Significance
Medical castration or interference with androgen receptor (AR) function is the principal treatment for advanced prostate cancer. However, progression is universal, and therapies following the emergence of castration resistance do not offer durable control of the disease. Lysine-specific demethylase 1 (LSD1) is an important regulator of gene expression, including in cancer. Here, we show that LSD1 is highly expressed in tumors of patients with lethal castration-resistant prostate cancer (CRPC) and that LSD1 promotes AR-independent survival in CRPC cells in a noncanonical, demethylase-independent manner. We determined that the drug SP-2509 acts as an allosteric inhibitor of LSD1–blocking demethylase-independent functions. Our demonstration of tumor suppression with this inhibitor in CRPC preclinical models provides the rationale for clinical trials with LSD1 inhibitors.
Keywords: LSD1, prostate cancer, castration resistance, ZNF217
Abstract
Medical castration that interferes with androgen receptor (AR) function is the principal treatment for advanced prostate cancer. However, clinical progression is universal, and tumors with AR-independent resistance mechanisms appear to be increasing in frequency. Consequently, there is an urgent need to develop new treatments targeting molecular pathways enriched in lethal prostate cancer. Lysine-specific demethylase 1 (LSD1) is a histone demethylase and an important regulator of gene expression. Here, we show that LSD1 promotes the survival of prostate cancer cells, including those that are castration-resistant, independently of its demethylase function and of the AR. Importantly, this effect is explained in part by activation of a lethal prostate cancer gene network in collaboration with LSD1’s binding protein, ZNF217. Finally, that a small-molecule LSD1 inhibitor―SP-2509―blocks important demethylase-independent functions and suppresses castration-resistant prostate cancer cell viability demonstrates the potential of LSD1 inhibition in this disease.
Metastatic prostate cancer is incurable and is the second leading cause of cancer-related death in the United States (1). Androgen-deprivation therapy (ADT), or castration, that disables the function of the androgen receptor (AR) is the principal treatment for men with metastatic prostate cancer. However, cancer control with ADT is of limited duration, and in many men there is no benefit at all, underscoring the urgent need to identify new targets and treatments for lethal prostate cancer (2, 3).
Several recent studies have clarified that the transcriptomes of lethal cancers, including prostate cancers, are enriched with cell-cycle, mitosis, and embryonic stem cell (ESC) identity ontologic categories or gene sets that contribute to tumor progression (4–7). Moreover, recent work with the master regulator (MR) inference algorithm (MARINa) identified transcription factors that are activated in lethal prostate cancer tumor models and patient samples (8). However, few of these MRs are directly targetable, demonstrating the importance of identifying upstream activators of these MRs and lethal prostate cancer gene networks. Targets of potential interest include proteins that are most highly expressed in lethal prostate cancer and that appear to be necessary and sufficient to activate lethal prostate cancer gene networks. Here we report that the lysine-specific demethylase 1 (LSD1) meets that standard.
LSD1 (also known as “KDM1A”) is a histone demethylase and a regulator of gene expression in ESCs, hematopoietic stem cells, and cancer (9–17). LSD1 was initially identified as a transcriptional repressor via demethylation of active histone H3 marks [monomethyl lysine 4 (H3K4me1) and di-methyl lysine 4 (H3K4me2)] (18). Prior work also suggested that LSD1-induced gene activation occurs through histone demethylation of monomethyl lysine 9 (H3K9me1) and di-methyl lysine 9 (H3K9me2), including at AR target genes (19–21), even though these marks are not direct LSD1 substrates in biochemical reactions (18). However, LSD1’s role in prostate cancer, particularly in lethal prostate cancer, and the mechanisms that contribute to LSD1’s effects are not well appreciated.
In this report, we describe a distinct role of LSD1 as a driver of proliferation and survival of prostate cancer cells independently of the AR. Specifically, LSD1 activates a lethal prostate cancer gene network comprised of cell-cycle, mitosis, and ESC identity genes and MRs that are enriched in lethal prostate cancers—genes that we find not to be activated by androgens. Importantly, our global epigenomic studies and biochemical studies demonstrate that LSD1’s demethylase function is dispensable for activating this lethal prostate cancer gene network. Rather, LSD1 activates this network and promotes prostate cancer cell survival in collaboration with its binding protein ZNF217. Finally, a reversible LSD1 inhibitor called “SP-2509” blocks key demethylase-independent functions of LSD1, demonstrating a promising approach to target LSD1 in prostate cancer.
Results
LSD1 Is Up-Regulated in the Majority of Castration-Resistant Prostate Cancer and Promotes AR-Independent Prostate Cancer Cell Survival.
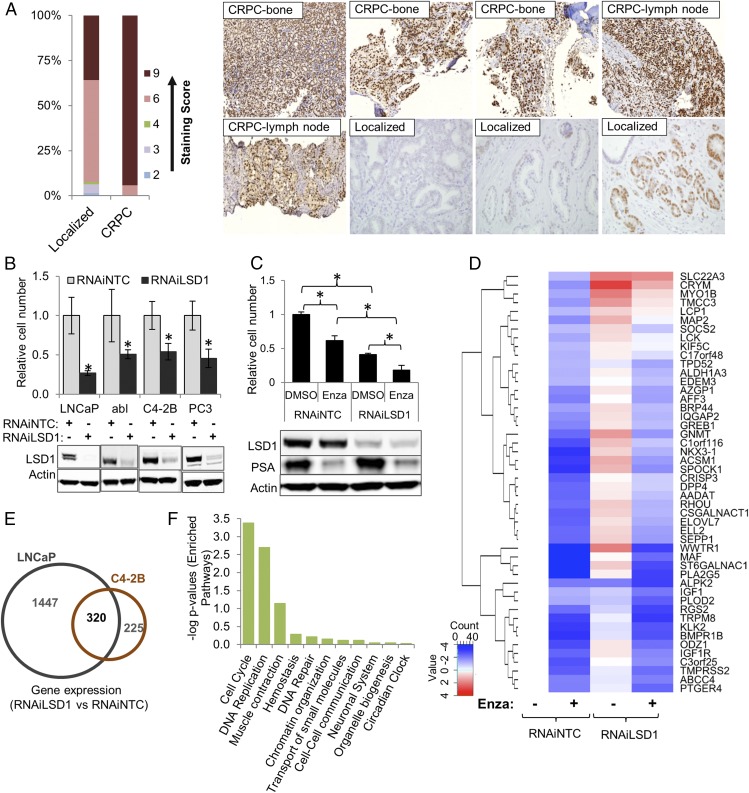
Prior reports demonstrate that LSD1 up-regulation in primary prostate cancers is associated with an increased risk of recurrence (22). However, the frequency of LSD1 up-regulation in metastatic castration-resistant prostate cancer (CRPC) tumors was unknown. Therefore, we quantified LSD1 expression with immunohistochemistry (IHC) in primary and metastatic human CRPC tumors. Intense and diffuse nuclear LSD1 expression was seen in 16 of 17 (94%) of CRPC cases that are described in Fig. S1A but in only 36% of the primary tumors (Fig. 1A). We confirmed that a second antibody showed similar LSD1 staining and that staining was absent from cells depleted of LSD1 by RNAi (Fig. S1 B and C).
Fig. 1.
LSD1 is up-regulated in majority of CRPC samples and promotes AR-independent prostate cancer cell survival by regulating cell-cycle and ESC gene sets. (A, Left) Proportions of intensity scores of LSD1 IHC staining in metastatic human CRPC (n = 17) and castration-naive primary prostate tumors (n = 223) are shown. Median scores of staining intensity are significantly different between the two groups of samples (two-sided P value < 0.001, Wilcoxon rank sum test). (Right) Representative images of LSD1 expression. See also Fig. S1 A and B. (B, Upper) LSD1 RNAi suppresses prostate cancer cell viability. LNCaP cells were grown in medium containing full serum. abl, C4-2B, and PC3 cells were grown in medium containing charcoal-stripped medium. Cells were transfected with 50 nM NTC or LSD1 RNAi oligonucleotides. Cell number was determined 6 d later with the Trypan Blue exclusion method (n = 3). *P < 0.01 for RNAiLSD1 versus RNAiNTC, two-tailed unpaired t test. (Lower) Immunoblotting was performed with the indicated antibodies. See also Fig. S1C. (C, Upper) Combined LSD1 suppression and enzalutamide (Enza) treatment is more effective than either intervention alone. C4-2B cells were transfected with 50 nM of NTC or LSD1 RNAi oligonucleotides grown in full serum for 72 h. Then, DMSO vehicle or 10 µM enzalutamide was added to the medium. Cell number was quantified 72 h later with the Trypan Blue exclusion method (n = 3). Both RNAi and enzalutamide treatments are significant main effects (P value < 0.001; two-way ANOVA); significant pairwise comparisons are indicated in the graph (*P < 0.05). (Lower) Immunoblotting was performed with the indicated antibodies. (D) Gene-expression changes after RNAiLSD1, enzalutamide treatment, or the combination. C4-2B cells were transfected with 50 nM of NTC or LSD1 RNAi oligonucleotides for 72 h. Then, DMSO vehicle or 10 µM enzalutamide was added to the medium. Cells were harvested 24 h later. Extracted RNA was reverse-transcribed into cDNA, which was then applied to a TaqMan qRT-PCR array comprised of randomly selected androgen-activated genes from prior microarrays after DHT stimulation in C4-2B cells (n = 3) (see also Dataset S1 and Fig. S1D). Genes underwent unsupervised clustering, resulting in the dendrogram on the left side of the figure. The log2 expression level is depicted relative to the vehicle/RNAiNTC treatment. Blue represents lower relative expression, and red represents higher relative expression. (E) Venn diagram showing integration of gene-expression profiles based on microarray results in LNCaP and C4-2B cells after introducing NTC or LSD1 RNAi (n = 3). There was significant overlap among differentially expressed genes (up- and down-regulated) after LSD1 suppression in LNCaP and C4-2B cells (OR = 26.6, P < 0.0001) with 320 genes conserved between the two cell lines. (F) Pathways enriched in the REACTOME analysis of the conserved 320 differentially expressed genes (Dataset S2). The cell-cycle pathway was the most enriched. See also Figs. S1 and S2 and Datasets S2 and S3.
To further examine the role of LSD1 in prostate cancer cell survival, we suppressed LSD1 with RNAi in multiple cell lines, including LNCaP cells and LNCaP CRPC derivatives (C4-2B and abl cells) that proliferate well over the long term in androgen-depleted conditions (23, 24). In each case, LSD1 suppression reduced cell viability (Fig. 1B and Fig. S1D). Examination of the Project Achilles Database (https://portals.broadinstitute.org/achilles) also demonstrated that LSD1 suppression reduced the viability of VCaP cells and AR-null NCI-H660 cells, corroborating LSD1’s importance for prostate cancer cell survival.
LSD1 is a putative AR cofactor, but we sought to determine if LSD1 promotes AR-independent survival. In keeping with this notion, LSD1 suppression in AR-null PC3 CRPC cells reduced cell viability (Fig. 1B and Fig. S1D). To further demonstrate LSD1’s AR independence, we suppressed LSD1 alone or in combination with the AR antagonist enzalutamide in C4-2B cells. Either LSD1 RNAi or enzalutamide treatment reduced cell viability (Fig. 1C). However, the combination was more effective (Fig. 1C). Importantly, LSD1 RNAi did not reduce the expression of KLK3 or most other androgen-activated AR target genes we examined (Fig. 1 C and D). Indeed, LSD1 RNAi increased the expression of a subset of these AR targets, many of which are markers of prostatic epithelial differentiation (Fig. 1D). Finally, we grew C4-2B cells in androgen-depleted serum and suppressed LSD1 with RNAi or stimulated with the AR ligand dihydrotestosterone (DHT) followed by gene-expression microarrays. Virtually none of the DHT-activated genes was shared with the LSD1-activated genes (Fig. S1E and Dataset S1), further demonstrating an important AR-independent role for LSD1 in prostate cancer progression.
LSD1 Activates the Expression of Functionally Important Target Genes That Are Enriched in Lethal Prostate Tumors.
To identify target genes that contribute to LSD1’s effects on promoting prostate cancer cell survival, we compared microarray results after suppressing LSD1 in LNCaP or C4-2B cells. There were 320 common differentially expressed genes between these cell lines (Fig. 1E), and there was significant overlap among differentially expressed genes (up- and down-regulated) [odds ratio (OR) = 26.6, P < 0.0001]. The overlap was even stronger for LSD1-activated genes (down-regulated after LSD1 RNAi) (OR = 63.8, P < 0.0001). In examining the conserved LSD1-activated target genes, cell-cycle and mitosis―gene sets that are enriched in lethal prostate cancer patient tumors (6)―were the top enriched Reactome pathways in each cell line (Fig. 1F and Dataset S2).
LSD1 is a key regulator of gene expression in ESCs, and ESC gene sets are also enriched in lethal cancers (4, 5, 7, 12, 13, 25). Enrichment analysis determined that all but one of these previously described lethal cancer ESC gene sets (4, 5, 7, 25) were enriched among the LSD1-activated genes (Fig. S1F and Dataset S3). Enrichment remained significant even after genes with a cell-cycle functional annotation were removed (Fig. S1F), demonstrating that enrichment of these gene sets is not driven by proliferation-induced changes in gene expression. Importantly, RNAi-mediated suppression of many of these target genes recapitulated the effect of LSD1 RNAi (Fig. S2). In contrast to the LSD1-activated genes, adult tissue stem cell gene sets of differentiated cell types were enriched among the conserved LSD1-repressed target genes (Fig. S1F and Dataset S3).
LSD1 Regulates Gene Expression Independently of Its Canonical Demethylase Function.
LSD1 is a histone demethylase. However, it was not known whether LSD1’s demethylase function was critical for LSD1-induced gene regulation—particularly for genes comprising lethal cancer gene sets—and for the survival of prostate cancer cells.
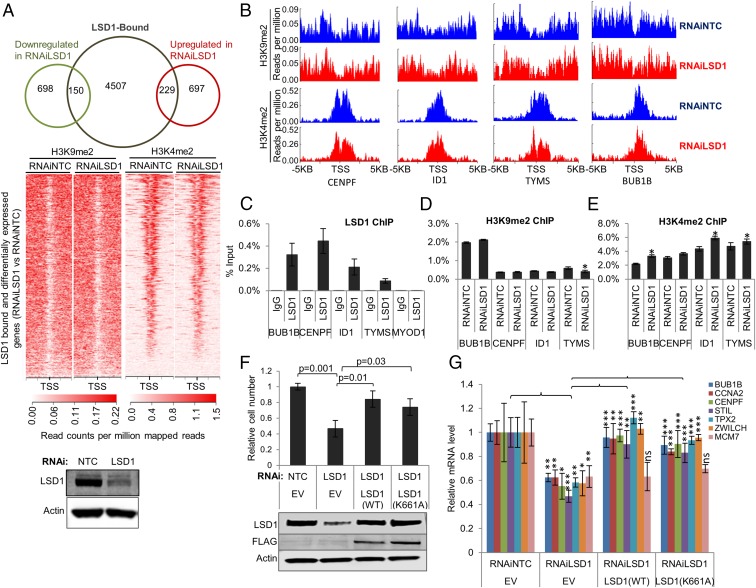
To clarify this, we performed an integrative analysis of the genes that were differentially expressed with LSD1 RNAi in LNCaP cells and published LSD1 ChIP-sequencing (ChIP-seq) (21) in LNCaP cells. Only a minority of the differentially expressed genes were directly LSD1-bound (Fig. 2A). Next, to determine if H3K4me2 and H3K9me2 changes accounted for the gene-expression changes, we suppressed LSD1 with RNAi in LNCaP cells and then performed H3K4me2 or H3K9me2 ChIP-seq. H3K4me2 or H3K9me2 changes were uncommon after LSD1 RNAi for LSD1-bound, differentially expressed genes (Fig. 2 A and B and Fig. S3 A and B). Indeed, decreased H3K4me2 occurred at a higher frequency than increased H3K4me2 on the LSD1-bound promoter regions after LSD1 suppression (Fig. S3 A and B). H3K9me2 changes were virtually absent at the LSD1-bound regions after LSD1 suppression (Fig. 2A and Fig. S3 A and B). One caveat concerning our H3K9me2 ChIP-seq data is that we observed broad peaks for this mark (Fig. 2 A and B), which matches a prior report in a different cell model (26). Genome browser snapshots for specific LSD1-bound regions of target genes are shown in Fig. 2B. Next, we performed ChIP-PCR studies to confirm these results (Fig. 2 C–E). Importantly, like our ChIP-seq results, there were no significant changes in H3K9me2 after LSD1 RNAi (Fig. 2 D and E). We did observe increased H3K4me2 levels at some LSD1-activated genes, although the changes were quite small (Fig. 2E).
Fig. 2.
LSD1 regulates gene expression independently of its demethylase function. (A) Integrative analysis of differentially expressed genes with RNAiLSD1 in LNCaP cells. (Top) The Venn diagram shows the integration of genes down-regulated or up-regulated after LSD1 RNAi with the genes bound by LSD1 in ChIP-seq studies in LNCaP cells published previously (21). (Middle) H3K9me2 and H3K4me2 ChIP-seq peaks [5 kb upstream and downstream of the transcription start site (TSS)] for the genes bound by LSD1 in published ChIP-seq in LNCaP cells (21) and that were differentially expressed after LSD1 RNAi are shown. (Bottom) Immunoblotting was performed to confirm LSD1 suppression with LSD1 RNAi. See also Fig. S3 A and B. (B) Representative genome browser chromosome regions for selected LSD1-bound and activated genes. (C) LSD1 ChIP-PCR showing LSD1 occupancy at selected genes in LNCaP cells. The MYOD1 gene promoter was used as a negative control region (n = 3). See Fig. S3C. (D and E) ChIP-PCR in LNCaP cells showing H3K9me2 (D) or H3K4me2 (E) enrichment after RNAiLSD1 vs. RNAiNTC. Enrichment was calculated as a percentage of input (n = 3). *P < 0.05 for enrichment in RNAiLSD1 vs. RNAiNTC. (F and G) Complementation with an ectopic wild-type or mutant K661A LSD1 expression plasmid abrogates the effect of LSD1 RNAi. LNCaP cells were sequentially transfected with 50 nM NTC or LSD1 RNAi oligonucleotides targeting the 3′ UTR of LSD1 plus RNAi-resistant wild-type LSD1 expression vector, RNAi-resistant K661A catalytically deficient mutant LSD1 expression vector, or empty vector (EV). (F, Upper) Cell viability was determined 96 h later using the Promega Aqueous One MTS assay and was normalized to an initial time point for each condition (n = 3). P values are indicated. (Lower) Immunoblotting was performed with the indicated antibodies. (G) mRNA expression of selected LSD1 target genes was measured using TaqMan assays (n = 3). Data are reported as ± SD. In D–G, a two-tailed unpaired t test was performed; *P < 0.05, **P < 0.01, ***P < 0.001.
LSD1 is also capable of demethylating nonhistone substrates (15, 27, 28). To clarify whether LSD1’s demethylase function was critical for promoting prostate cancer cell survival and the expression of lethal prostate cancer genes, we suppressed endogenous LSD1 with RNAi targeting the 3′ UTR of LSD1 mRNA and then complemented cells with ectopic wild-type LSD1 or with the catalytically deficient K661A mutant LSD1 (29). Overexpression of either construct abrogated the effects of LSD1 RNAi on reducing cell survival or the expression of lethal prostate cancer genes (Fig. 2 F and G), demonstrating the importance the noncanonical demethylase-independent functions of LSD1.
LSD1 Activates an MR Transcriptional Network Activated in Lethal Human Prostate Cancer Patient Tumors.
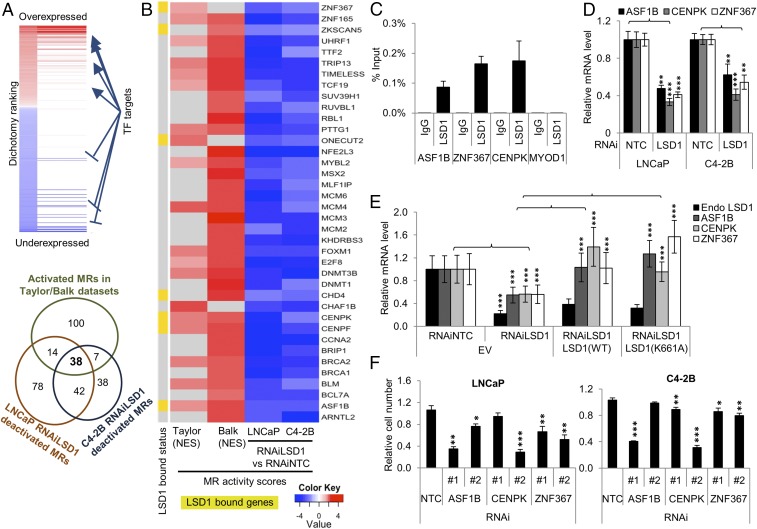
Many of the genes that change in expression with LSD1 RNAi are not directly LSD1-bound (Fig. 2A). To identify key, direct target genes regulated by LSD1 that might contribute to the gene-expression changes, we used MARINa, focusing on differentially expressed genes with LSD1 RNAi in LNCaP and C4-2B cells. We focused on LSD1-activated MRs whose function was predicted to be suppressed with LSD1 RNAi. Next, we integrated the list of LSD1-activated MRs with MRs previously shown to be activated in aggressive human prostate cancer patient datasets by Aytes et al. (8). Thirty-eight MRs met this standard (Fig. 3A). We confirmed that several of these MRs were LSD1 direct targets (Fig. 3 B–D). Importantly, LSD1’s demethylase function was dispensable for activating these MRs’ expression (Fig. 3E and Fig. S4). Notably, RNAi-mediated suppression of several of these MRs recapitulated the effects of LSD1 RNAi, demonstrating these MRs’ importance (Fig. 3F). Finally, we confirmed that the majority of these direct LSD1 MRs were highly expressed in lethal human CRPC samples in an independent dataset (30) not used by Aytes et al., further demonstrating their clinical relevance (Fig. S5).
Fig. 3.
LSD1 activates an MR transcriptional network activated in tumors from patients with lethal human prostate cancer. (A, Upper) Schema representing MARINa analysis in gene-expression datasets. (Lower) Venn diagram showing the overlap between the MRs activated in prostate cancer patient samples in Aytes et al. (8) and the MRs deactivated by RNAiLSD1 based on microarray analysis of LNCaP and C4-2B prostate cancer cell lines. Thirty-eight MRs met that standard. (B) Heatmap showing activity scores of the 38 MRs in two prostate cancer datasets: Taylor (aggressive vs. indolent primary tumors) and Balk (castration-resistant metastatic tumors vs. hormone-naive primary tumors) analyzed by Aytes et al. (8) previously and in RNAiLSD1 vs. RNAiNTC conditions in LNCaP and C4-2B cells. Highlighted in yellow are the MRs that we determined to be LSD1-bound in published LSD1 ChIP-seq data in LNCaP cells (21). NES, normalized enrichment score. (C) LSD1 ChIP-PCR demonstrating LSD1 enrichment at several MR genes. The MYOD1 gene promoter was used as a negative control region (n = 4). See also Fig. S4. (D) qRT-PCR showing the mRNA expression of the MR genes with RNAiLSD1 vs. RNAiNTC in LNCaP and C4-2B cell lines (n = 3). (E) qRT-PCR quantifying expression of the MRs and endogenous (endo) LSD1 after LSD1 suppression with RNAi followed by ectopic expression of either wild-type or mutant K661A LSD1 (n = 3). (F) Cell viability was measured after suppressing each of the three selected MRs, ASF1B, CENPK, and ZNF367. Cells were transfected with 50 nM NTC or two independent RNAi oligonucleotides targeting each MR. Cell viability was measured using CellTiter-Glo 96 h after the transfection (n = 4). Data are reported as ±SD. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired t test. See also Fig. S5.
The LSD1-Binding Protein ZNF217 Contributes to the Activation of Lethal Prostate Cancer Gene Networks.
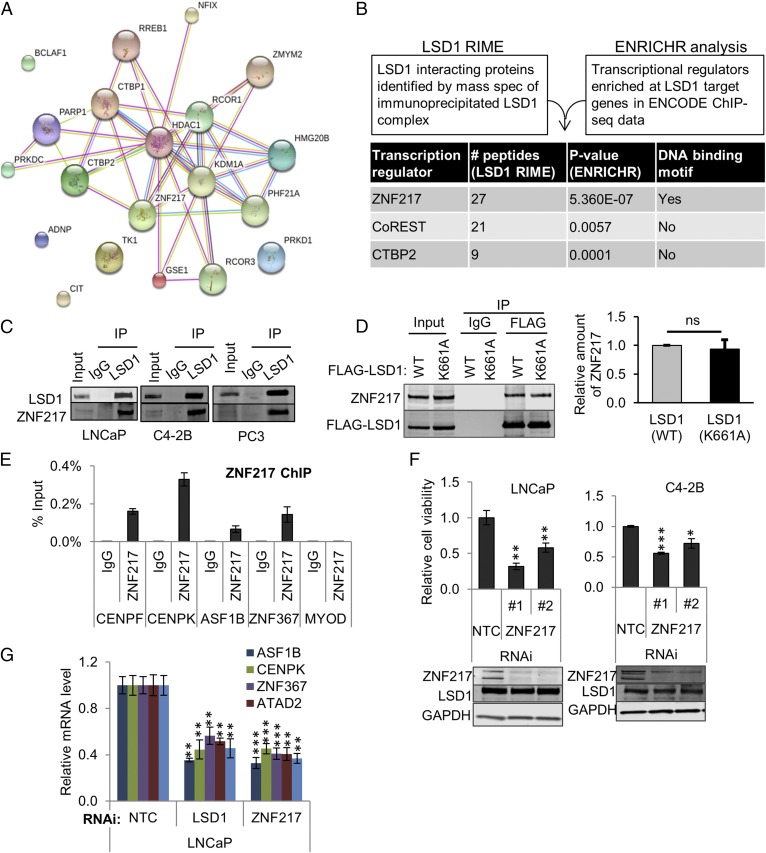
Because we determined that LSD1’s demethylase function was not critical for the regulation of its key target genes, we sought to identify important proteins that might complex and cooperate with LSD1. First, we performed rapid immunoprecipitation (IP) mass spectrometry of endogenous proteins (RIME) (31). RIME identified 72 unique proteins that were enriched with LSD1 IP in C4-2B cells (Fig. 4A and Dataset S4). One of the most enriched proteins was ZNF217 that was previously shown to interact with LSD1 and that has previously been implicated in gene repression (32, 33). Next, we sought to clarify proteins enriched at LSD1 target genes at chromatin. Therefore, we compared the list of LSD1-bound and -regulated target genes in LNCaP cells (Fig. 2A) with ENCODE ChIP-seq results (Fig. 4B and Dataset S5). ZNF217, CoREST, and CTBP2 were the top proteins identified (Fig. 4B).
Fig. 4.
The LSD1-binding protein ZNF217 contributes to activation of lethal CRPC gene networks. (A) Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis of the LSD1-interacting proteins identified by RIME is shown. See Dataset S4. (B) Schema for prioritizing LSD1-interacting proteins. The list of genes bound by LSD1 in LSD1 ChIP-seq (21) that were also differentially expressed after RNAiLSD1 in LNCaP cells (our microarray data) was integrated with transcription factor or chromatin-modifying enzyme ChIP-seq data from multiple cell types through the ENCODE project using the ENRICHR tool (amp.pharm.mssm.edu/Enrichr/) (44). See Dataset S5. RIME and ENRICHR analyses identified three conserved proteins: ZNF217, CoREST, and CTBP2. For each protein, the number of peptides identified in RIME and P value for enrichment in ENRICHR analysis are shown. (C) IP immunoblots showing LSD1’s interaction with ZNF217 in the indicated cell lines. Lysates from LNCaP, C42B, and PC3 cells were immunoprecipitated using an anti-LSD1 antibody or normal IgG. The eluates were analyzed by immunoblots using antibodies against LSD1 and ZNF217. (D) ZNF217 interacts with both wild-type LSD1 and catalytically deficient K661A mutant LSD1. LNCaP cells overexpressing wild-type FLAG-LSD1 (WT) or mutant FLAG-LSD1 (K661A) were subjected to affinity enrichment using immobilized anti-FLAG antibody or control IgG. The eluate was immunoblotted for FLAG and ZNF217. The intensity of the coimmunoprecipitated ZNF217 band was normalized to the immunoprecipitated FLAG-LSD1 band intensity Three independent immunoprecipitations were performed. (E) ZNF217 binds to LSD1-bound regions for MRs. ChIP-PCR showing the enrichment of ZNF217 at LSD1-bound regions of specific MR target genes. Enrichment of ZNF217 or IgG was calculated as percentage of input. The MYOD1 gene promoter was used as a negative control region (n = 4). (F, Upper) Cell viability after ZNF217 suppression. LNCaP and C4-2B cells were transfected with 50 nM NTC RNAi or two independent RNAi oligonucleotides targeting ZNF217. Cell viability was measured using the CellTiter-Glo assay 96 h after transfection (n = 4). (Lower) Immunoblotting was used to quantify ZNF217 and LSD1. (G) qRT-PCR measuring the effect of RNAiZNF217 vs. RNAiNTC on mRNA expression of LSD1-regulated MR and cell-cycle genes (n = 3). Data are reported as ±SD. *P < 0.05, **P <0.01, ***P < 0.001, two-tailed unpaired t test; ns, no statistical significance. See also Fig. S6.
For follow-up studies, we focused on ZNF217 because it was implicated in our ENCODE ChIP-seq analysis, because it was among the most enriched proteins in RIME, and because its roles in prostate cancer and in the activation of LSD1-dependent pathways were poorly defined. First, we confirmed that LSD1 interacted with ZNF217 in multiple cell lines (Fig. 4C) and that LSD1’s catalytic function was not necessary for its interaction with ZNF217 (Fig. 4D). Next, we confirmed by ChIP that ZNF217 occupied the same promoter regions of important MR target genes (Fig. 4E) that were bound by LSD1 (Fig. 3C). Furthermore, ZNF217 suppression phenocopied the effect of LSD1 suppression in reducing cell survival (Fig. 4F) and target gene expression (Fig. 4G). Importantly, neither ZNF217 nor LSD1 suppression affected the expression of the other protein (Fig. 4F and Fig. S6), suggesting this interaction is not critical for LSD1 or ZNF217 protein stability, unlike other LSD1-binding proteins (34).
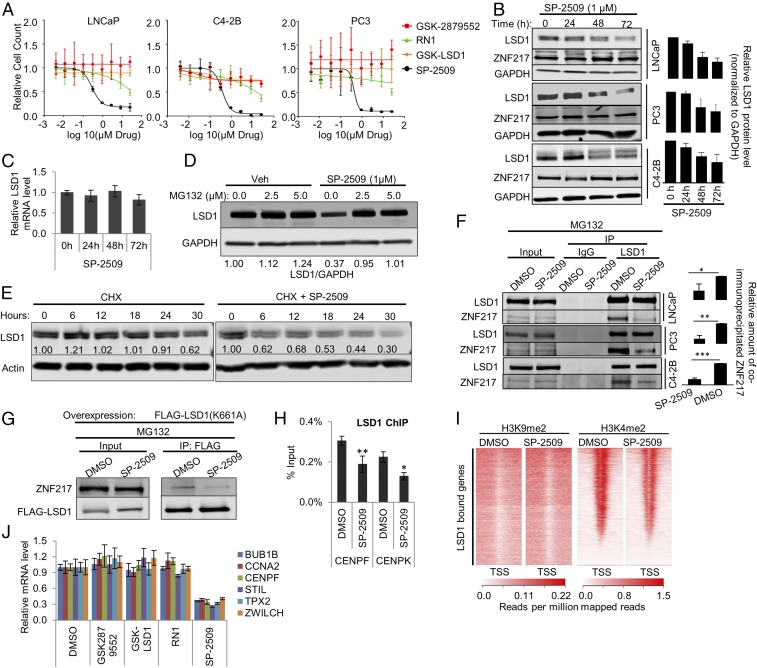
Because we confirmed that LSD1 was a target of interest in lethal prostate cancer, we sought to identify drugs to block its function. Our results demonstrated that LSD1’s demethylase activity was dispensable for promoting prostate cancer cell survival and activation of lethal prostate cancer network genes. Not surprisingly, treatment with catalytic inhibitors that interfere with FAD binding to LSD1―GSK2879552, GSK-LSD1, and RN1 (35, 36)―failed to suppress the survival of cell models sensitive to LSD1 RNAi (Figs. 1C and 5A and Fig. S7A). This matches prior work in prostate cancer with GSK2879552 (35). In contrast, treatment with SP-2509 (37, 38), a reversible inhibitor that does not interfere with FAD binding to LSD1, did reduce cell viability in the models tested (Fig. 5A) and induced apoptosis as indicated by increased caspase activity (Fig. S7B).
Fig. 5.
Treatment with SP-2509, a reversible LSD1 inhibitor, recapitulates the effect of LSD1 RNAi. (A) Cell growth rate of LNCaP, C4-2B, and PC3 cells treated with dose escalation of the LSD1 inhibitors GSK-LSD1, GSK2879552, RN-1, and SP-2509. The relative number of viable cells was measured by staining live cells in situ with Hoechst 33342 and NucGreen Dead dyes (Life Technologies) and capturing one 4× fluorescent field per well using a BioTek Cytation 5 imager (BioTek Instruments, Inc.) Cell-growth curves were fitted by nonlinear regression. See also Fig. S7A. (B, Left) Immunoblot showing a time-dependent effect of SP-2509 on LSD1 and ZNF217 protein levels in LNCaP, C4-2B, and PC3 cell lines. The cells were treated with 1 µM SP-2509. The lysates were prepared at the indicated time points after treatment and were immunoblotted with the indicated antibodies. (Right) The relative intensity of LSD1 bands normalized to the intensity of GAPDH bands in each sample (n = 3). Data are reported as ±SD. (C) qRT-PCR results showing the relative LSD1 mRNA levels after 24, 48, or 72 h treatment with 1 µM SP-2509. The data were normalized to actin mRNA levels. Data are reported as ±SD. (D) Immunoblot showing the effect of MG-132 treatment on SP-2509–induced LSD1 protein depletion. The cells were treated with either DMSO or 1 µM SP-2509. Twenty-four hours later, cells were treated with vehicle or the indicated dose of MG132. Cell lysates were prepared 16 h after MG132 treatment and were immunoblotted using the indicated antibodies. (E) Immunoblot showing the effect of SP-2509 on the half-life of LSD1 protein. LNCaP cells were treated with 10 µg/mL cycloheximide (CHX) alone or in combination with 1 µM SP-2509. The cell lysates were prepared at 0, 6, 12, 18, 24, and 30 h after the treatment and were immunoblotted with the indicated antibodies. (F, Left) SP-2509 disrupts the interaction of LSD1 with ZNF217. The cells were treated with DMSO vehicle or 1 µM SP-2509 for 24 h. Then 2.5 µM MG132 was added to the medium. Lysates were prepared 16 h later for IP analyses using immobilized anti-LSD1 antibody. The eluates were immunoblotted for LSD1 and ZNF217. (Right) The relative intensity of the coimmunoprecipitated ZNF217 band normalized to the intensity of immunoprecipitated FLAG-LSD1 band. Three independent immunoprecipitations/immunoblots were performed. (G) SP-2509 disrupts the interaction of catalytically deficient K661A LSD1 with ZNF217. LNCaP cells were transfected with FLAG-tagged catalytically deficient K661A LSD1 plasmid. Twenty-four hours after the transfection, the cells were treated with either DMSO vehicle or 1 µM SP-2509. Twenty-four hours later 2.5 µM MG132 was added to the medium. Cells lysates were prepared 48 h after SP-2509 treatment. FLAG-tagged LSD1 (K661A) was immunoprecipitated using immobilized antibody against FLAG, and the eluates were analyzed by immunoblots using antibodies against ZNF217 and FLAG. (H) SP-2509 depletes LSD1 from target gene promoters. LNCaP cells were treated with DMSO vehicle or 1 µM SP-2509. LSD1 ChIP-PCR was performed 24 h after the treatment. Enrichment was calculated as percentage of input (n = 4). The data are reported as ±SD. (I) H3K4me2 and H3K9me2 ChIP-seq peaks after SP-2509 are shown around the TSS of the genes previously shown to be bound by LSD1 (21). (J) qRT-PCR results for LSD1-regulated target genes after 72 h of treatment with the LSD1 inhibitors GSK-LSD1, GSK2879552, RN-1, and SP-2509. In B, C, F, H, and J, the data are reported as ±SD; *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired t test.
To understand SP-2509’s unique effects, we performed in silico computational docking studies with LSD1 and SP-2509. Our analysis showed that SP-2509 docks favorably into the H3 pocket region of LSD1 adjacent to the basal helix Sα1 with a docking score of −10.82, suggesting that SP-2509 binds the same region occupied by a previously described substrate-like peptide inhibitor (Fig. S7 C and D) (39). Interestingly, the H3 pocket of LSD1 that interacts with SP-2509 functions as an allosteric site and regulates the rotation of the amino oxidase domain with respect to the tower domain (40). This suggests that SP-2509 may act as an allosteric inhibitor by binding to LSD1.
We next sought to understand the demethylase-independent LSD1 functions blocked by SP-2509. First, SP-2509 treatment alone reduced LSD1 protein expression in a time-dependent manner without deceasing LSD1 RNA (Fig. 5 B and C and Fig. S7E). That LSD1 protein depletion was reversed by cotreatment with a proteasome inhibitor suggests that SP-2509 leads to LSD1 protein instability (Fig. 5D). Further, SP-2509 reduced the half-life of LSD1 measured after cycloheximide treatment, supporting the conclusion that SP-2509 treatment reduces LSD1 protein stability (Fig. 5E). We next examined LSD1’s interaction with ZNF217. SP-2509 treatment blocked the LSD1–ZNF217 interaction, an effect not achievable with catalytic inhibitors (Fig. 5F and Fig. S7F). Importantly, this effect was also observed on the interaction of the ectopic K661A mutant LSD1 with ZNF217 (Fig. 5G).
To confirm whether LSD1 or ZNF217 was responsible for the antitumor effects of SP-2509, we introduced nontargeted control (NTC), LSD1, or ZNF217 RNAi and treated cells with SP-2509. Depletion of ZNF217 or LSD1 abrogated the antitumor activity of SP-2509, suggesting that interference with the LSD1–ZNF217 complex contributes to the antitumor effects of this drug (Fig. S7G). Further, we determined the effect of SP-2509 on LSD1 chromatin occupancy and histone methylation. SP-2509 treatment depleted LSD1 from chromatin without changing levels of H3K4me2 and H3K9me2, recapitulating the effects of LSD1 RNAi (Fig. 5 H and I and Fig. S7 H and I). Importantly, only SP-2509 reduced the expression of genes from the lethal prostate cancer gene network that LSD1 activates independently of its demethylase function (Fig. 5J).
To determine the global effect of SP-2509 on gene-expression changes, we performed RNA-sequencing (RNA-seq) in LNCaP, C4-2B, and PC3 cells after SP-2509 treatment. There were 591 differentially expressed genes shared across these cell lines after SP-2509 treatment, and 507 genes showed coordinate expression changes (same direction of change) across all three cell lines (Fig. S8A). Similar to LSD1 RNAi, cell-cycle and ESC gene sets of lethal cancer (4, 5, 7, 25) were enriched among the conserved LSD1-activated transcripts (the transcripts down-regulated with SP-2509 treatment) (Fig. S8B). As was the case with LSD1 RNAi, the ESC signatures remained intact even after subtracting cell-cycle genes from the data analysis. These results further demonstrate that SP-2509 recapitulates the effects of LSD1 suppression with RNAi.
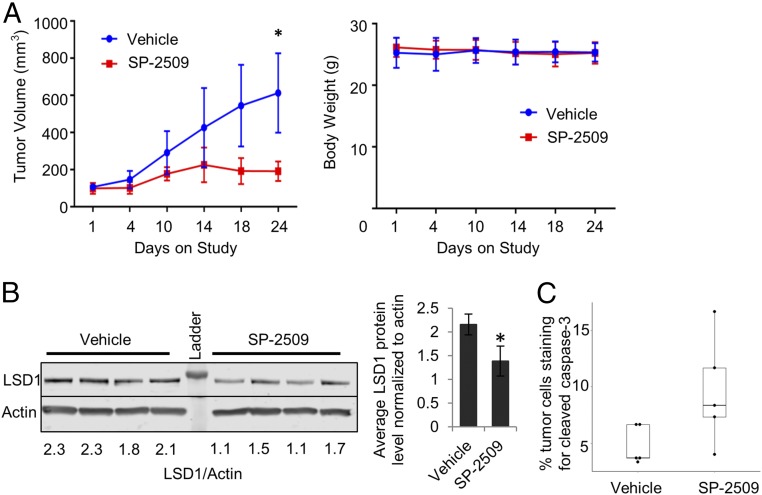
Finally, to determine the antitumor activity of SP-2509 in vivo, we treated AR-independent PC3 CRPC xenografts implanted in mice. SP-2509 significantly reduced tumor growth without changes in animal weight (Fig. 6A). Consistent with in vitro experiments (Fig. 5B), SP-2509–treated tumors had lower levels of LSD1 protein (Fig. 6B) and increased levels of caspase-3 staining vs. control tumors (Fig. 6C).
Fig. 6.
SP-2509 inhibits tumor growth in vivo. (A) Tumor growth of PC3 xenografts was measured as a function on time. Tumor volume (Left) and body weight (Right) are shown as mean ± SEM. A two-tailed t test with unequal variance was performed at the end point (day 24). (B, Left) Immunoblot shows LSD1 protein levels in xenograft tumor tissues from mice treated with SP-2509 vs. vehicle. LSD1 band intensity was normalized to the intensity of actin. (Right) The average LSD1 levels normalized to actin in control vs. SP-2509–treated tumor samples. The data are reported as ±SD; *P < 0.05, two-tailed unpaired t test. (C) Results of IHC staining for cleaved caspase-3. The percentage of tumor cells staining positive for cleaved caspase-3 was significantly different in vehicle and SP-2509–treated tumors (P < 0.05, Wilcoxon two-sided rank sum test).
Discussion
LSD1 is a key regulator of gene expression, and this role has been attributed to its demethylase function (9–11). In prostate cancer, specifically, LSD1 had been shown to contribute to androgen regulation of AR target gene expression in androgen-dependent models (19–21, 41). However, LSD1’s role in lethal prostate cancer and the mechanisms by which LSD1 promotes prostate cancer cell survival were largely unknown. In this study, we demonstrate that LSD1 promotes the survival of lethal prostate cancer cell models independently of its demethylase function and identify a drug, SP-2509, that blocks demethylase-independent functions.
Significantly, we determined that LSD1 up-regulation is ubiquitous in lethal CRPC samples (Fig. 1A). A hallmark of lethal prostate cancer tumors is resistance to AR interference. We confirmed that LSD1 suppression reduces survival of prostate cancer cells that are grown without androgens, that are resistant to the AR antagonist enzalutamide, or that do not even express AR (Fig. 1 B and C), demonstrating the importance of LSD1 in conferring AR-independent CRPC cell survival.
A previous study by Cai et al. (41) reported that supplementation of prostate cancer cells with high doses of androgen ligands leads to the recruitment of an AR–LSD1 complex to regulatory elements that lead to suppression of transcription of the AR itself and other target genes. As pointed out by the authors, the androgen concentrations required for the formation of this AR-repressive complex were much higher than those commonly found in CRPC metastases, suggesting that this complex may not be active under physiological conditions (41). In this study, we sought to clarify androgen and AR-independent functions of LSD1. It is noteworthy that we did not stimulate cells with high doses of supplemental androgens when conducting experiments to measure gene-expression changes after LSD1 suppression using RNAi or SP-2509. Therefore, we cannot rule out the possibility that additional LSD1 target genes may be coregulated by LSD1 and AR under high-androgen conditions like those identified by Cai et al. (41).
The critical genes regulated by LSD1 that promote the survival of prostate cancer cells were poorly defined. A recent study showed that cell-cycle gene sets were the top pathways regulated by LSD1 in prostate cancer, including the gene CENPE (42). In agreement with those results, we determined that genes in the ontological categories of cell-cycle/mitosis and ESC maintenance, including CENPE, that are enriched in profiling studies of lethal prostate cancer patients (6, 43) are among the top categories of LSD1-activated genes (Fig. 1F, Fig. S1F, and Datasets S2 and S3). Significantly, virtually none of the LSD1-activated genes we identified are androgen-responsive (Fig. 1D, Fig. S1D, and Dataset S1). These results clarify key AR-independent pathways activated by LSD1 in prostate cancer.
Prior work with MARINa identified important transcriptional regulators that are important for human prostate cancer aggressiveness and lethality (8). Our results demonstrating that LSD1 regulates the expression and activity of many of these clinically relevant MRs―including some that were direct LSD1 target genes, such as ASF1B, CENPK, and ZNF367―suggests that LSD1 acts at the top of a hierarchy for many of these MRs (Fig. 3 and Figs. S4 and S5). Based on our MARINa results, it is possible that patients with high expression and activity of the LSD1-regulated MRs identified herein may be ideally suited to LSD1-targeting approaches.
It is likely that regulation of other ontologic categories of genes also contributes to LSD1’s effects in lethal prostate cancer. Indeed, in contrast to the LSD1-activated gene program, we observed a strong enrichment for adult tissue stem cell gene sets of cellular differentiation in the LSD1-repressed genes from our microarray analysis of prostate cancer cells (Fig. S1F). This is consistent with LSD1’s known role in regulating cell fate and differentiation through gene repression in ESCs, hematopoietic stem cells, and cancers such as leukemia (11–17).
Factors that cooperate with LSD1 to regulate prostate cancer cell survival, especially in CRPC cells, were not well established. Our integrative analysis implicated ZNF217 as an important LSD1-binding protein. Importantly, despite prior work describing ZNF217 primarily as a transcriptional repressor (32, 33), we determined that ZNF217, like LSD1, was important for the activation of the lethal prostate cancer gene network and prostate cancer cell survival, demonstrating ZNF217’s importance. Thus, our work sets the stage for further studies to characterize the LSD1–ZNF217 transcriptional activator complex.
Several lines of complementary evidence demonstrate that noncanonical, demethylase-independent mechanisms contribute to LSD1’s activation of the lethal prostate cancer gene network and promotion of prostate cancer cell survival (Fig. 2). First, LSD1 RNAi did not increase levels of H3K4me2 or H3K9me2 at the great majority of LSD1-bound and -regulated genes by ChIP-seq (Fig. 2 A–E). This suggests that demethylation of these histone substrates is not necessary for gene-expression changes of most LSD1 direct target genes and matches a prior report in leukemia in which H3K4me2 and H3K9me2 histone methylation changes were not seen with LSD1 inhibition (11). Importantly, like that prior report in leukemia, our conclusions that LSD1 regulates the expression of the majority of its targets genes independently of H3K4me2 and H3K9me2 histone demethylation in the prostate cancer models we examined are based on ChIP-seq. This technique enables one to gain a much deeper appreciation of levels of histone mark peaks than obtained with ChIP-PCR that evaluates only a small region of interest that may not be representative of the entire peak region. Second, complementation with wild-type or mutant K661A LSD1 abrogates the effects of RNAi-mediated LSD1 suppression on reducing prostate cancer cell survival and the expression of lethal prostate cancer network genes (Figs. 2 F and G and 3E). Further, LSD1’s catalytic function is not important for interaction with its important binding protein ZNF217 (Fig. 4D). Finally, in keeping with other reports demonstrating lack of antitumor activity of catalytic LSD1 inhibitors in prostate cancer models (35), we demonstrated that irreversible catalytic LSD1 inhibitors do not recapitulate the effects of LSD1 RNAi (Fig. 5 and Fig. S7). These results strongly argue that LSD1’s canonical demethylase activity is not critical for promoting survival of the prostate cancer models we studied.
In contrast to results with LSD1 catalytic inhibitors, treatment with the drug SP-2509 recapitulated the effects of LSD1 RNAi on reducing cell survival and suppressing prostate cancer network genes. Importantly, our molecular docking studies suggest that SP-2509 binds to the H3 pocket of LSD1, a region distinct from the LSD1-binding site of catalytic inhibitors. There are several demethylase inhibition-independent mechanisms achieved with this drug that may explain these effects, including blocking LSD1’s interaction with the important coactivator ZNF217, reduced LSD1 protein stability, and LSD1 depletion from chromatin. That these effects are achieved without increases in H3K4me2 and H3K9me2 at LSD1 target genes further demonstrates the demethylase inhibition-independent mechanisms of action of this inhibitor. Finally, that SP-2509 treatment in vivo using AR-null PC3 CRPC cells suppressed tumor growth demonstrates the importance of blocking the demethylase-independent functions of LSD1 in prostate cancer. (Fig. 5).
These data demonstrate that LSD1 promotes prostate cancer survival by activating a lethal prostate cancer gene network independently of its demethylase function. However, this does not rule out the possibility that LSD1’s demethylase function is important for regulating other cancer hallmarks or gene networks other than those examined here. Nonetheless, our work identifies previously unappreciated, noncanonical, and AR-independent functions of LSD1 in prostate cancer. Importantly, our work suggests that targeting LSD1 protein stability or targeting important coactivators such as ZNF217, rather than LSD1’s demethylase function, is a rational strategy to control prostate cancer above and beyond what may be achievable with currently approved therapies.
Materials and Methods
We collected human metastatic CRPC tissues through an Oregon Health and Science University (OHSU) Institutional Review Board (IRB)-approved tissue-collection protocol. All experiments on human tissues were approved by the IRB, and all subjects provided informed consent. All animal experiments were performed under an Institutional Animal Care and Use Committee (IACUC)-approved protocol. Details of experimental procedures, immunohistochemistry, cell culture, transfection, qRT-PCR, cell-viability assays, immunoblotting, IP, RIME, molecular docking, in vivo experiments, microarray, RNA-seq, ChIP-seq, pathway-enrichment analysis, MARINa analysis, and statistical analyses are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ms. Mandy Burns and Ms. Shanta Green for help with submitting the manuscript. This research was supported by NIH/National Cancer Institute (NCI) Grants R01 CA178610, R01 CA100743, and R01 CA169172; Pacific Northwest Prostate Cancer Specialized Programs of Research Excellence/NCI Grant P50 CA097186; NIH/NCI Cancer Center Support Grants P30 CA069533-16 and P30 CA051008-16; Oregon Clinical and Translational Research Institute Grant UL1TR000128 from the National Center for Advancing Translational Sciences, a component of the NIH and NIH Roadmap for Medical Research; and a Department of Defense (DOD) Synergistic Idea Award W81XWH-16-1-0597. Other support includes a Medical Research Foundation New Investigator Award, Kuni Foundation Scholar Award, a Prostate Cancer Foundation (PCF) Young Investigator Award, Anna Fuller Fund, and Stand Up To Cancer-PCF Prostate Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409 made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research (7465sc). RNA and ChIP-sequencing were performed in the OHSU Massively Parallel Sequencing Shared Resource. Affymetrix microarrays were performed in the OHSU Gene Profiling Shared Resource. Mass spectrometry was performed in the OHSU Proteomics Shared Resource. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or DOD.
Footnotes
Conflict of interest statement: S.S. and T.M.B. have an equity interest in Salarius Pharmaceuticals, which developed SP-2509. Salarius Pharmaceuticals had no role in the design or conduct of this study.
This article is a PNAS Direct Submission.
Data deposition: Microarray data for microarray sample preparation and expression analysis have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE61630). Sequence data for RNA-seq library preparation and expression analysis have been deposited in the GEO database (accession no. GSE59009). ChIP-seq data have been deposited in the GEO database (accession no. GSE77762.)
See Commentary on page 4530.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719168115/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Scher HI, et al. AFFIRM Investigators Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, et al. COU-AA-301 Investigators Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 7.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aytes A, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulte JH, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: Implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 11.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamo A, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 14.Sprüssel A, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 16.Foster CT, et al. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol. 2010;30:4851–4863. doi: 10.1128/MCB.00521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerenyi MA, et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 20.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 21.Cai C, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9:1618–1627. doi: 10.1016/j.celrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahl P, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 23.Culig Z, et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer. 1999;81:242–251. doi: 10.1038/sj.bjc.6690684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thalmann GN, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54:2577–2581. [PubMed] [Google Scholar]
- 25.Assou S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald OG, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49:367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 28.Xie Q, et al. Methylation-mediated regulation of E2F1 in DNA damage-induced cell death. J Recept Signal Transduct Res. 2011;31:139–146. doi: 10.3109/10799893.2011.552914. [DOI] [PubMed] [Google Scholar]
- 29.Lee MG, et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammed H, et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 32.Banck MS, et al. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics. 2009;4:100–106. doi: 10.4161/epi.4.2.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thillainadesan G, et al. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol. 2008;28:6066–6077. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi YJ, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Mohammad HP, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 36.McGrath JP, et al. Pharmacological inhibition of the histone lysine demethylase KDM1A suppresses the growth of multiple acute myeloid leukemia subtypes. Cancer Res. 2016;76:1975–1988. doi: 10.1158/0008-5472.CAN-15-2333. [DOI] [PubMed] [Google Scholar]
- 37.Fiskus W, et al. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia. 2014;28:2155–2164. doi: 10.1038/leu.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theisen ER, et al. Reversible inhibition of lysine specific demethylase 1 is a novel anti-tumor strategy for poorly differentiated endometrial carcinoma. BMC Cancer. 2014;14:752. doi: 10.1186/1471-2407-14-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 40.Baron R, Vellore NA. LSD1/CoREST is an allosteric nanoscale clamp regulated by H3-histone-tail molecular recognition. Proc Natl Acad Sci USA. 2012;109:12509–12514. doi: 10.1073/pnas.1207892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai C, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang Y, et al. LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res. 2017;77:5479–5490. doi: 10.1158/0008-5472.CAN-17-0496. [DOI] [PubMed] [Google Scholar]
- 43.Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci USA. 2011;108:21276–21281. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuleshov MV, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]