum with strong and near-exclusive depletion under drought, some members of the order Chromatiales and the family Myxococcaceae are known to possess additional or unusual layers within their peptidoglycan cell wall (54, 55); correspondingly, we observe a strong enrichment under drought for both. Similarly, OTUs belonging to Nitrospira, an atypical diderm phylum that contains a triple-layered cell wall, also exhibit enrichment under drought (56). Finally, members of the phylum Chloroflexi, which have recently been shown to contain a mixture of lineages with and without cell walls (57, 58), showed approximately equal numbers of enriched and depleted genera. These observations suggest that while the drought-induced enrichment within the root microbiome largely follows the boundary between monoderm and diderm lineages, the true discriminating factors may be the presence, thickness, and structure of the cell wall, rather than the presence or absence of an outer membrane. As the structure of the bacterial cell wall has only been thoroughly characterized for a few bacterial taxa (28), further exploration of the cell wall composition within diverse lineages is needed. Indeed, our data may suggest a potential screen for identifying genera for which cell wall architecture may differ from close relatives as candidates for further cell wall analysis.
Recent evidence has shown that exudation of specific plant metabolites can influence root microbiome composition, and that normal developmentally driven shifts in exudation profiles are correlated with changes in the plant-associated microbiome (59, 60). One possible cause of the monoderm enrichment under drought that is related to cell wall architecture is the overproduction of G3P by the plant host (Fig. 6). The absolute enrichment of Actinobacteria observed after short periods of drought in our inoculation experiments, in which roots are only exposed to a single microbial strain, is consistent with some form of positive selection. While the drought-induced increases in the production of most of the other amino acids and sugars do not offer a clear explanation for the specific increase in actinobacterial lineages, G3P is an important precursor to peptidoglycan biosynthesis, which is the primary component of the bacterial cell wall. Given the increased average thickness of the peptidoglycan layer in monoderms (57), it has been suggested that the availability of G3P may act as a positive regulator of growth rate in monoderm lineages (61). We noted that among the ABC transporters and metabolic pathways associated with G3P in the rhizosphere microbiome that showed increased activity during drought, the majority of these (98%) belonged specifically to Actinobacteria. Based on these results, it is conceivable that overproduction of G3P within the plant root may lead to exudation of this metabolite into the rhizosphere and apoplast, where it is utilized by Actinobacteria to foster their growth.
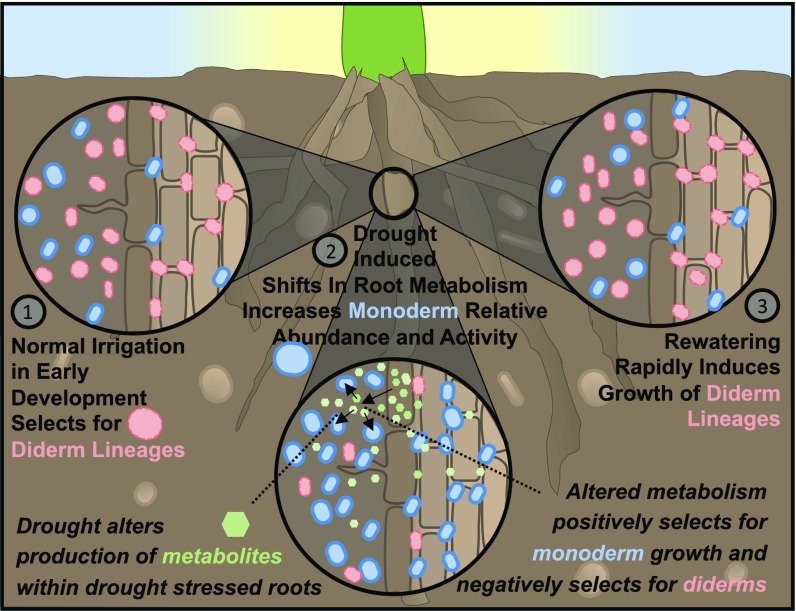
Fig. 6.
Proposed model for selection of monoderm lineages during drought in the root-associated microbiome. (1) Early plant root development selects for diderm lineages under normal irrigation. (2) Drought induces shifts in plant root metabolism, including increases in a range of carbohydrates, secondary metabolites, and amino acids. These shifts lead to exudation of these and possibly other metabolites, which, in turn, supports growth of specific lineages. Additionally, negative selection through an as yet unknown mechanism causes decreases in the absolute abundance of all bacteria, but with greater selection against cells with diderm cell wall characteristics. (3) Rewatering leads to a release from metabolite-mediated and other unknown selective pressures, allowing for rapid growth of diderms and a return to the pattern of root microbiome development and activity observed under control conditions.
However, we expect that positive selective pressures alone are unlikely to fully explain the relative increase in monoderm abundance for two reasons. First, the increase in G3P-related transcripts was only observed in Actinobacteria, and thus cannot account for the increase in relative abundance of other monoderm lineages (namely, Firmicutes and Chloroflexi). Second, our experimental evidence from the field with native soil communities suggests that while drought leads to greater absolute losses in diderm lineages compared with monoderm lineages, decreases are, in fact, observed for all bacterial lineages we tested. Based on these observations, it is plausible that the enrichment in monoderms is driven, in part, by negative selection of diderm lineages by compounds produced by the host or by other microbes. Actinobacteria themselves are known to produce a wide variety of antimicrobials, and many of these compounds are induced after specific environmental cues (62). Indeed, we observe that drought leads to increased activity of polyketide synthetase and cytochrome P450 genes (SI Appendix, Table S14), both of which are involved in the production of secondary metabolites such as antibiotics (63). However, the majority of antibiotics work by inhibiting the biosynthesis of cell walls and typically have larger negative impacts on the growth of monoderm lineages, which lack an outer membrane to protect them (64). Alternatively, plant tissues are known to undergo significant increases in reactive oxygen species (ROS) under drought, and monoderms have been shown to exhibit greater tolerance to the presence of ROS than diderms (50). The susceptibility of the outer membrane of diderms to oxidative damage and the relatively greater protection afforded by the thicker cell wall in monoderms (28) have been suggested as potential causes of this phenomenon (52). In support of this hypothesis, the rapid increase in diderms following rewatering seems to us to suggest relief from a negative selection with a relatively short half-life, and ROS typically degrade within short periods of time (65). Further experimentation with plants impaired in or enhanced for ROS scavenging could help to test this hypothesis.
A recent study on the effect of drought on the microbiomes of 30 angiosperms has observed a significant correlation between the relative abundance of the genus Streptomyces in plant roots and host drought tolerance (38). In accordance with this, we observed that drought stress not only increases colonization of Streptomyces isolates within the root and rhizosphere but that this colonization can increase root development during drought. In our study, inoculation was not correlated with shifts in shoot fresh weight (SI Appendix, Fig. S31), demonstrating a microbially induced increase in relative root-to-shoot resource allocation that has been shown in some studies to be beneficial for overall fitness during drought stress (66). However, additional experimentation with a wide range of monoderm lineages, and across a variety of soil environments, will be necessary to establish the extent to which these trends are generalizable.
Conclusions
Manipulation of the crop microbiome represents a promising strategy for addressing many of the challenges drought poses to agricultural productivity. However, to capitalize on this potential, an improved understanding of the causes, consequences, and timing of drought-induced shifts in the soil and crop microbiome is needed. This study demonstrates that drought significantly restructures root microbiome composition and functionality, especially in early development, and highlights the importance of temporal sampling when studying plant-associated microbiomes.
Material and Methods
Microbial community analysis was conducted as in the study by Naylor et al. (13) on soil, rhizosphere, and root endosphere samples of two sorghum cultivars collected from field-grown plants in Parlier, California (36.6008°N, 119.5109°W). Metatranscriptome profiling, metabolomics, qPCR, HPLC analysis, bacterial isolation and conjugation, inoculation experiments, and confocal microscopy were performed as described in SI Appendix, Material and Methods. A combined assembly of the metatranscriptome can be accessed and downloaded via integrated microbial genomes (IMG)/expert review (ER) (https://img.jgi.doe.gov/) using the IMG ID 3300017790.
Supplementary Material
Acknowledgments
We thank Siwen Deng, Judith Owiti, Ilsa Zhang, Rachel Bosnyak, Kristy Cheng, Daniel Caddell, Yixin Cai, and Heidi Wipf for sample collection and DNA preparation; Vineetha Zacharia for sharing the mCherry plasmid; and Samuel Leiboff for bioinformatics support. This research was funded, in part, by grants from the Department of Energy (DOE) (DE-FOA-0001207) and US Department of Agriculture (CRIS 2030-21430-008-00D), and through the Joint BioEnergy Institute, a facility sponsored by the DOE (Contract DE-AC02-05CH11231) between Lawrence Berkeley National Laboratory and the US Department of Energy. Research was performed using Environmental Molecular Sciences Laboratory, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All scripts used in this study are available at GitHub (https://github.com/dcolemanderr/EPICON-Drought-Study). The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, www.ncbi.nlm.nih.gov/ (accession nos. PRJNA435634, PRJNA435642, and PRJNA435643).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717308115/-/DCSupplemental.
References
- 1.Nie M, Bell C, Wallenstein MD, Pendall E. Increased plant productivity and decreased microbial respiratory C loss by plant growth-promoting rhizobacteria under elevated CO2. Sci Rep. 2015;5:9212. doi: 10.1038/srep09212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goh C-H, Veliz Vallejos DF, Nicotra AB, Mathesius U. The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J Chem Ecol. 2013;39:826–839. doi: 10.1007/s10886-013-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman K, van der Heijden MG, Roussely-Provent V, Walser J-C, Schlaeppi K. Deciphering composition and function of the root microbiome of a legume plant. Microbiome. 2017;5:2. doi: 10.1186/s40168-016-0220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki A, et al. Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS One. 2016;11:e0164533. doi: 10.1371/journal.pone.0164533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner MR, et al. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun. 2016;7:12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitter EK, de Freitas JR, Germida JJ. Bacterial root microbiome of plants growing in oil sands reclamation covers. Front Microbiol. 2017;8:849. doi: 10.3389/fmicb.2017.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santhanam R, et al. Specificity of root microbiomes in native-grown Nicotiana attenuata and plant responses to UVB increase Deinococcus colonization. Mol Ecol. 2017;26:2543–2562. doi: 10.1111/mec.14049. [DOI] [PubMed] [Google Scholar]
- 10.Zgadzaj R, et al. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc Natl Acad Sci USA. 2016;113:E7996–E8005. doi: 10.1073/pnas.1616564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estendorfer J, et al. The influence of land use intensity on the plant-associated microbiome of Dactylis glomerata L. Front Plant Sci. 2017;8:930. doi: 10.3389/fpls.2017.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreote FD, Gumiere T, Durrer A. Exploring interactions of plant microbiomes. Sci Agric. 2014;71:528–539. [Google Scholar]
- 13.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017;11:2691–2704. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos-Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio. 2017;8:e00764-17. doi: 10.1128/mBio.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production. Nature. 2016;529:84–87. doi: 10.1038/nature16467. [DOI] [PubMed] [Google Scholar]
- 16.Schwalm CR, et al. Global patterns of drought recovery. Nature. 2017;548:202–205. doi: 10.1038/nature23021. [DOI] [PubMed] [Google Scholar]
- 17.Ngumbi E, Kloepper J. Bacterial-mediated drought tolerance: Current and future prospects. Appl Soil Ecol. 2016;105:109–125. [Google Scholar]
- 18.Rolli E, et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol. 2015;17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 19.Marulanda A, Barea J-M, Azcón R. Stimulation of plant growth and drought tolerance by native microorganisms (AM fungi and bacteria) from dry environments: Mechanisms related to bacterial effectiveness. J Plant Growth Regul. 2009;28:115–124. [Google Scholar]
- 20.Coleman-Derr D, et al. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016;209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kron AP, Souza GM, Ribeiro RV. Water deficiency at different developmental stages of Glycine max can improve drought tolerance. Bragantia. 2008;67:43–49. [Google Scholar]
- 22.Angel R, et al. The root-associated microbial community of the world’s highest growing vascular plants. Microb Ecol. 2016;72:394–406. doi: 10.1007/s00248-016-0779-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh J, Byrd AL, Park M, Kong HH, Segre JA. NISC Comparative Sequencing Program Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89, and erratum (2016) 17:117. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redford AJ, Fierer N. Bacterial succession on the leaf surface: A novel system for studying successional dynamics. Microb Ecol. 2009;58:189–198. doi: 10.1007/s00248-009-9495-y. [DOI] [PubMed] [Google Scholar]
- 26.Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngara R, Ndimba BK. Model plant systems in salinity and drought stress proteomics studies: A perspective on Arabidopsis and Sorghum. Plant Biol (Stuttg) 2014;16:1029–1032. doi: 10.1111/plb.12247. [DOI] [PubMed] [Google Scholar]
- 28.Tocheva EI, Ortega DR, Jensen GJ. Sporulation, bacterial cell envelopes and the origin of life. Nat Rev Microbiol. 2016;14:535–542. doi: 10.1038/nrmicro.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Shaughnessy SA, Evett SR, Colaizzi PD, Howell TA. A crop water stress index and time threshold for automatic irrigation scheduling of grain sorghum. Agric Water Manage. 2012;107:122–132. [Google Scholar]
- 30.DeJonge KC, Taghvaeian S, Trout TJ, Comas LH. Comparison of canopy temperature-based water stress indices for maize. Agric Water Manage. 2015;156:51–62. [Google Scholar]
- 31.Hanin M, et al. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal Behav. 2011;6:1503–1509. doi: 10.4161/psb.6.10.17088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halder T, Upadhyaya G, Ray S. YSK2 type dehydrin (SbDhn1) from Sorghum bicolor showed improved protection under high temperature and osmotic stress condition. Front Plant Sci. 2017;8:918. doi: 10.3389/fpls.2017.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eriksson S, Eremina N, Barth A, Danielsson J, Harryson P. Membrane-induced folding of the plant stress dehydrin Lti30. Plant Physiol. 2016;171:932–943. doi: 10.1104/pp.15.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood AJ, Goldsbrough PB. Characterization and expression of dehydrins in water-stressed Sorghum bicolor. Physiol Plant. 1997;99:144–152. [Google Scholar]
- 35.Fierer N, Nemergut D, Knight R, Craine JM. Changes through time: Integrating microorganisms into the study of succession. Res Microbiol. 2010;161:635–642. doi: 10.1016/j.resmic.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Morton JT, et al. Uncovering the horseshoe effect in microbial analyses. mSystems. 2017;2:e00166-16, and erratum (2018) 3:e00006-18. doi: 10.1128/mSystems.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitzpatrick CR, et al. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci USA. 2018;115:E1157–E1165. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keijser BJF, et al. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J Bacteriol. 2007;189:3624–3634. doi: 10.1128/JB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocha DJP, Santos CS, Pacheco LGC. Bacterial reference genes for gene expression studies by RT-qPCR: Survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685–693. doi: 10.1007/s10482-015-0524-1. [DOI] [PubMed] [Google Scholar]
- 41.Liao XL, Wen GQ, Liu QL, Wu XYLMX, Pan YZ. 2016. The drought-stress response of a drought resistant impatiens processed in PEG-6000 solution in a simulation test. arXiv:1611.04425 Q-Bio.
- 42.Vellend M. Conceptual synthesis in community ecology. Q Rev Biol. 2010;85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 43.Descamps-Julien B, Gonzalez A. Stable coexistence in a fluctuating environment: An experimental demonstration. Ecology. 2005;86:2815–2824. [Google Scholar]
- 44.Adler PB, HilleRisLambers J, Kyriakidis PC, Guan Q, Levine JM. Climate variability has a stabilizing effect on the coexistence of prairie grasses. Proc Natl Acad Sci USA. 2006;103:12793–12798. doi: 10.1073/pnas.0600599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naylor D, Coleman-Derr D. Drought stress and root-associated bacterial communities. Front Plant Sci. 2018;8:2223. doi: 10.3389/fpls.2017.02223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hueso S, García C, Hernández T. Severe drought conditions modify the microbial community structure, size and activity in amended and unamended soils. Soil Biol Biochem. 2012;50:167–173. [Google Scholar]
- 47.Alster CJ, German DP, Lu Y, Allison SD. Microbial enzymatic responses to drought and to nitrogen addition in a southern California grassland. Soil Biol Biochem. 2013;64:68–79. [Google Scholar]
- 48.Hartmann M, et al. A decade of irrigation transforms the soil microbiome of a semi-arid pine forest. Mol Ecol. 2017;26:1190–1206. doi: 10.1111/mec.13995. [DOI] [PubMed] [Google Scholar]
- 49.Bouskill NJ, et al. Belowground response to drought in a tropical forest soil. I. Changes in microbial functional potential and metabolism. Front Microbiol. 2016;7:525. doi: 10.3389/fmicb.2016.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shade A, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell BJ, Kirchman DL. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J. 2013;7:210–220. doi: 10.1038/ismej.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao S, et al. Micromonospora parathelypteridis sp. nov., an endophytic actinomycete with antifungal activity isolated from the root of Parathelypteris beddomei (Bak.) Ching. Int J Syst Evol Microbiol. 2017;67:268–274. doi: 10.1099/ijsem.0.001614. [DOI] [PubMed] [Google Scholar]
- 53.Shashkov AS, et al. Teichuronic acid of the cell wall of Actinoplanes brasiliensis. Biochim Biophys Acta. 1994;1201:333–338. doi: 10.1016/0304-4165(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 54.Harris WF, Scriven LE. Function of dislocations in cell walls and membranes. Nature. 1970;228:827–829. doi: 10.1038/228827a0. [DOI] [PubMed] [Google Scholar]
- 55.Dworkin M, Kaiser D. 1993 Myxobacteria II (American Society for Microbiology, Washington, DC). Available at agris.fao.org/agris-search/search.do?recordID=US201300168615. Accessed September 21, 2017.
- 56.Kristjansson JK, Stetter KO. Thermophilic Bacteria. CRC; Boca Raton, FL: 1992. [Google Scholar]
- 57.Gupta RS. Origin of diderm (Gram-negative) bacteria: Antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek. 2011;100:171–182. doi: 10.1007/s10482-011-9616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutcliffe IC. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010;18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem. 2013;288:4502–4512. doi: 10.1074/jbc.M112.433300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaparro JM, et al. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One. 2013;8:e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemieux MJ, Huang Y, Wang D-N. Glycerol-3-phosphate transporter of Escherichia coli: Structure, function and regulation. Res Microbiol. 2004;155:623–629. doi: 10.1016/j.resmic.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 62.Behie SW, Bonet B, Zacharia VM, McClung DJ, Traxler MF. Molecules to ecosystems: Actinomycete natural products in situ. Front Microbiol. 2017;7:2149. doi: 10.3389/fmicb.2016.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aparicio JF, Caffrey P, Gil JA, Zotchev SB. Polyene antibiotic biosynthesis gene clusters. Appl Microbiol Biotechnol. 2003;61:179–188. doi: 10.1007/s00253-002-1183-5. [DOI] [PubMed] [Google Scholar]
- 64.Schaalje J. 2018 Medical terminology: Gram positive vs. Gram negative bacteria. Available at info.achs.edu/blog/bid/282924/Medical-Terminology-Gram-Positive-vs-Gram-Negative-Bacteria. Accessed April 6, 2018.
- 65.Taverne YJHJ, Bogers AJJC, Duncker DJ, Merkus D. Reactive oxygen species and the cardiovascular system. Oxid Med Cell Longev. 2013;2013:862423. doi: 10.1155/2013/862423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang Y, Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci. 2015;72:673–689. doi: 10.1007/s00018-014-1767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kasim WA, et al. Control of drought stress in wheat using plant-growth-promoting bacteria. J Plant Growth Regul. 2013;32:122–130. [Google Scholar]