Significance
We demonstrate that ATRX maintains ribosomal DNA (rDNA) heterochromatin formation and stability. ATRX-depleted cells suffer extensive rDNA copy loss, resulting in a reduced ribosomal RNA transcription output and an increased sensitivity to RNA polymerase I (Pol I) inhibition. Supporting these data, we have also detected reduced rDNA copy in ATRX mutated ALT-positive human primary tumor samples and increased sensitivity of human ALT cancer cell lines to RNA Pol I transcription inhibitor. Our study highlights the therapeutic potential of Pol I transcription inhibitors for the treatment of ATRX-mutated cancers.
Keywords: ribosomal DNA, telomeres, ALT, ATRX, H3.3
Abstract
ATRX (alpha thalassemia/mental retardation X-linked) complexes with DAXX to deposit histone variant H3.3 into repetitive heterochromatin. Recent genome sequencing studies in cancers have revealed mutations in ATRX and their association with ALT (alternative lengthening of telomeres) activation. Here we report depletion of ATRX in mouse ES cells leads to selective loss in ribosomal RNA gene (rDNA) copy number. Supporting this, ATRX-mutated human ALT-positive tumors also show a substantially lower rDNA copy than ALT-negative tumors. Further investigation shows that the rDNA copy loss and repeat instability are caused by a disruption in H3.3 deposition and thus a failure in heterochromatin formation at rDNA repeats in the absence of ATRX. We also find that ATRX-depleted cells are reduced in ribosomal RNA transcription output and show increased sensitivity to RNA polymerase I (Pol I) transcription inhibitor CX5461. In addition, human ALT-positive cancer cell lines are also more sensitive to CX5461 treatment. Our study provides insights into the contribution of ATRX loss of function to tumorigenesis through the loss of rDNA stability and suggests the therapeutic potential of targeting Pol I transcription in ALT cancers.
ATRX (alpha thalassemia/mental retardation X-linked) is a member of the SWI/SNF family of helicase/ATPases. Mutations in ATRX result in developmental abnormalities including α-thalassaemia, severe mental retardation, facial dysmorphism, and urogenital abnormalities (1). ATRX acts together with death domain-associated protein (DAXX) as a histone chaperone for the histone H3 variant H3.3 (2, 3). The ATRX/DAXX/H3.3 complex is important for heterochromatin assembly at repetitive DNA sequences, such as retrotransposons, and pericentric and telomeric repeats (2–8). A number of studies have shown that specific mutations in H3.3, ATRX, and DAXX are initiating events in a range of cancers including brain cancers, pancreatic neuroendocrine tumors, chondroblastomas, and osteosarcomas (9, 10). In these cancers, ATRX mutations are linked to a telomerase-negative, ALT (alternative lengthening of telomeres) phenotype (9–12). ALT is an aberrant DNA recombination mechanism that drives telomere DNA elongation independent of telomerase activity, resulting in enormously long and damaged telomeres (11, 13). ALT activities give rise to the formation of extrachromosomal telomeric DNA byproducts that localize with telomere binding proteins within large PML (promyelocytic leukemia) bodies known as ALT-associated PML bodies (APBs) and some consist of single-stranded C-rich strands and are known as C-circles (14).
While the role of ATRX in directing heterochromatin assembly at telomeres is established (4, 15), it has also been observed that this protein binds to ribosomal RNA gene (rDNA) repeats. RNA polymerase I (Pol I)-driven transcription of rDNA produces the 47S ribosomal RNA (rRNA) precursor, which is processed to form the 18S, 5.8S, and 28S rRNAs. rRNA transcription is essential for ribosome biogenesis and cell viability. In mammalian cells, ∼15–200 copies of rRNA genes are arranged in tandem and organized into clusters. Only a fraction of the genes are transcriptionally active at any given time. In yeast, the silent rDNA copies play an essential role in maintaining the genetic stability of the rDNA repeats; however, in mammals, the role of the silent rDNA fraction is not clear (16, 17). Given their repetitive structure, rDNA repeat clusters are ideal substrates for genomic rearrangement (18). Indeed, genomic instability at the rDNA loci has been reported in human diseases including Bloom syndrome and ataxia-telangiectasia diseases, and it has been linked with increased cancer predisposition in these diseases (16, 17). In addition, the rDNA repeats show some of the most commonly observed chromosomal alterations in human cancers (17, 19, 20). However, the molecular mechanisms underlying rDNA instability in cancer are unclear.
This study aimed to determine the function of ATRX in rDNA chromatin assembly and the effect of ATRX mutation on the integrity of rDNA repeat loci in cancers. Previous studies demonstrated a reduced level of DNA methylation at rDNA repeats in primary peripheral blood mononuclear cells from patients with ATRX syndrome (4, 15), suggesting a role of ATRX in regulating heterochromatin assembly at rDNA repeats, and therefore the potential disruption of rDNA chromatin integrity in ATRX-depleted cells and ALT cancers. To investigate these hypotheses, we used a mouse ES cell model system to determine the effect of ATRX loss on chromatin assembly, repeat stability, and Pol I transcription activity at rDNA loci. In ATRX knockout (KO) ES cells, we detected a substantial reduction in rDNA repeat copy. This was the result of a loss of ATRX-mediated H3.3 deposition and heterochromatin assembly at rDNA loci. Supporting this, we detected a substantial reduction in rDNA copy number in ATRX-mutated ALT cancers including in primary human sarcoma tissue samples. This provides a further link of ATRX loss to rDNA instability and indicates rDNA copy loss to be a feature of ALT cancers. In addition, we detected reduced binding of chromatin remodeling factor UBF and Pol I at rDNA loci, resulting in a decrease in rRNA transcription and increased sensitivity of ATRX-depleted cells to Pol I transcription inhibitor CX5461. Our study provides important insights into ATRX-related genomic abnormalities in human cancers and suggests the therapeutic potential of targeting Pol I transcription in the treatment of ATRX-mutated ALT cancers.
Results
Loss of ATRX Leads to rDNA Copy Loss.
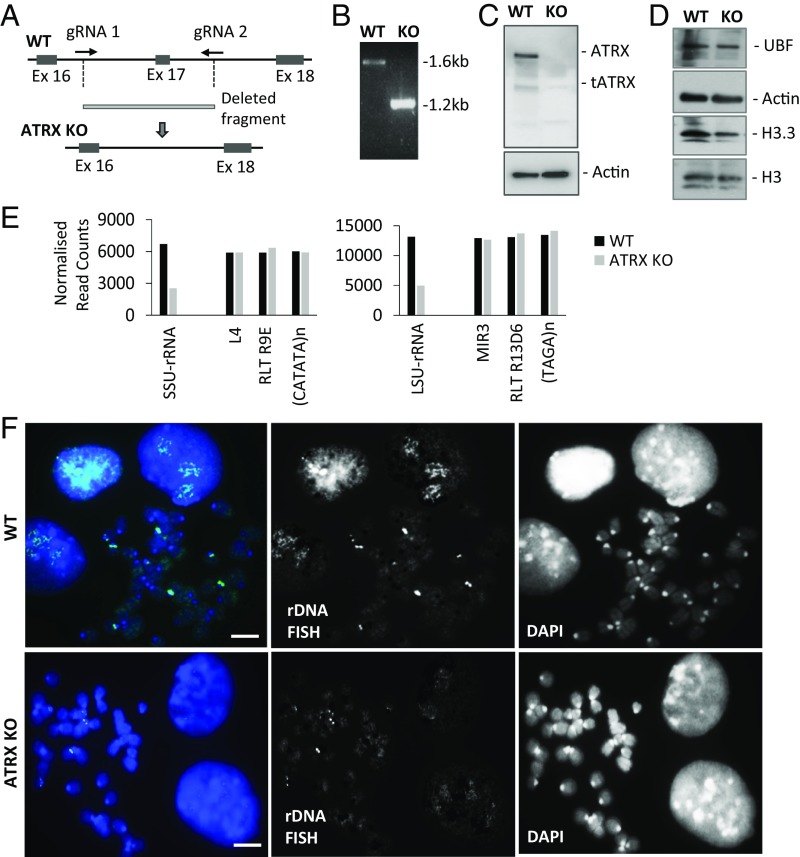
A number of studies have described presence of ATRX at rDNA repeat loci (4, 15, 21). To investigate the role of ATRX at rDNA repeats, we mapped the ChIP-sequencing datasets of ATRX and its interacting partner DAXX (4, 5, 21) and showed enriched binding of both ATRX and DAXX at the rDNA repeat loci (Fig. S1). We then used the CRISPR-Cas9–mediated DNA editing system to knock out the Atrx gene in mouse ES cells (Fig. 1 A and B). ES cells provide a valuable cell model for functional analysis of oncogenic mutations in a genetically defined normal background. They allow study of epigenetic changes associated with specific mutations in a clean background without other confounding mutated factors. Immunoblot (Fig. 1 C and D and Fig. S2A) and immunofluorescence assays (Fig. S2B) confirmed the loss of ATRX protein expression. Short-read sequencing was performed on genomic DNA prepared from whole cell extracts of WT and ATRX KO cells (Fig. 1E and Fig. S3). Repeat copy numbers were estimated by counting reads which aligned to annotated repeats and normalizing for total read counts. Representative repeats from different classes with equivalent copy numbers are shown for comparison. A greater than 50% copy-number decrease was found in both SSU-rRNA (small subunit rRNA; 18S) and LSU-rRNA (large subunit rRNA; 28S) in ATRX KO cells compared with WT cells, while no changes were seen in the content of other repetitive DNA sequences (Fig. 1E and Fig. S3). To confirm the loss of rDNA repeats in ATRX KO cells, we carried out FISH analysis and detected a reduction in FISH signal at rDNA repeats (Fig. 1F). Unlike the rDNA repeats, the stability of centromeric satellite DNA repeats was not affected in ATRX KO cells (Fig. S2C). These data indicate a selective loss of rDNA copy number in ATRX KO cells, and this variance was specific to the rDNA loci as other repetitive elements were unchanged (Fig. S3). In support of this, qPCR analyses in two additional ATRX KO ES cell clones showed a consistent loss of ∼50% of rDNA copy number without a change in UBF protein levels (Fig. S4 A and B), further supporting the link of ATRX loss to rDNA repeat instability and copy loss.
Fig. 1.
ATRX KO cells show reduced copies of rDNA. (A) Locations of guide RNAs for CRISPR-Cas9–directed KO of exon 17 in the Atrx gene in mouse ES129.1 cells. (B) Successful deletion of exon 17 leading to a shorter PCR product when primers outside exon 17 were used in PCR analysis. (C) Western blot analysis showing a loss of ATRX protein (275 KDa) in the KO cells (ATRX KO #1) using a rabbit polyclonal antibody. The faint truncated ATRX isoform (190 kDa) is also shown (33). Actin was used as loading control. (D) Western blot analysis showing equal UBF, H3.3, and H3 protein levels. Actin was used as loading control. (E) Genomic DNA reads from WT and ATRX KO cells was aligned to selected genomic repeats and normalized to total read counts (Fig. S3). In ATRX KO cells, rDNA copy number (LSU-RNA and SSU-RNA) is reduced while other repetitive DNA elements remain unchanged. (F) DNA FISH analyses showing clear and discrete rDNA foci in WT ES cells, while staining intensities were significantly reduced in ATRX KO cells. Split images are shown. (Scale bars: 5 μm.)
DNA Loss at rDNA Loci in ATRX-Mutated Human ALT Cancers.
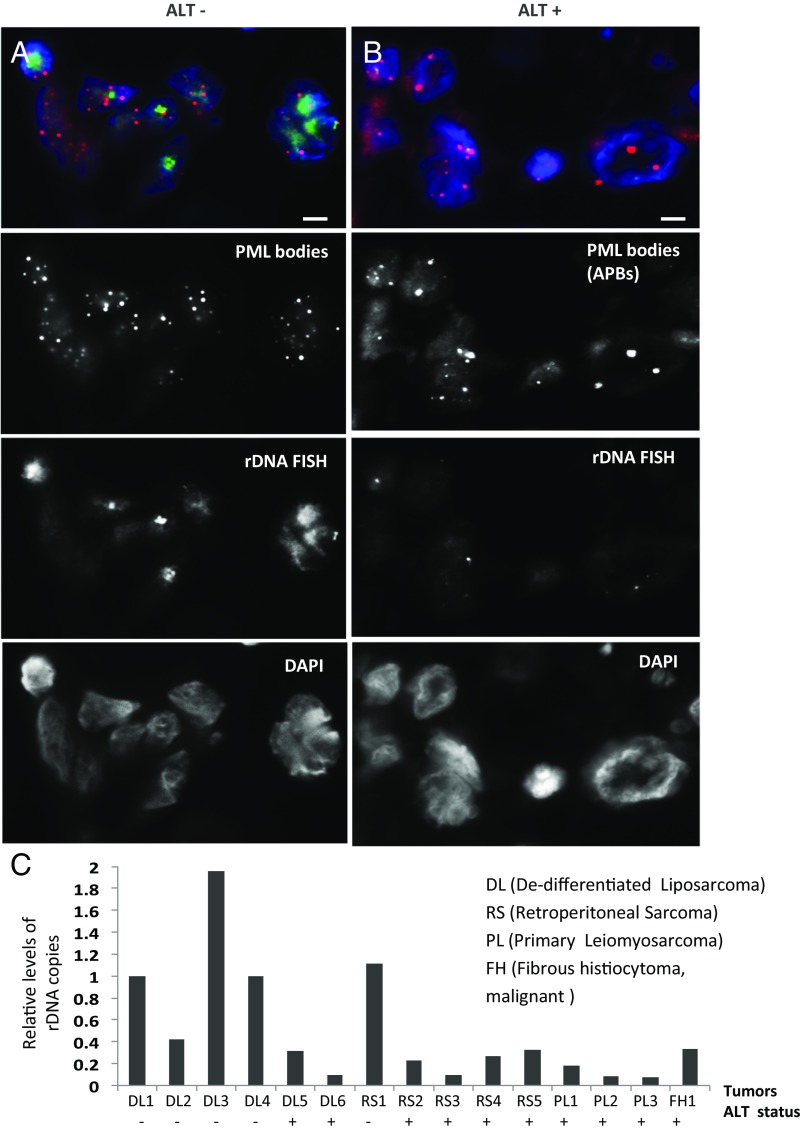
Recent studies have shown a strong association of ATRX mutations with ALT activation in human cancers including osteosarcoma, pancreatic neuroendocrine tumors, and glioblastomas (9–11, 22). Greater than 90% of ALT cancers cell lines and tumors show ATRX mutations (9–11, 22). To further investigate whether ATRX loss affects rDNA stability, we evaluated rDNA integrity in ALT-positive human cancers. FISH and qPCR analyses were performed to assess rDNA repeat copy in primary human tumor samples including dedifferentiated liposarcoma, retroperitoneal sarcoma, and leiomyosarcoma, which show a high rate of ALT activation (23). The ALT activation status of these tumors was determined by staining for the presence of large PML bodies (APBs; Table S1). Considering that ALT-positive cancers show the presence of C-circle production (14), we also validated the ALT status of the human tumor samples by assessing the levels of C-circle products (Fig. S5). FISH analyses showed a reduction in rDNA FISH signals in ALT-positive tumor samples (Fig. 2 A and B). Consistent with this finding, qPCR analyses also showed a lower rDNA copy number in ALT-positive samples, compared with those in ALT-negative samples (Fig. 2C). These data further illustrate the link between ATRX loss and rDNA repeat instability and suggest rDNA repeat copy loss as a feature of ALT-positive tumors.
Fig. 2.
ATRX-null ALT cancers showed rDNA copy loss. (A and B) Immunofluorescence analysis was performed using antibody against PML. ALT-negative (ALT−) tumors show the presence of small PML bodies, while ALT-positive (ALT+) tumors showed staining of large APBs. ALT status was determined by the presence of APB bodies and increased C-circle formation (Fig. S5 and Table S1). DNA FISH analyses showed prominent rDNA foci on APB/ALT-negative tumors, whereas the signal intensity was significantly reduced in APB/ALT-positive tumors. (Scale bars: 5 μM.) (C) DNA was extracted from human dedifferentitaed liposarcoma (DL), retroperitoneal sarcoma (RS), primary leiomyosarcoma (PL), and fibrous histocytoma (FH) tissue samples, followed by qPCR analyses using primers against ETS (of human rDNA repeat) and GAPDH housekeeping gene. Values of rDNA copy number were normalized against GAPDH DNA copy number for each sample. Overall, the ALT-positive samples show lower levels of rDNA copies compared with ALT-negative samples.
ATRX Is Required for Maintaining rDNA Chromatin Integrity.
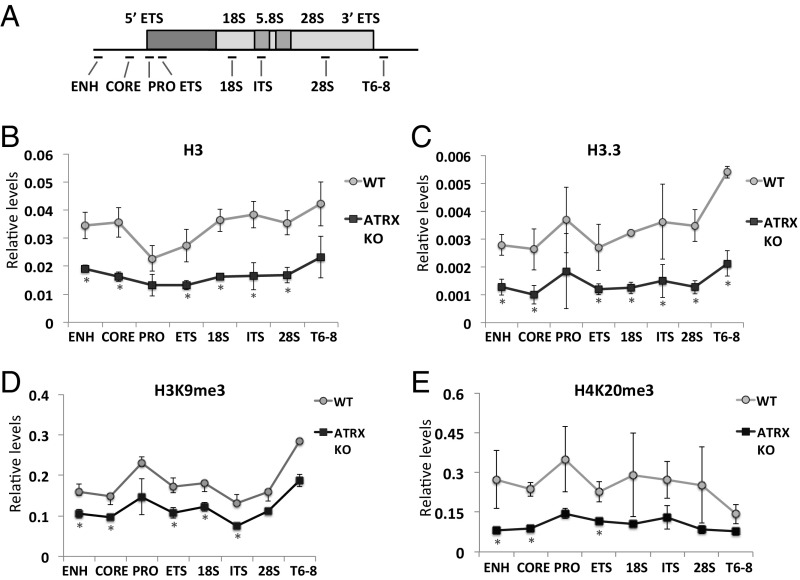
Because ATRX plays a role in directing heterochromatin assembly at repeats such as the telomeres (2, 3), we hypothesized that chromatin status at rDNA repeats may be affected in ATRX KO cells. To determine this, ChIP/qPCR analyses were performed on the WT and ATRX KO cells (Fig. 3). As controls (2, 4, 7), we detected reduced levels of H3.3, H3K9me3, and H4K20me3 at the telomeres in the ATRX KO cells (Fig. S6A). In these cells, we also detected significant reductions (approximately half) in the levels of H3 and H3.3 at the rDNA repeats (Fig. 3 B and C). These reductions in the levels of H3 and H3.3 at rDNA repeats were likely attributed to the loss in rDNA copy number in the absence of ATRX and thus H3/H3.3 occupancy per rDNA repeat is unaltered in ATRX KO cells. To determine the possible changes in histone modification patterns on the remaining rDNA chromatin in the ATRX KO cells, ChIP/qPCR analyses were performed using antibodies against two heterochromatin marks, H3K9me3 and H4K20me3, and normalized to the total level of H3 at rDNA repeats (Fig. 3 D and E). In the ATRX KO cells, we found decreases in H3K9me3 and H4K20me3 at a subset of loci within the rDNA repeats (Fig. 3 D and E), suggesting that ATRX loss affects heterochromatin formation at these repeats. Next, we performed ChIP analyses to assess for potential increase in DNA damage and detected an increased γH2Ax level at the rDNA repeat loci (Fig. S6C). These findings suggest that ATRX loss affects heterochromatin assembly and rDNA chromatin integrity.
Fig. 3.
rDNA chromatin state is changed in ATRX KO cells. (A) A schematic diagram of rDNA cluster, showing the locations of various PCR fragments across the region. (B and C) ChIP/qPCR analyses of H3 (B) and H3.3 (C) in WT and ATRX KO (clone KO #1) mouse ES cells showing substantial losses of H3 and H3.3 at the rDNA repeats in the absence of ATRX. (D and E) To determine the levels of heterochromatin present at the rDNA repeats, H3K9me3 (D) and H4K20me3 (E) ChIP/qPCR analyses were performed and normalized to the H3 ChIP/qPCR values, and reductions in both heterochromatin marks, H3K9me3 and H4K20me3, appeared to be significant at a subset of the rDNA repeat loci (error bars represent SD, n = 4, *P < 0.05).
ATRX-Depleted Cells Show Reduced rRNA Transcription Output and Increased Sensitivity to RNA Pol I Transcription Inhibition.
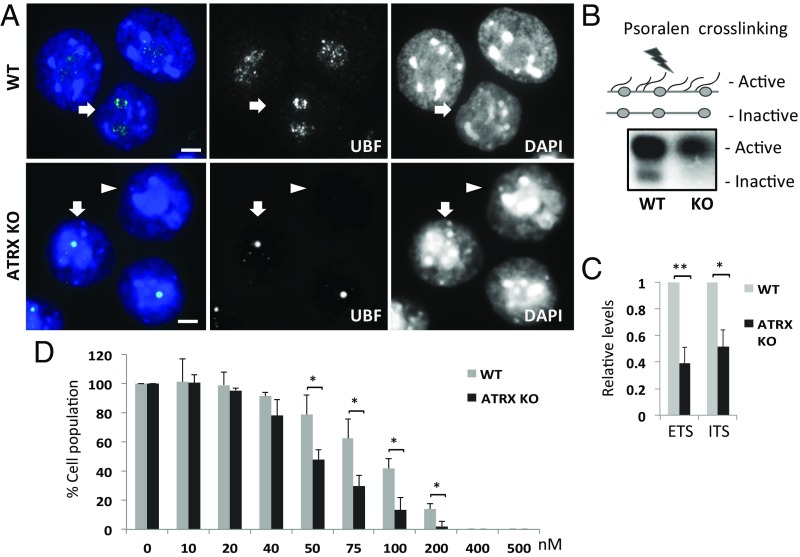
Next, we predicted that the altered rDNA copy in the absence of ATRX might affect rRNA transcription. We therefore examined the effect of ATRX loss on the binding of UBF, which is essential for establishing and maintaining a decondensed rDNA chromatin state permissive for Pol I transcription (24). We detected a striking change in UBF staining from a prominent extensive pattern in WT cells to a tight focal pattern with an overall reduced intensity in ATRX KO cells (Fig. 4A), suggesting that UBF binding was affected in the absence of ATRX. Consistent with this, ChIP/qPCR analyses showed reduced levels of binding of UBF and Pol I at rDNA repeat (Fig. S6 D and E). We then analyzed the ratio of active to inactive rDNA chromatin by performing psoralen cross-linking combined with Southern blotting, which distinguishes psoralen-accessible open rDNA chromatin structures from inaccessible and closed, silent rDNA (24). The analysis revealed the almost complete loss of the silent rDNA pool (Fig. 4B) in ATRX KO cells and the data were consistent with the loss of heterochromatic silencing in the absence of ATRX (Fig. 3 D and E). In addition, the active rDNA fraction was also decreased in ATRX KO cells compared with WT (Fig. 4B), in line with the reduction in rDNA copy (Fig. 1 and Figs. S3 and S4) and overall decreases in the binding of UBF and Pol I to rDNA repeats (Fig. S6 D and E) upon ATRX depletion. As a consequence, a significant reduction (close to 50%) in the level of 47S/45S rRNA precursor transcripts was detected in ATRX KO cells (Fig. 4C). Based on these findings, we propose that ATRX mediated H3.3 deposition and heterochromatin formation is required to maintain stable rRNA synthesis. To further explore the importance of H3.3 deposition at rDNA repeat loci, we investigated the impact of the loss of function of DAXX, which acts in complex with ATRX to deposit H3.3 on the total rRNA synthesis (Fig. S7). Like the ATRX KO cells, a similar pattern of reduced rRNA transcription output was found in DAXX KO ES cells (25) (Fig. S7 A and B). This reduction in the total rRNA synthesis in DAXX KO cells is associated with the altered UBF binding and reduced rDNA copy as shown by immunofluorescence, DNA FISH, and qPCR analyses (Fig. S7 C–E).
Fig. 4.
ATRX KO cells showed altered UBF binding pattern and reduced rRNA transcription. (A) Immunofluorescence showing a strong accentuated speckle-like UBF staining distribution in WT ES cells, while ATRX KO (clone #1) cells showed a dense focal UBF staining pattern. Split images are shown. (Scale bars: 5 μM.) (B) Psoralen cross-linking of rDNA chromatin followed by DNA fragmentation and Southern blot analysis. Compared with WT cells, ATRX KO cells showed reduced level of inactive rDNA chromatin over the active chromatin. In addition, there was a noticeable decrease in the active fraction. (C) RT-PCR analyses showing that ATRX KO cells were reduced in rRNA transcript levels compared with WT cells (error bars represent SD, n = 5, *P < 0.05, **P < 0.001). Levels of transcripts were normalized against the level of Actin transcript. (D) Equal numbers of WT and ATRX KO ES cells were treated with CX5461 (ranging from 0 to 500 nM, x axis) for 48 h. The y axis shows the percentage of the cell population in comparison with the cell population in untreated groups (error bars represent SD, n = 3 in every treatment group, *P < 0.05).
Because we found that rDNA transcription output decreased upon the loss of ATRX, we predicted this could lead to increased sensitivity of cells to inhibition of Pol I transcription. We treated WT and ATRX KO cells with CX5461 (26), an inhibitor that selectively inhibited Pol I transcription, and showed that ATRX KO cells were more sensitive to growth inhibition by CX-5461 compared with the WT cells (Fig. 4D). To further investigate this, we also treated ALT-positive sarcoma cell lines with CX-5461 and found that ALT-positive U2OS and SAOS2 cells were also more sensitive to CX-5461 treatment compared with ALT-negative MG63 and B143 cells (Fig. S8). We also tested drug sensitivity with α-amanitin RNA Pol II inhibitor and found no increased sensitivity of ATRX KO and ALT-positive U2OS toward treatment with α-amanitin (Fig. S9). Taken together, we propose that the disruption in heterochromatin formation and rDNA copy loss lead to reduced total rRNA transcription output and increased sensitivity to Pol I inhibition in ATRX-depleted cells. This suggests the potential therapeutic utility of Pol I transcription inhibitors for treating ALT cancers.
Discussion
A number of studies have documented the importance of ATRX-mediated H3.3 deposition in controlling heterochromatin formation at repetitive DNA elements (4–6). The close association of ATRX mutation with ALT activation, characterized by highly unstable and damaged telomeres, also illustrates this. Here we show that ATRX is required for the maintenance of another important class of DNA repeats in the genome, rDNA repeats. Like at the telomeres, ATRX directs heterochromatin formation at rDNA loci through recruitment of H3.3 and trimethylation of H3K9 and H4K20 (Fig. 3). The loss of ATRX leads to the disruption of ATRX-mediated heterochromatin assembly and thus affects rDNA stability and copy number, as shown by DNA sequencing and FISH analyses (Fig. 1). Additionally, our findings that ALT-positive primary sarcoma tumors exhibit substantial loss of rDNA copy (Fig. 2) and the high frequency of ATRX mutations in ALT-activated cancers (9–11) further support the importance of ATRX for the maintenance of rDNA chromatin integrity and rDNA loci stability. Based on these findings, we propose that rDNA instability/copy loss is a hallmark of ATRX-related genomic abnormalities and provides a biomarker and molecular indicator for ALT tumors.
Recent studies have highlighted the critical role of Pol I in driving rDNA transcription and ribosomal biogenesis for survival of cancer cells. Importantly, it has been reported that rDNA transcription can be therapeutically targeted to selectively kill cancer cells in in vivo models of lymphoma and leukemia (26). Here we show that the reductions in UBF and RNA Pol I binding at rDNA repeats, and the overall decrease in rRNA transcription output, also lead to the increased sensitivity of ATRX KO cells to Pol I transcription inhibitor CX5461. This indicates the therapeutic potential of Pol I transcription inhibitors such as CX5461 for treatment of ALT cancers that show a high mutation rate in ATRX. The reduced pool of active rDNA and decreased rate of rDNA transcription may render ATRX mutant cancers sensitive to inhibitors of Pol I transcription.
Our data reinforce ATRX’s role as a guardian that protects the integrity and stability of repetitive DNA in the genome (6) and suggest that rDNA instability is driven by a similar mechanism that promotes telomeric DNA rearrangement in the absence of ATRX through a defective heterochromatin assembly pathway. In support of our findings, work in Drosophila has demonstrated the importance of heterochromatin organization for maintenance of nucleolar structure and regulation of rDNA transcription. Loss of H3K9 histone methyltransferase Su(var)3-9, a driver of H3K9 trimethylation, causes fragmentation of nucleolus and formation of extrachromosomal rDNA circles. This effect is attributed to aberrant recombination of rDNA repeat sequences, resulting in instability of the loci (27). This has been attributed to aberrant recombination of rDNA repeat sequences, resulting in instability of the loci. Given the close interplay between ATRX and SUVAR39H1/H2 (2–7), we propose that ATRX maintains rDNA heterochromatin through SUVAR3-9H1/H2–mediated K9 trimethylation of H3.3. Consequently, loss of ATRX leads to a failure to form heterochromatin and, in turn, an increase in damage at rDNA loci. Importantly, a recent study in Caenorhabditis elegans has shown that H3K9 methylation is required for suppressing R-loop–associated repeat instability (28). R-loops are three-stranded nucleic acid structures that comprise nascent RNA hybridized with the DNA template. rDNA repeats that are highly GC-rich in sequence have a high tendency to form these structures, and a recent study has shown a role of ATRX in suppressing R-loop formation at telomeric sequences (29). It is plausible that ATRX-mediated heterochromatin assembly may also prevent R-loop formation at rDNA loci. In the absence of ATRX, these structures may persist, leading to DNA damage, as shown by the up-regulated levels of γ-H2Ax at rDNA repeats in ATRX KO cells (Fig. S6C). As a consequence, the increased damage may then lead to recombination-mediated repair at rDNA loci and hence reductions in DNA copy and transcription output (Figs. 1 and 3). Future investigations are needed to address these important mechanistic questions.
Here we have demonstrated a strong correlation between ATRX mutations and the loss of rDNA integrity, and that rDNA instability is a hallmark of ALT cancers. We propose a model in which loss of ATRX function impairs heterochromatin assembly at rDNA repeats. This in turn leads to rDNA repeat destabilization and copy loss. rDNA copy loss has been shown to be a driving force for global genomic instability in yeast, although the link is not clear in mammalian cells. In human ALT cancers that are frequently mutated in ATRX (>90%), disruption in heterochromatin silencing and reductions in rDNA content could directly affect global genome stability and increase recombination rates to further promote cancer development. Finally, we propose that rDNA instability in ATRX mutated ALT cancers may provide a vulnerability to selective inhibitors of Pol I transcription, which are currently undergoing phase I clinical trials for cancer treatment (26). Further, reduced rDNA copy number may function as a biomarker or molecular indicator for sensitivity of ALT cancers to Pol I transcription inhibitors. Our study potentially unveils a novel therapeutic strategy for ATRX-mutated tumors.
Methods
Cell Culture and Drug Treatment.
Human colon cancer cell lines B143 and SW620 and osteosarcoma cell lines MG63, U2OS, and SAOS2 were cultured in RPMI supplemented with 10% heat-inactivated FCS, 100 U/mL penicillin, and 10 mg/mL streptomycin in a humidified 5% CO2/95% air atmosphere at 37 °C. WT and DAXX KO (25) mouse ES cells were cultured in DMEM supplemented with 15% FCS, 103 units/mL leukemia inhibitory factor, and 0.1 mM β-mercaptoethanol. For drug treatment, WT and ATRX KO ES cells (1 × 105) were seeded onto 12-well plates. After 12 h, cells were treated with either with CX5461 (0–500 nM) or α-amanitin (0–40 μg/mL) for 48 h. Human B143, MG63, U2OS, and SAOS2 cells were treated with CX5461 ranging from 0 to 10 μM for 48 h. Cell number was then counted after treatment.
Human Sarcoma Tissue Samples.
Formalin-fixed paraffin-embedded human tumor tissues were obtained from the Health Science Alliance (HSA) Biobank, University of New South Wales Biorepository. The use of the human tissues was approved by Monash University Human Research Ethics Committee and HSA Biobank. To purify DNA from tissue blocks, slides were immersed in 100% xylene overnight, rinsed in 100% and 70% ethanol, and air-dried. Lysis buffer [100 mM Tris⋅Cl, pH 8.0, and 50 mM EDTA; 1% (wt/vol) SDS] was added to remove tissue off the slides, followed by incubation with Proteinase K at 55 °C overnight. The samples were then subjected to phenol/chloroform and ethanol precipitation.
C-Circle Assay.
Rolling circle amplification of C-circle was performed with 20 ng genomic DNA, 0.2 µg/µL BSA, 0.1% Tween, 4 mM DTT, 1.2 mM each of dATP, dCTP, dGTP, dTTP, and φ29 DNA polymerase (φ29; 3.75 U/16 ng DNA) (NEB) in 1× φ29 buffer (14). The reaction was incubated at 30 °C for 4 h then at 70 °C for 20 min. For each sample, the assay was done with and without φ29. The assay products were diluted for qPCR analyses using telomeric DNA and single-copy gene primer (GAPDH) to determine the level by measuring the increase in total telomeric DNA. C-circle level was reported as the relative telomeric content of the C-circle assay products relative to a single-copy gene (GAPDH).
Genomic DNA Preparation and Illumina HiSeq Sequencing.
Genomic DNA from WT and ATRX KO cells were prepared for ChIP input. In brief, cells were fixed with 1% formaldehyde for 10 min at room temperature. Genomic DNA was released through sequential lysis with 0.2% Igepal and 1% SDS and sonicated with a Bioruptor (Diagenode) to obtain chromatin fragments <500 bp. Sample concentrations were determined and 20 ng of DNA was used as starting material. ChIP libraries were prepared with Nugen Ovation Ultralow System V2 (Nugen protocol M01379v1, 2014) with 11 cycles of amplification. Libraries quality was assessed by Bioanalyzer (Agilent) and qPCR, and a single equimolar pool was made based on size-adjusted qPCR quantitation. Following denaturation, 12 pM of library pools were used for hybridization and cluster generation (Illumina protocol 15006165 v02, February 2016), and samples were sequenced on an Illumina HiSEq. 1500 rapid mode (50-bp SR sequencing; Illumina protocol 15035788 Rev D, April 2014).
Repeats Analysis.
Fastq files were aligned to genomic repeats using Repeat Enrichment Estimator v1.0 (30) with mouse mm9 repeat assembly based on RepBase annotations (31). Data were normalized for total read counts. For comparison of genomic sequencing, repeats with greater than 5,000 reads in the WT sample were log2-transformed and plotted with heatmap.2 (32) in R (v 3.3.2). Published GEO datasets of ATRX (21) (GSE22162 and GSM551138) and DAXX (5) (GSE59189 and GSM1429920) ChIP-seq were mapped to repeats as described above and compared with respective input tracks (GSM639380 and GSM1429923). Data were normalized for total read counts.
Additional experimental procedures are provided in Supporting Information.
Supplementary Material
Acknowledgments
We thank HSA Biobank (University of New South Wales) for providing the tumor tissue samples. This work was supported by the Norwegian Cancer Society and the Research Council of Norway (to P.C.); an Australia Research Council Future Fellowship award (to L.H.W.); National Health and Medical Research Council Program Grant 1053792 (to R.B.P. and R.D.H.), senior research fellowships (to R.B.P. and R.D.H.), and a project grant (to L.H.W.); and a Cure Brain Cancer Foundation Australia project grant (to L.H.W. and H.P.J.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720391115/-/DCSupplemental.
References
- 1.Gibbons RJ, Suthers GK, Wilkie AO, Buckle VJ, Higgs DR. X-linked alpha-thalassemia/mental retardation (ATR-X) syndrome: Localization to Xq12-q21.31 by X inactivation and linkage analysis. Am J Hum Genet. 1992;51:1136–1149. [PMC free article] [PubMed] [Google Scholar]
- 2.Wong LH, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong LH, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law MJ, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 5.Elsässer SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voon HP, Wong LH. New players in heterochromatin silencing: Histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res. 2016;44:1496–1501. doi: 10.1093/nar/gkw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Udugama M, et al. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015;43:10227–10237. doi: 10.1093/nar/gkv847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang FT, et al. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 2013;41:4447–4458. doi: 10.1093/nar/gkt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 11.Lovejoy CA, et al. ALT Starr Cancer Consortium Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voon HPJ, Collas P, Wong LH. Compromised telomeric heterochromatin promotes ALTernative lengthening of telomeres. Trends Cancer. 2016;2:114–116. doi: 10.1016/j.trecan.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 14.Henson JD, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons RJ, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 16.Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 17.Diesch J, Hannan RD, Sanij E. Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease. Cell Biosci. 2014;4:43. doi: 10.1186/2045-3701-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McStay B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016;30:1598–1610. doi: 10.1101/gad.283838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stults DM, et al. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009;69:9096–9104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
- 20.Xu B, et al. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017;13:e1006771. doi: 10.1371/journal.pgen.1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voon HP, et al. ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 2015;11:405–418. doi: 10.1016/j.celrep.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaphy CM, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanij E, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelson JS, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bywater MJ, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeller P, et al. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat Genet. 2016;48:1385–1395. doi: 10.1038/ng.3672. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen DT, et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017;18:914–928. doi: 10.15252/embr.201643078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day DS, Luquette LJ, Park PJ, Kharchenko PV. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 2010;11:R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warnes GR, et al. 2016 Package ‘gplots’: Various R programming tools for plotting data. R version 4(4):1. Available at https://cran.r-project.org/web/packages/gplots/gplots.pdf.
- 33.Garrick D, et al. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]