Significance
Quorum sensing is a communication system that allows bacteria to coordinate their activities, and these systems are critical for virulence in several bacteria, including Pseudomonas aeruginosa. There is a significant gap in knowledge about how quorum sensing proceeds during infection, particularly how spatial organization of the infecting microbial community impacts signaling. Using a model that recapitulates the biogeographical properties of P. aeruginosa infection of the cystic fibrosis lung, we discovered that communication primarily occurs within P. aeruginosa aggregates and that communication between aggregates is only observed for very large aggregates containing ≥5,000 cells. This study identifies a critical role for spatial distribution and bacterial phenotypic heterogeneity in bacterial signaling during infection, and provides a platform for future ecological and evolutionary studies.
Keywords: Pseudomonas aeruginosa, aggregate, quorum sensing, cystic fibrosis, spatial distribution
Abstract
Quorum sensing (QS) is a bacterial communication system that involves production and sensing of extracellular signals. In laboratory models, QS allows bacteria to monitor and respond to their own cell density and is critical for fitness. However, how QS proceeds in natural, spatially structured bacterial communities is not well understood, which significantly hampers our understanding of the emergent properties of natural communities. To address this gap, we assessed QS signaling in the opportunistic pathogen Pseudomonas aeruginosa in a cystic fibrosis (CF) lung infection model that recapitulates the biogeographical aspects of the natural human infection. In this model, P. aeruginosa grows as spatially organized, highly dense aggregates similar to those observed in the human CF lung. By combining this natural aggregate system with a micro-3D–printing platform that allows for confinement and precise spatial positioning of P. aeruginosa aggregates, we assessed the impact of aggregate size and spatial positioning on both intra- and interaggregate signaling. We discovered that aggregates containing ∼2,000 signal-producing P. aeruginosa were unable to signal neighboring aggregates, while those containing ≥5,000 cells signaled aggregates as far away as 176 µm. Not all aggregates within this “calling distance” responded, indicating that aggregates have differential sensitivities to signal. Overexpression of the signal receptor increased aggregate sensitivity to signal, suggesting that the ability of aggregates to respond is defined in part by receptor levels. These studies provide quantitative benchmark data for the impact of spatial arrangement and phenotypic heterogeneity on P. aeruginosa signaling in vivo.
Chronic infections are a major health problem in the United States and throughout the world. These infections are often caused by complex, multispecies biofilm communities (1). Biofilms are high-density microbial clusters frequently attached to surfaces that are encased in an extracellular polymeric matrix and often highly resistant to antimicrobial treatment. Due to their negative impact in the clinical environment, significant attention has been devoted to biofilm research. Development of in vitro biofilms has been proposed to follow a precise developmental program that results in communities often composed of “mushroom”-like structures containing millions of cells (2, 3). While these in vitro models are likely a good platform for studying many types of biofilms, those found in some chronic infections, including those found in the cystic fibrosis (CF) lung, have recently been shown to be composed of small, dense bacterial “aggregates” (∼101–104 cells) that are spatially organized (1, 4–7). Both in vitro and murine models of infection show that aggregates display similar phenotypes to in vivo biofilms, including enhanced antimicrobial tolerance (4, 8).
Microbes, including those growing as biofilms and aggregates, are highly social and utilize dedicated systems that have evolved for communication. Quorum sensing (QS) is a common microbial communication system that involves self-produced extracellular signals, which can accumulate in a local environment and promote transcription of specific genes (9). The genetics and biochemistry of many QS systems are well described, and these systems control a range of microbial behaviors important for in vivo fitness (9, 10). However, there is a significant gap in our understanding of how QS proceeds in chronic CF infections. Do aggregates quorum sense, and if so, how does aggregate size impact QS? Furthermore, how does the physical distribution of aggregates at the microscale impact signaling within and between aggregates?
Previous studies have utilized a mixture of wet laboratory and modeling approaches to begin to explore these questions. By patterning engineered aggregates on a glass surface, our laboratory previously demonstrated that an aggregate of the bacterium Pseudomonas aeruginosa containing ∼2,000 QS signal-producing cells could induce a detectable response from a neighboring aggregate 8 µm away (11). Weigert and Kümmerli (12) demonstrated that the reach of a diffusible public good can be visualized among attached aggregates in an in vitro agar model. In this model, aggregates responded to siderophores produced by a neighboring aggregate at a maximum distance of 100 µm, via the production of their own secreted product. In addition, the spatial positioning of coinfecting oral bacteria has been shown to be critical for fitness and disease severity in an abscess model of infection (13). These studies and others have focused on the spatial structure of microbial communities and have provided new insights into how spatial distribution impacts aggregate interactions (14); however, many of these systems do not recapitulate the natural growth environment of the infection site nor have the ability to precisely manipulate aggregate spatial structure. Thus, there is a need to further develop an experimental system that addresses these challenges.
In this study, we further developed an in vitro model based on chronic P. aeruginosa infection of the CF lung. This system utilizes a synthetic CF sputum (SCFM2), designed to mimic the lung secretions of individuals with CF, which importantly promotes natural formation of P. aeruginosa aggregates (5). Combining this with high-resolution microscopy and micro-3D printing (15, 16), we defined both how aggregate size and geographical location impacts intra- and interaggregate QS communication. Specifically, we found that aggregates containing 2,000 cells display intra- but not interaggregate signaling, while aggregates ≥5,000 cells can signal neighboring aggregates 120–180 µm away. Aggregates also displayed differential sensitivity to signal, and this heterogeneity could be reduced by increasing transcription of the signal receptor.
Results
SCFM2 Has Viscoelastic Properties Similar to Expectorated CF Sputum.
The goal of this study was to develop a versatile experimental system that captured relevant biological aspects of chronic infection and use this system to study intra- and interaggregate signaling. The system we chose was P. aeruginosa growth in a synthetic CF sputum medium (SCFM2). There have been two formulations of this medium developed by the M.W. laboratory (SCFM and SCFM2). SCFM was initially developed to study how the nutritional environment of CF sputum impacts P. aeruginosa growth and virulence (17, 18), and was subsequently modified by adding relevant levels of DNA, lipid, and mucin to SCFM to more closely mimic the physical properties of expectorated CF sputum. Previous work has shown that the genes important for P. aeruginosa growth in SCFM2 and authentic sputum are highly similar (19), and importantly for this study, SCFM2 promotes the formation of natural P. aeruginosa aggregates with sizes similar to those observed in the CF lung (5).
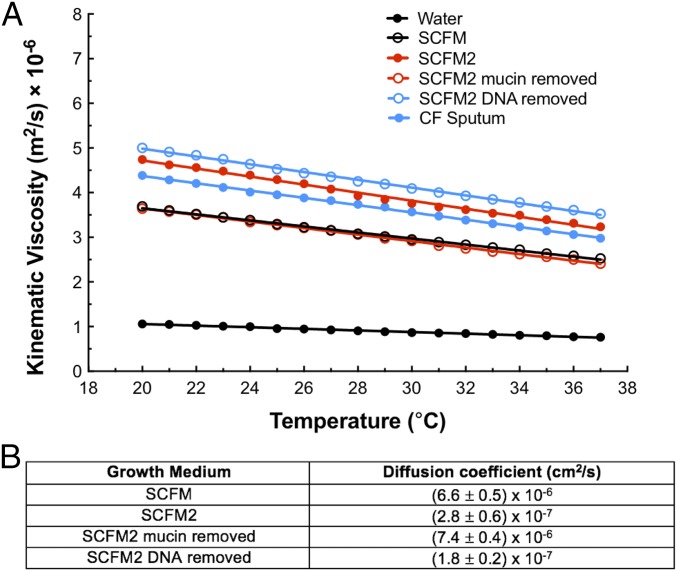
While previous studies provide support for SCFM2 as a relevant model system to study P. aeruginosa chronic CF infection, we reasoned that physical factors might also impact the relevance of aggregate studies in SCFM2. For example, the viscoelastic properties of CF sputum are believed to play a supportive role in a developing biofilm, including the aggregate stage (20, 21). Thus, we quantified the kinematic viscosity of SCFM2 using a viscometer and compared it with authentic expectorated CF sputum from patients, SCFM, and water (Fig. 1A). Both SCFM2 and authentic sputum had significantly higher average viscosities than SCFM and water across a range of temperatures (Fig. 1A). At 37 °C, SCFM2 had a viscosity of 3.2 × 10−6 m2/s, which was slightly more viscous than expectorated sputum [2.9 × 10−6 m2/s (P = 0.01)]. This viscosity is similar to that of the commonly used 30–40% glycerol laboratory solution. To determine which polymer in SCFM2 contributed most to viscosity, we removed mucin or DNA from the medium. Removing mucin decreased the viscosity by ∼25% to 2.4 × 10−6 m2/s at 37 °C (P = 0.001, compared with SCFM2), essentially identical to the original SCFM formulation, while removing DNA had little effect (Fig. 1A). These data reveal that the viscosity of SCFM2 is more similar to that of authentic sputum than to SCFM, and mucin addition is the most significant contributor to this increased viscosity.
Fig. 1.
SCFM2 has a similar viscosity to expectorated CF sputum, and the presence of mucin impacts viscosity and diffusion. (A) Kinematic viscosity measurements of SCFM2 (red) compared with SCFM, SCFM2 with mucin removed, SCFM2 with DNA removed, expectorated CF sputum and water. Viscosity is measured as the kinematic viscosity (m2/s × 10−6) across a temperature range of 20–37 °C. Statistical comparisons at 37 °C using a two-tailed t test indicate that SCFM2 has a similar viscosity to expectorated CF sputum (P = 0.01), that the removal of mucin changes viscosity (P = 0.0001) and is more viscous than water (P = 0.0001). Data represent the mean value of three replicates ± SEM. Error bars are too small to be seen. (B) Diffusion coefficients of pyocyanin in SCFM, SCFM2, SCFM2 with mucin removed, and SCFM2 with DNA removed. Diffusion coefficients were measured at 25 °C using chronoamperometry. Significant differences using a two-tailed t test were determined for: SCFM and SCFM2 (P = 0.0002); SCFM2 and SCFM2 mucin removed (P < 0.0001); SCFM2 and SCFM2 DNA removed (P = 0.002). As a control, the diffusion coefficient of ferrocene methanol in SCFM2 was also assessed and determined to be (2.1 ± 0.4) × 10−7 cm2/s. Data represent the mean of three replicates ± SEM.
The significant change in viscosity in SCFM2 led us to next ask whether diffusion of QS signals is impacted in SCFM2. Ideally we would measure diffusion of the P. aeruginosa signal 3-oxo-dodecanoyl homoserine lactone (3OC12-HSL), as this model QS system is the focus of our study. However, diffusion of this molecule has been notoriously difficult to study due to the lack of effective analytical techniques for detection in the absence of chemical modification; thus, we focused on another QS signal molecule (pyocyanin) to assess signal diffusion in SCFM2 and the impact of polymers on diffusion. We propose that pyocyanin is a reasonable surrogate for 3OC12-HSL, as it has similar molecular weight (PYO MW = 210.08 g/mol, 3OC12-HSL MW = 283.21 g/mol), and has been shown in agar to have a similar diffusion coefficient as another homoserine lactone, octanoyl homoserine lactone (MW = 227.3 g/mol) (22, 23). Moreover, because it is redox active, biologically relevant pyocyanin concentrations can be detected with high sensitivity using electroanalytical methods, thus allowing for accurate measurement of diffusion (11, 22, 24, 25). Fig. 1B shows the apparent diffusion coefficients of pyocyanin in SCFM, SCFM2, SCFM2 with mucin removed, and SCFM2 with DNA removed. The diffusion coefficient of pyocyanin in SCFM was reduced compared with SCFM2 (6.6 × 10−6 cm2/s to 2.8 × 10−7 cm2/s), demonstrating that the addition of polymers to SCFM slows diffusion. The removal of mucin from SCFM2 produced a similar diffusion coefficient to SCFM (7.4 × 10−6 cm2/s), while removal of DNA had a small but statistically significant effect (1.8 × 10−7 cm2/s). These data indicate that as for viscosity, mucin is likely the primary polymer impacting diffusion of pyocyanin in SCFM2.
Developing a Robust Experimental System to Study QS in P. aeruginosa Aggregates.
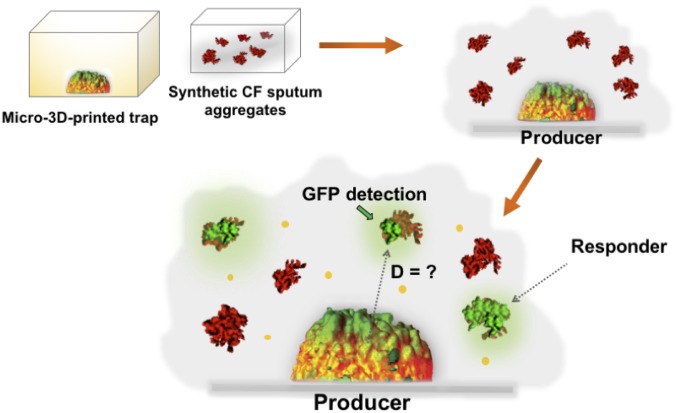
Our results that mucin not only impacts P. aeruginosa aggregate formation but also viscosity and diffusion rates prompted us to next assess inter- and intraaggregate QS in P. aeruginosa SCFM2 communities. To accomplish this, we developed a system that involved surrounding a QS signal-producing P. aeruginosa aggregate (“producer”) with P. aeruginosa aggregates that are able to respond to, but not produce, QS signals (“responder”) (Fig. 2). The goal was then to determine how size of the producer aggregate and geographical location of the responder cells/aggregates impacted their ability to communicate via QS.
Fig. 2.
Experimental design for studying intra- and interaggregate P. aeruginosa signaling. Schematic representation of the experimental design used to study aggregate signaling. P. aeruginosa responder aggregates are formed in SCFM2 then overlain on micro-3D–printed producer aggregates. “GFP detection” refers to responder aggregates that responded to signal from the producer. “D” indicates distance.
The first step in developing this system was construction of a P. aeruginosa responder strain. To accomplish this, a plasmid (pSEDQS) was created that constitutively expresses mCherry and places the GFP-encoding gene gfp under control of the promoter from the P. aeruginosa QS regulated gene rsaL. Transcription of rsaL is controlled by the QS transcriptional regulator LasR when it is bound to the QS signal 3OC12-HSL, and the rsaL promoter has previously been used as a sensitive measure for LasR/3OC12-HSL QS activation (26). To construct the responder strain, pSEDQS was introduced into P. aeruginosa PA14 ΔlasIΔrhlI, which is unable to produce QS signals but is able to respond to them.
The next step in studying aggregate interactions in SCFM2 was to establish a single wild-type P. aeruginosa producer aggregate of specified size and location that could be surrounded with responder aggregates. To achieve this, we used a micro-3D–printing technique developed by the J.B.S. laboratory to enclose a P. aeruginosa producer cell in a picoliter-sized dome-shaped enclosure (referred to as a “trap”) (Fig. 2 and Fig. S1) (8, 11, 15, 16). This technique involves temporarily immobilizing P. aeruginosa cells within a thermally set gel (containing gelatin, BSA, and photosensitizer), then constructing a picoliter-sized trap around a single bacterium by covalently photocross-linking a matrix comprised of gelatin and BSA using a layer-by-layer, laser-scanning micro-3D–printing process. Unreacted thermally set gel is melted and removed following trap construction by washing with warmed media, leaving gelatin-BSA walls that confine the bacterium but provide facile diffusion of numerous solutes including signals (16). Using this technique, we constructed traps of varying sizes (volumes of 2, 5, 10, and 20 pL), each around a single P. aeruginosa cell capable of producing QS signals (Fig. 2 and Fig. S1, dome-shaped structures). Individual cells were allowed to divide and multiply until a clonal population filled the trap (maximum capacity of ∼103 cells/pL). Hundreds of responder aggregates, naturally formed in SCFM2 as previously described (5), were then overlaid on top of the filled trap, and the system was imaged at multiple time points using confocal laser-scanning microscopy (CLSM). This experimental method provided several advantages. First, the natural aggregate formation supported by SCFM2 allows us to assess the response of a large number of aggregates simultaneously. Second, the aggregates are spatially supported in the z axis (vertical axis) by the viscosity of SCFM2 and are therefore exposed to the extracellular environment on all surfaces. Third, the use of micro-3D–printed aggregates allows us to finely tune the number of cells present in the producer community by manipulating the size of the trap.
P. aeruginosa Aggregates Containing ≥5,000 Cells Engage in Interaggregate Signaling.
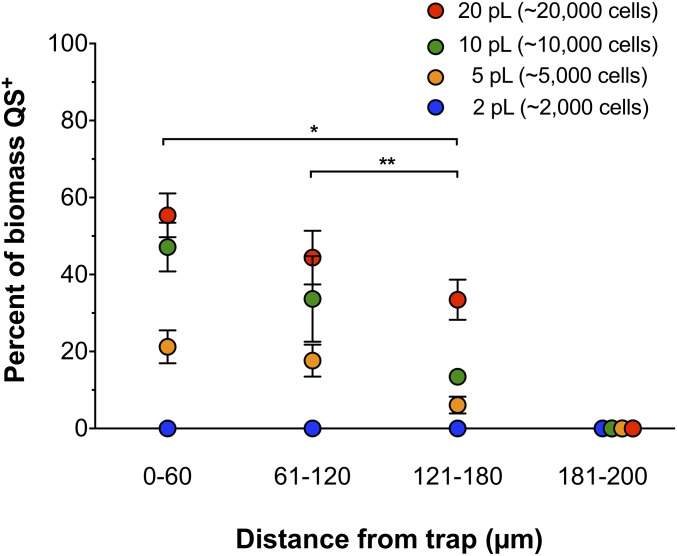
We previously showed that traps containing as few as 500 P. aeruginosa cells initiate QS when confined in 2-pL traps (11), thus it was not surprising that producer communities confined in 2-, 5-, 10-, and 20-pL traps (2,000–20,000 cells) displayed 3OC12-HSL–mediated gene expression (Fig. S2A). To determine if these producer communities could signal neighboring aggregates, trapped producer communities were then overlaid with responder aggregates that had been formed in SCFM2 for 2.5 h by exponentially growing P. aeruginosa (5). The total volume of QS+ P. aeruginosa responder biomass surrounding an individual trap was then determined after 1.5 h (Fig. 2, Table 1, and Figs. S1 and S2B). This time was used as it was sufficient for inducing the maximum QS response in responder communities (Fig. S2B) and oxygen levels, which are critical for P. aeruginosa growth and GFP folding, were above 2–5% as P. aeruginosa had not yet induced the high-affinity oxidase cbb3-2 (Fig. S2C). Our results reveal that 2-pL traps (∼2,000 cells) were unable to induce QS in neighboring aggregates, even in those as close as 5 µm; however, 5-pL traps induced a mean response of 18% of the P. aeruginosa responder biomass, and 20-pL traps induced the highest mean response (∼48%) (Table 1 and Dataset S1). These results indicate that 2-pL aggregates engage in intra- but not interaggregate 3OC12-HSL–mediated QS, while larger aggregates engage in both. Although over 90% of the P. aeruginosa responder biomass used in these experiments were in aggregates, the data allowed us to examine whether aggregates or planktonic (free-swimming) cells were differentially responsive to signal from trapped producer cells. We found that both aggregates (objects ≥5.0 µm3) and planktonic cells (objects ≥0.5 µm3 and <5.0 µm3) were equally sensitive to 3OC12-HSL produced by the trapped community (Table 1 and Dataset S1). Based on these data, and the fact that the majority of biomass is aggregated, the fraction of total biomass that is QS+ is reported in all subsequent experiments.
Table 1.
Aggregates containing ≥5,000 producer P. aeruginosa engage in interaggregate signaling
| Trap size, pL | Percent QS+, % | ||
| Biomass | Aggregate | Planktonic | |
| 2 | ND | ND | ND |
| 5 | 19.2 ± 7.3* | 19.3 ± 7.6* | 22.2 ± 10.4 |
| 10 | 41.4 ± 8.6† | 39.2 ± 8.2† | 51.7 ± 12.6† |
| 20 | 45.25 ± 11.2 | 44.8 ± 11.1 | 45.7 ± 10.3 |
QS+ responder total biomass, aggregate biomass and planktonic biomass surrounding traps of increasing size. QS+ response was calculated as the total volume of GFP+ voxels/total number of mCherry voxels. Data are presented as the mean value of six replicates ± SEM. Analysis by ANOVA following arcsine transformation revealed a significant difference between all datasets for total biomass [F(3, 20) = 14.37 (P ≤ 0.0005)], aggregate biomass [F(3, 20) = 14.18 (P ≤ 0.0005)], and planktonic biomass [F(3, 20) = 9.83 (P ≤ 0.005)].
A statistically significant increase in QS+ response between 5 and 20 pL traps for total biomass (P = 0.044) and aggregate biomass (P = 0.044).
A statistically significant increase in QS+ response between 10- and 5-pL traps for total biomass, aggregate biomass, and planktonic biomass (for all three, P = 0.05).
Geographical Location of an Aggregate Impacts the Response to 3OC12-HSL.
While the analysis above focused on the cumulative response of all responder cells surrounding a trap, the experimental set-up also allowed us to assess if the ability of responder cells to initiate QS was impacted by geographical location. To begin to resolve the “calling distance” of quorum signals arising from the trapped producer aggregate, the distance of all responding planktonic cells and aggregates was determined in 60-µm intervals originating at the trap wall (Fig. 3 and Figs. S1, S3, and S4 and Dataset S2). Our results reveal that distance from the trap impacted the number of responding cells, with fewer cells responding in the 121- to 180-µm interval than in the 0- to 60-µm interval for all trap sizes ≥5 pL (Fig. 3 and Dataset S2). However, trap size did not impact the “calling distance” of QS signals, as some cells were shown to respond in the 121- to 180-µm interval for all trap sizes but none were shown to respond in the outermost interval of our experimental setup (181–200 µm). While our data indicate that geographical location impacts the frequency of QS response in SCFM2, we were not able to define a precise maximum calling distance for each aggregate size, as it was necessary to bin the data for statistical purposes, although it is clear that the maximum calling distance of each aggregate size is between 121 and 180 µm. Of note, the furthest distance a responder aggregate was shown to initiate QS was 176 µm from the outer edge of a 20-pL trap (Fig. S3 and Dataset S2).
Fig. 3.
P. aeruginosa response to QS signals varies with geographical location. QS+ P. aeruginosa responder biomass surrounding traps of increasing size. Response is grouped into 60-µm intervals from the edge of the trap to 180 µm and an outermost interval of 181–200 µm (the furthest interval imaged). Data are expressed as the percentage of total biomass at each distance interval that is QS+. Error bars are SEM. All values are from six replicates, except 10-pL data has three replicates omitted due to the absence of biomass at the 60- to 120-µm interval. Analysis by ANOVA following arcsine transformation indicated a significant difference between distance intervals surrounding a 5-pL trap [F(3, 20) = 18.65 (P ≤ 0.00005)], 10-pL trap [F(3, 20) = 11.86 (P ≤ 0.005)] and 20-pL trap [F(3, 20) = 47.20 (P ≤ 0.00000005)]. An asterisk (*) indicates a single trap size induced a statistically significant difference in QS response at the 0- to 60-µm interval compared with the 121- to 180-µm interval (P = 0.0008 for 5 pL, and 0.05 for 20 pL). Two asterisks (**) indicate the 5-pL trap induced a statistically significant difference in QS response at the 61- to 120-µm interval compared with the 121- to 180-µm interval (P = 0.04). Statistical significance was determined by two-tailed t test following arcsine transformation.
P. aeruginosa Aggregates Have Differential Sensitivities to 3OC12-HSL.
We next sought to understand why only a maximum of 48% of the responder biomass surrounding traps initiated QS (Table 1 and Dataset S1). We hypothesized that aggregates have differential sensitivities to 3OC12-HSL and that nonresponsive cells simply require higher levels of 3OC12-HSL to initiate QS. To test this hypothesis, we prepared aggregates of the responder strain, then added different concentrations of 3OC12-HSL and quantified QS-responsive biomass after 1.5 h. The results revealed a linear response to increasing concentrations of 3OC12-HSL from 0.25 µM to 5 µM, with over 80% of the responder biomass QS+ at 5 µM (Fig. S4A). A decrease in QS biomass was observed with 10 µM 3OC12-HSL, indicating that higher concentrations of signal inhibit the response. These data indicate that SCFM2 aggregates have a range of sensitivities to 3OC12-HSL and over 80% are capable of responding to 5 µM 3OC12-HSL under our experimental conditions.
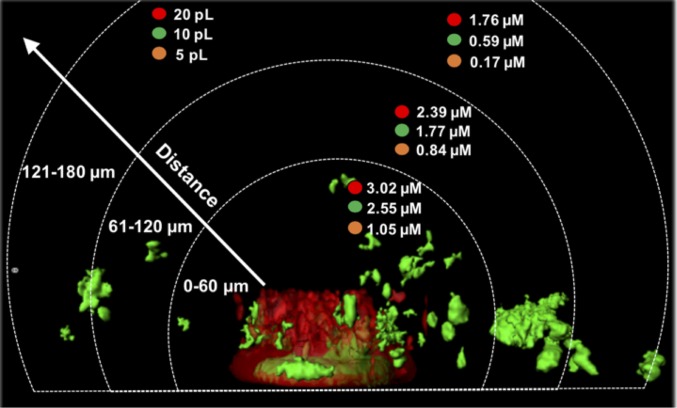
We next took advantage of the linear response of responder cells to 3OC12-HSL concentrations below 5 µM (Fig. S4A) to calculate the concentration of signal surrounding each trap. This was performed for each 60-µm interval surrounding each trap to complement the data in Fig. 3. To calculate concentrations, we used the biomass response (Fig. 3) and the dose–response plot (Fig. S4A) to estimate concentrations at different distances from the outermost walls of 5-, 10-, and 20-pL traps. As expected, with increasing trap size, an increasing amount of signal was produced, with 5-pL traps producing the lowest levels of 3OC12-HSL and 20-pL traps producing the highest (Fig. 4). In addition, 3OC12-HSL levels decreased at further distances from the trap, with outermost levels ∼three- to fourfold lower. While we were unable to calculate the levels of 3OC12-HSL produced by 2-pL aggregates because no biomass surrounding these traps showed a QS response, our dose–response curve indicates that addition of as little as 0.5 µM 3OC12-HSL is sufficient to induce QS in ∼5% of responder cells (Fig. S4A), thus the levels of 3OC12-HSL produced nearest the 2-pL aggregates is likely below this value. Collectively, these data indicate that aggregates have a range of activation thresholds to 3OC12-HSL, and that 1.5 µM is sufficient to induce QS in neighboring aggregates. However, some aggregates do not respond because they require higher concentrations of 3OC12-HSL than that present, and this level is defined by their location relative to the trap.
Fig. 4.
Larger producer aggregates synthesize a higher concentration of 3OC12-HSL. 3OC12-HSL concentrations at 60-µm intervals from the outer edge of 5-, 10-, and 20-pL traps. 3OC12-HSL concentrations were determined by calculating the percentage of QS+ biomass within each interval followed by interpolation onto the dose–response curve in Fig. S4A.
Aggregates Are Clonal and Overexpression of lasR Sensitizes Aggregates to 3OC12-HSL.
Why are some aggregates more responsive to 3OC12-HSL than others? One hypothesis is that aggregates arise from clonal expansion of individual cells that possess differential sensitivities. To begin to test this hypothesis, we first asked whether aggregates formed in SCFM2 are clonal (i.e., arising from a single cell) or if they are formed by the merger of multiple cells/aggregates. To do this, we inoculated SCFM2 with two strains of wild-type P. aeruginosa, one expressing GFP and the other expressing cyan fluorescent protein (CFP), allowed them to form aggregates, then imaged them with CLSM. By assessing whether aggregates contained GFP+ cells, CFP+ cells, or both GFP+ and CFP+ cells, one could assess whether aggregates form by clonal expansion or mixing of cells. The data reveal that 19–27% of aggregates were nonclonal (Table S1), indicating that most aggregates are clonal. In addition, aggregates containing both CFP- and GFP-expressing cells were often observed at a position that marked the boundaries of two aggregates, suggesting that aggregates considered nonclonal by our method likely arise from the merging of two aggregates.
Why do clonal aggregates display differential sensitivities to 3OC12-HSL? One common mechanism for bacterial differential sensitivity to external stimuli is heterogeneity in transcriptional regulator levels in responding cells (27). This heterogeneity can often be overcome by overexpression of the transcriptional regulator in the responding population. To test this in our system, we formed aggregates of the P. aeruginosa responder strain that overexpressed lasR (Fig. 5) and assessed the sensitivity of these aggregates to 3OC12-HSL. Overexpression of lasR resulted in increased sensitivity to concentrations of 3OC12-HSL below 5 µM, with the largest increases in sensitivity occurring at concentrations below 1 µM (Fig. S4B). Interestingly, overexpression of lasR did not increase the overall sensitivity of the population to high levels of 3OC12-HSL, as the maximum QS response remained at 70–80% of the responder biomass.
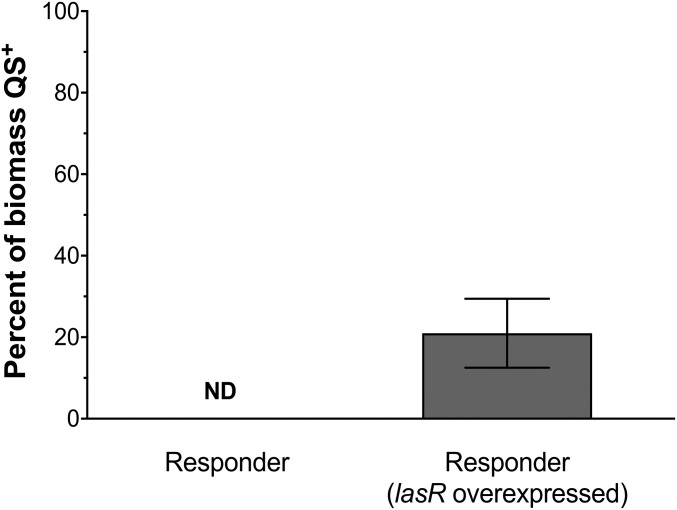
Fig. 5.
Overexpression of lasR increases the response to 3OC12-HSL. The percent of QS+ P. aeruginosa biomass overexpressing lasR surrounding a 2-pL trap was calculated by dividing QS+ (GFP+) biomass by the total (mCherry+) biomass. Compared with a 2-pL trap containing wild-type cells (from Table 1) that induced no detectable response (ND), a significant increase in response was detected in biomass surrounding a trap containing cells overexpressing lasR (P < 0.05). Data represent the mean of six replicates ± SEM.
We next tested whether lasR overexpression would allow 2-pL producer communities, which we showed were unable to signal with surrounding aggregates (Table 1), to engage in interaggregate signaling. Our results revealed that overexpression of lasR sensitized responder aggregates to signal produced by surrounding 2-pL producer traps, with ∼20% of the biomass QS+ (Fig. 5). By plotting this biomass response onto the dose–response curve for the lasR overexpressing responder (similar to the methodology in Fig. 4), we calculated the concentration of 3OC12-HSL produced by the 2-pL trap to be 0.43 µM (Fig. S4B). Collectively, these data indicate that P. aeruginosa aggregates are clonal and their differential sensitivities to 3OC12-HSL levels below 5 µM can be partially overcome by overexpressing lasR.
Discussion
One of the key findings of this study is that SCFM2 aggregates are mostly clonal and display a range of sensitivities to 3OC12-HSL (Fig. 3, Table 1, Table S1, and Datasets S1 and S2). From a population level, the response to increasing 3OC12-HSL is graded with a maximum of ∼80% of aggregates initiating QS when exposed to a 5-µM signal (Fig. S4A). The lack of complete (100%) response to added signal could be biologically relevant or a result of the stringent parameters we used to classify cells as QS+. We suggest the former, because overexpression of lasR increased the sensitivity of P. aeruginosa to 3OC12-HSL levels below 5 µM, but did not increase the maximum response (Fig. S4B). Thus, it is likely that those cells may simply be “blind” to biologically relevant levels of signal (between 0.5 and 1.0 µM). Of course, these nonresponsive cells could be dead or dormant “persisters,” although they are at least grossly structurally intact as they can be identified by the presence of intracellular mCherry, and at least some are metabolically active as we observed nonresponding planktonic cells swimming during microscopic examination. These data indicate that aggregate response is not simply binary, but instead a dynamic response, in which the response of subpopulations of cells lies within a gradient, similar to that observed for single cells (28, 29). In addition, the volumes of individual nonresponsive and QS+ aggregates were similar (Table S2), indicating that there is no threshold volume for a positive response to quorum signals in SCFM2. Our discovery of a range of aggregate sensitivities to 3OC12-HSL likely has important implications for P. aeruginosa fitness and evolution during chronic infection, as well as the emergent properties of P. aeruginosa communities that are impacted by aggregate interactions.
The observation that 2-pL–sized producer communities do not induce QS in neighboring aggregates as close as 5 µm (Fig. 3, Table 1, and Datasets S1 and S2) contradicted our previous studies that trapped communities of this size induced QS in a responder population 8 µm away (11). The primary difference in these studies was that Connell et al. (11) used a common laboratory media (LB), while in this study we used SCFM2. In light of the significant impact of SCFM2 on diffusion (Fig. 1B), it is not surprising that interaggregate communication is impacted by this new growth environment. Mucin had the most significant impact on diffusion and viscosity (Fig. 1), and it has previously been shown to reduce QS induction in vitro (13, 30–32), thus the lack of interaggregate signaling observed in SCFM2 is likely due to the presence of this polymer.
It should also be noted that 2-pL P. aeruginosa aggregates are twice as large as the maximum size observed in CF lung tissue (∼1,000 cells) (5), suggesting that most aggregates within the CF lung are not of sufficient size to engage in interaggregate signaling in the CF lung. This finding has important consequences for understanding the evolution and ecology of P. aeruginosa in the CF lung, as social interactions including cooperation and cheating have been implicated as important (33), yet our data indicate that these interactions may be largely confined to individual aggregates. One caveat to our study is that the autoinduction (signal amplification) loop was removed in the responder cells as the goal was to assess 3OC12-HSL–mediated interactions originating from a single aggregate. This likely has a profound effect on aggregate response and calling distance as bacteria that are situated on the outer edges of an aggregate have the potential to respond first and QS induction of interior cells (and more distant aggregates) may be initiated by this response and feedback. Studying autoinduction is challenging experimentally, and future studies likely will require new experimental approaches to fully answer this question. Finally, while one of the primary strengths of our system is the ability to study a single signal-producing aggregate, a CF sputum sample containing 108 cells will have ∼105 signal producing aggregates. Thus, in addition to size, the geographical location of aggregates in relation to one another will no doubt impact communication.
While our results support a model in which P. aeruginosa 3OC12-HSL signaling is primarily an intraaggregate phenomenon in CF based on the aggregate sizes observed in CF lung tissue, aggregate size may of course vary with ecology. Indeed, an examination of P. aeruginosa aggregate sizes in a murine surgical wound model during coinfection with Staphylococcus aureus revealed that P. aeruginosa aggregates are considerably larger than 2,000 cells (∼7,000 cells) and formed distinct monospecies aggregates, indicating that aggregate sizes sufficient for interaggregate communication in vitro are observed in some infection models (Fig. S5). While it is possible that the physical characteristics of each infection setting (e.g., viscosity, chemical composition) may impact aggregate communication in unique ways, our aggregate size data suggests that the role of QS in coordinating activities may be different in some ecological settings. In the future we plan to adapt the methodology used here to study intra- and interaggregate QS signaling in animal models of P. aeruginosa infection.
Our observation of a 3OC12-HSL “calling distance” that was similar for all trapped communities indicates that producer traps greater than 5 pL are able to signal to aggregates at distances (120–180 µm) that are at the outermost perimeters of our experimental system (200 µm). We were unable to define a calling distance more precisely, as it was necessary to bin the data into 60-µm intervals for statistical analysis (Fig. 3 and Dataset S2). It should be noted that Fig. 3 does not imply a defined point at which communication stops, as responding aggregates were found throughout the 120- to 180-µm distance interval (Fig. S3 and Dataset S2), although we can say with confidence that the outermost aggregate that responded in our experimental set-up was a single aggregate in one replicate at 176-µm outside a 20-pL trap. We predict that larger traps would signal at greater distances within the 120- to 180-µm region, and our results provide benchmark data for future experiments and modeling studies. It is important to point out that the CF lung presents a vast landscape in which populations of cells can develop, and considering the numbers of cells commonly retrieved from an expectorated sputum sample, it is not unfounded to consider that aggregates may be distributed hundreds of microns apart (34), thus communication over this distance is likely biologically important in the CF lung.
Materials and Methods
Strains, Plasmids, and Media.
Wild-type P. aeruginosa strain PA14 and PA14 ΔlasIΔrhlI carrying either pSEDQS or pSEDQS2 (construction described in SI Materials and Methods) were used in this study. pSEDQS allows constitutive expression of mCherry and contains an rsaL:gfp transcriptional fusion. pSEDQS2 is pSEDQS carrying a lasR overexpression construct. Overnight cultures were grown in tryptic soy broth (TSB) at 37 °C with shaking at 200 rpm. All washing steps were performed with 1× PBS, pH 7.0.
SCFM2 Viscosity and Diffusion Methods.
Viscosity measurements were performed using a Sine-Wave Vibro Viscometer SV-10 and Haake temperature-controlled water bath, and electrochemical measurements were performed using a three-electrode cell system at room temperature (25 °C) as described in SI Materials and Methods.
Combining Micro-3D–Printed Aggregates with SCFM2 Aggregates.
Micro-3D printing was performed as described previously (16), and SCFM2 aggregates of P. aeruginosa PA14 ΔlasIΔrhlI carrying pSEDQS were formed using previous methods (5), with slight modifications described in SI Materials and Methods. All images were acquired and analyzed with a Zeiss LSM 700 confocal laser scanning microscope as previously described (5), with modifications fully described in SI Materials and Methods.
Statistical Analysis.
All graphs were generated using GraphPad Prism 7. Statistical tests performed using ANOVA, followed by a two-tailed t test, with Bonferroni correction.
Supplementary Material
Acknowledgments
We thank Dr. Kendra Rumbaugh’s laboratory for generating surgical wound images. This work was supported by National Institutes of Health Grant R01GM116547 (to M.W.), a grant from Human Frontiers Science (to M.W.), Cystic Fibrosis Foundation Grant WHITEL16G0 (to M.W.), Welch Foundation Grant F-1331 (to J.B.S.), Cystic Fibrosis Foundation Postdoctoral Fellowship DARCH16F0 (to S.E.D.), and a University of Texas at Austin Graduate School fellowship (to O.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719317115/-/DCSupplemental.
References
- 1.Bjarnsholt T, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Pamp SJ, Sternberg C, Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry A. 2009;75:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- 3.Roberts AEL, Kragh KN, Bjarnsholt T, Diggle SP. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 2015;427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Alhede M, et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PloS One. 2011;6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darch SE, et al. Phage inhibit pathogen dissemination by targeting bacterial migrants in a chronic infection model. MBio. 2017;8:e00240-17. doi: 10.1128/mBio.00240-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kragh KN, et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun. 2014;82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kragh KN, et al. Role of multicellular aggregates in biofilm formation. MBio. 2016;7:e00237-16. doi: 10.1128/mBio.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connell JL, et al. Probing prokaryotic social behaviors with bacterial “lobster traps”. MBio. 2010;1:e00202-10. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci USA. 2012;109:8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell JL, Kim J, Shear JB, Bard AJ, Whiteley M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc Natl Acad Sci USA. 2014;111:18255–18260. doi: 10.1073/pnas.1421211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigert M, Kümmerli R. The physical boundaries of public goods cooperation between surface-attached bacterial cells. Proc Biol Sci. 2017;284:20170631. doi: 10.1098/rspb.2017.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci USA. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drescher K, et al. Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc Natl Acad Sci USA. 2016;113:E2066–E2072. doi: 10.1073/pnas.1601702113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielson R, Kaehr B, Shear JB. Microreplication and design of biological architectures using dynamic-mask multiphoton lithography. Small. 2009;5:120–125. doi: 10.1002/smll.200801084. [DOI] [PubMed] [Google Scholar]
- 16.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci USA. 2013;110:18380–18385. doi: 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer KL, Brown SA, Whiteley M. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J Bacteriol. 2007;189:4449–4455. doi: 10.1128/JB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landry RM, An D, Hupp JT, Singh PK, Parsek MR. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol. 2006;59:142–151. doi: 10.1111/j.1365-2958.2005.04941.x. [DOI] [PubMed] [Google Scholar]
- 22.Bellin DL, et al. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun. 2014;5:3256. doi: 10.1038/ncomms4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trovato A, et al. Quorum vs. diffusion sensing: A quantitative analysis of the relevance of absorbing or reflecting boundaries. FEMS Microbiol Lett. 2014;352:198–203. doi: 10.1111/1574-6968.12394. [DOI] [PubMed] [Google Scholar]
- 24.Bellin DL, et al. Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nat Commun. 2016;7:10535. doi: 10.1038/ncomms10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koley D, Ramsey MM, Bard AJ, Whiteley M. Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc Natl Acad Sci USA. 2011;108:19996–20001. doi: 10.1073/pnas.1117298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wessel AK, et al. Oxygen limitation within a bacterial aggregate. MBio. 2014;5:e00992. doi: 10.1128/mBio.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan DN, Tsao CY, Wu HC, Bentley WE. Quorum sensing desynchronization leads to bimodality and patterned behaviors. PLoS Comput Biol. 2016;12:e1004781. doi: 10.1371/journal.pcbi.1004781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anetzberger C, Schell U, Jung K. Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals. BMC Microbiol. 2012;12:209. doi: 10.1186/1471-2180-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez PD, Hagen SJ. Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One. 2010;5:e15473. doi: 10.1371/journal.pone.0015473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frenkel ES, Ribbeck K. Salivary mucins promote the coexistence of competing oral bacterial species. ISME J. 2017;11:1286–1290. doi: 10.1038/ismej.2016.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pammi M, Liang R, Hicks J, Mistretta T-A, Versalovic J. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol. 2013;13:257. doi: 10.1186/1471-2180-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AC, et al. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun. 2017;85:e00116-17. doi: 10.1128/IAI.00116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 34.Darch SE, Ibberson CB, Whiteley M. Evolution of bacterial “frenemies”. MBio. 2017;8:e00675-17. doi: 10.1128/mBio.00675-17. [DOI] [PMC free article] [PubMed] [Google Scholar]