Significance
Natural products biosynthesized by cryptic gene clusters represent a largely untapped source for drug discovery. However, mining of these products by promoter engineering is restricted by the lack of streamlined genetic tools, especially in nonmodel biosynthetic gene cluster (BGC)-rich bacteria. Here, we describe the discovery of a pair of bacteriophage recombinases and application of recombinase-assisted promoter engineering to rapidly identify and activate several cryptic biosynthetic gene clusters in two Burkholderiales strains that currently lack effective genetic tools. Construction of an efficient genome engineering platform in a natural product producer expedites mining of cryptic BGCs in their native backgrounds, and host melioration for yield or structure optimization. This strategy enables potentially scalable discovery of novel metabolites with intriguing bioactivities from many other bacteria.
Keywords: recombinases, genome mining, Burkholderiales, lipopeptide, natural product
Abstract
Bacterial genomes encode numerous cryptic biosynthetic gene clusters (BGCs) that represent a largely untapped source of drugs or pesticides. Mining of the cryptic products is limited by the unavailability of streamlined genetic tools in native producers. Precise genome engineering using bacteriophage recombinases is particularly useful for genome mining. However, recombinases are usually host-specific. The genome-guided discovery of novel recombinases and their transient expression could boost cryptic BGC mining. Herein, we reported a genetic system employing Red recombinases from Burkholderiales strain DSM 7029 for efficient genome engineering in several Burkholderiales species that currently lack effective genetic tools. Using specialized recombinases-assisted in situ insertion of functional promoters, we successfully mined five cryptic nonribosomal peptide synthetase/polyketide synthase BGCs, two of which were silent. Two classes of lipopeptides, glidopeptins and rhizomides, were identified through extensive spectroscopic characterization. This recombinase expression strategy offers utility within other bacteria species, allowing bioprospecting for potentially scalable discovery of novel metabolites with attractive bioactivities.
Bacteria represent an important source of medicines and pesticides (1, 2). Current genome sequencing projects have revealed a large number of biosynthetic gene clusters (BGCs), but their natural products have not yet been identified (3–5). The enzymes encoded by these unexploited biosynthetic genes may synthesize metabolites that could serve as a reservoir of novel scaffolds for drug screening (6). Two major genome mining strategies, featuring homologous and heterologous expression, were employed to mine cryptic BGCs (7). Homologous expression in the native producers ensures proper substrates, precursors, cofactors, and accessory factors for biosynthesis, which could maximize generation of cryptic BGC biosynthesis products. However, this strategy requires efficient genetic manipulation tools in the producers to meet the demand of high-throughput genome mining in the postgenomic era. Recently, yeast homologous recombination and CRISPR-Cas9 strategies were harnessed to activate silent BGCs by promoter engineering in metagenomic DNA and natural product proficient streptomycetes, respectively (8, 9). However, in other natural product producers, such as bacilli, Burkholderiales, myxobacteria, and cyanobacteria, scalable and efficient genome manipulation is still a frustrating challenge.
Red/ET recombineering is an in vivo homologous recombination-based genetic engineering method employed primarily in Escherichia coli by using short homology arms (∼50 bp) based on the expression of either Redα/Redβ from the Red operon of λ phage or the analogous RecE/RecT from Rac prophage, encoded in the E. coli chromosome (10–13). Full-length RecE and RecT mediate linear−linear homologous recombination (LLHR), which can be used to directly clone large DNA regions from genomic DNA into E. coli expression vectors (14), while Redα and Redβ promote homologous recombination between linear and circular DNA molecule [linear plus circular homologous recombination (LCHR)]. In addition, Redαβ recombineering is particularly relevant to bacterial genome engineering for genome mining, because the linear targeting DNA carrying short homology regions (conveniently generated by PCR) is sufficient for gene modification, and dedicated targeting plasmids are not required, unlike in single- or double-crossover mutagenesis (15). The Redαβ system from E. coli has been directly applied to several closely related Gram-negative bacteria for genome engineering, e.g., Salmonella enterica (16), Klebsiella pneumoniae (17), Agrobacterium tumefaciens (18), Pantoea ananatis (19), and naturally transformable Burkholderia species (20, 21). However, it is not functional in other Burkholderiales strains (20, 21). In more distant species, host-specific Redαβ or RecET homolog systems are mandatory (22–29).
The order Burkholderiales belongs to β-Proteobacteria and includes several pathogenic bacteria and several environmental species. A number of Burkholderiales genomes contain plentiful putative natural product BGCs (4, 5, 30, 31). The proportion of polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) in Burkholderia genomes is second only to that of actinobacteria and is higher than that of bacilli, cyanobacteria, and myxobacteria (32). Burkholderia species also exhibit great biosynthetic potential for production of siderophores and lipopeptides (31). Thus, Burkholderiales species have emerged as a new source of natural products with a plethora of BGCs still cryptic or silent (31–34). To access these BGCs, it is urgent to invent Burkholderiales specific homologous expression systems, particularly, novel Red/ET recombinases from Burkholderiales strains for efficient genome engineering.
This study invented a workflow for the discovery of novel recombinases and the associated transient expression system to facilitate in situ promoter insertion for cryptic BGC mining (Fig. 1). In particular, a pair of Redαβ homologs (Redαβ7029) in Burkholderiales strain DSM 7029 (35) was discovered, cloned for transient expression, and optimized for the efficient deletion of large chromosomal DNA fragments (up to 200 kb). The specialized recombinases-assisted in situ constitutive promoter insertion also facilitated mining of cryptic BGCs. Three cryptic NRPS-PKS BGCs were activated or mined in the native host. These specialized recombinases were also applied to other Burkholderiales strains that currently lack effective genetic tools and native Redαβ homologs, which resulted in the successful identification of two cryptic NRPS BGCs. Two of five identified BGCs were silent (i.e., products not detectable by liquid chromatography-mass spectrometry (LC-MS) analysis in their native hosts under the given conditions). In addition, two classes of lipopeptides, glidopeptins and rhizomides, were identified from DSM 7029 and Paraburkholderia rhizoxinica HKI 454 (36), respectively.
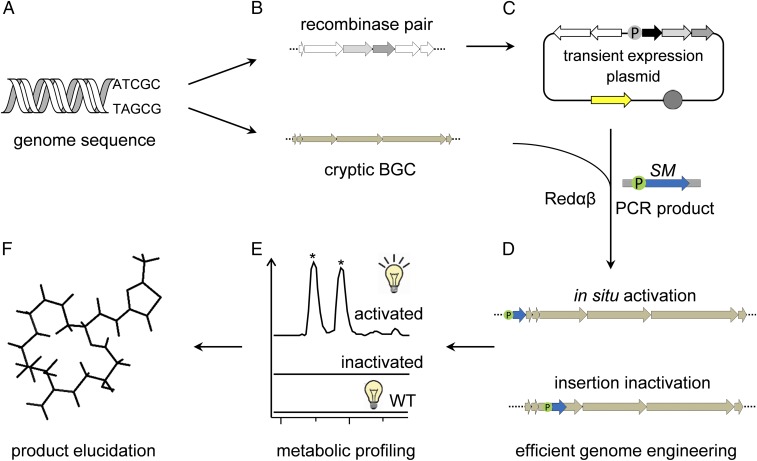
Fig. 1.
Workflow for the discovery of novel recombinases and a transient expression system to facilitate in situ promoter insertion for cryptic BGC mining. (A) Access to bacterial genome sequences. (B) Bioinformatics analysis to identify cryptic BGCs and also the phage recombinase pairs. (C) Construction of a recombinase transient expression system and verification of its function. (D) Use of a transient recombinase expression to generate BGC-inactivated mutants by gene replacement and BGC activation mutants by in situ promoter insertion. (E) Fermentation and comparative metabolic profiling analysis of mutants and wild-type strains to identify different peaks. The asterisks indicate activated products of a cryptic gene cluster. (F) Structural elucidation of potential products by HRMS and NMR measurements. P, promoter; SM, antibiotic selection marker.
Results
Host-Specific Phage-Derived Recombinase Pairs in Burkholderiales.
It had been shown that the Redαβ or RecET recombinases from E. coli could not be applied to nonnaturally transformable Burkholderia strains (21). This study also confirmed that Redαβ was not functional in the Burkholderiales strain DSM 7029 (Fig. 2). BlastP analysis using the amino acid sequences from E. coli Redαβ or RecET as queries revealed 7 Redαβ-like and 10 RecET-like recombinase pairs in ∼1,000 Burkholderiales strains (most of which belong to Burkholderia and Paraburkholderia genera) (SI Appendix, Table S1). A Redαβ-like operon in DSM 7029 contains a putative single-strand annealing protein AAW51_RS10370 (Redβ7029) that is functionally equivalent to Redβ, a neighboring 5′-3′ exonuclease AAW51_RS10365 (Redα7029) equivalent to Redα, a putative recombinase AAW51 RS10355 (H7029), and six hypothetical proteins (Fig. 2A). A pair of RecET-like recombinases (ET-BDU) in Burkholderia sp. BDU8 harbors a three-gene operon encoding a putative single-strand annealing protein WS71_RS13960 (RecT-BDU), an adjacent exonuclease WS71_RS13965 (RecE-BDU), and a putative inhibitor of the exonuclease WS71_RS13955 (H-BDU) which is similar to the E. coli exonuclease inhibitor Redγ (Fig. 2A). These two pairs of recombinases were chosen as representatives of the Redαβ and RecET systems for recombination assays in DSM 7029, because Redαβ7029 are the native recombinases and Redβ7029 shows a high identity to Redβ of E. coli, and RecET-BDU operon includes a putative redγ-like gene (Fig. 2A and SI Appendix, Table S1).
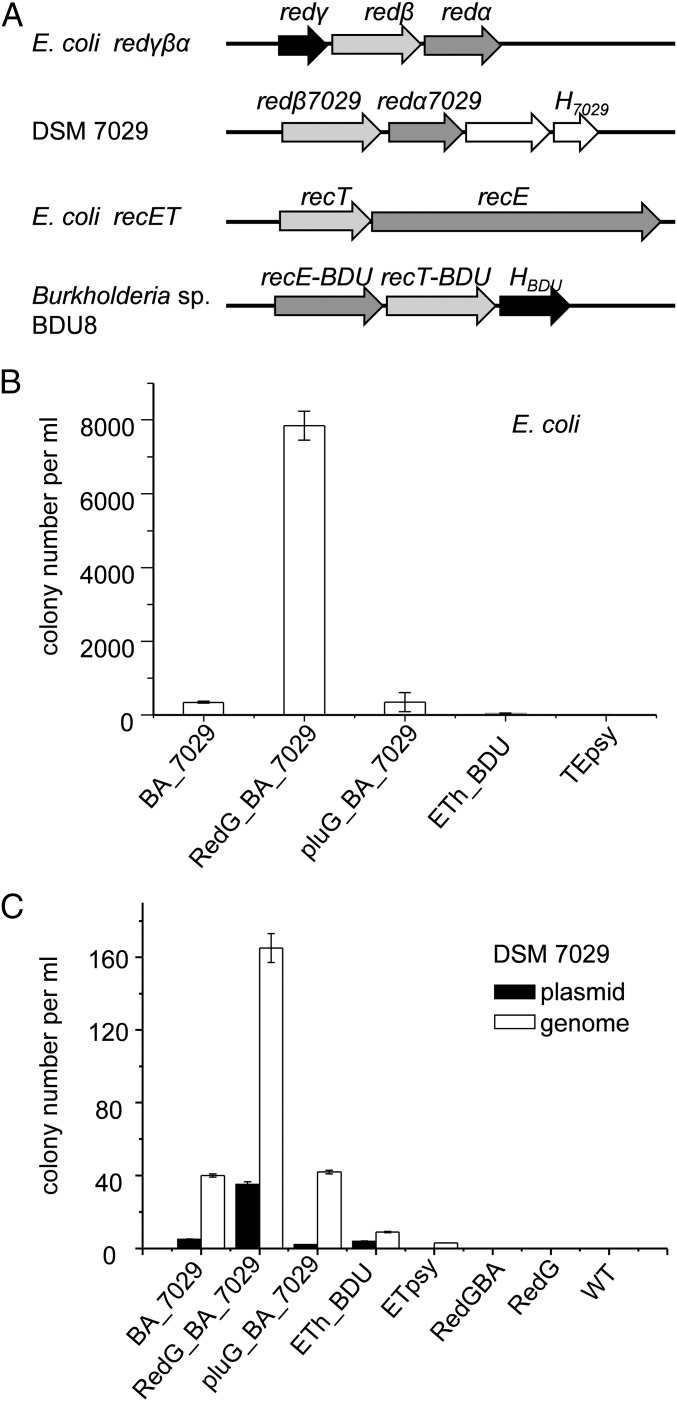
Fig. 2.
Selected Red/ET recombinase pairs in Burkholderiales and their functional characterization. (A) Recombinase pairs from E. coli and their homologs in DSM 7029 and Burkholderia sp. BDU8. Arrows with the same shading represent similar functional genes. (B) Functional characterization of recombinase combinations in E. coli using an LCHR assay. BA_7029, Redαβ7029; RedG_BA_7029, Redγ from E. coli combined with Redαβ7029; pluG_BA_7029, Pluγ from Photorhabdus luminescens combined with Redαβ7029; ETh_BDU, Rec ET homologs and a hypothetical protein of Burkholderia sp. BDU8; ETpsy, RecET homologs from P. syringae pv. tomato DC3000. (C) Functional characterization of recombinase combinations for plasmid and genome modification in DSM 7029. RedGBA, Redαβγ from E. coli; RedG, Redγ from E. coli. Error bars, SD; n = 3.
Functional Verification and Optimization of Recombinase Pairs.
To study their functions, we constructed a variety of combinations of recombinases (SI Appendix, Table S2), and two Redγ-like proteins (Redγ or Pluγ) that inhibit endogenous exonucleases and enhance homologous recombination mediated by the Redαβ and RecET systems are also included (28, 37). Transient high-level expression of the phage recombinases is important when manipulating DNA in vivo (13). As Redαβ7029 were transcriptionally silent in DSM 7029 strain (SI Appendix, Table S3), it is essential to place these phage recombinase genes under an inducible promoter, so that recombination can be induced when required and undesired recombination events can be temporally restricted (10). We first tested the efficiency of induction and stringency of four frequently used inducible promoters, the tetracycline-regulated promoter (tet) (38), the arabinose-regulated promoter (BAD) (39), the isopropyl thiogalactoside-regulated promoter (tac) (40), and the rhamnose-regulated promoter (Rha) (41), using the firefly luciferase gene in a pBBR1 plasmid as a reporter in DSM 7029 (SI Appendix, Fig. S1A) (42). The relative light unit (RLU) of the rhamnose-induced Rha promoter was at least 1.5 times higher than that of the other three inducible promoters with corresponding inducers, and the naive Rha promoter showed very weak RLU (SI Appendix, Fig. S1B). Although tet promoter is also stringent, previous reports indicated leaky expression during epothilone production in DSM 7029, rendering it unsuitable for our experiments (43). Therefore, Rha promoter was utilized to control recombinase expression in the subsequent work on the basis of its induction efficiency and stringency in DSM 7029.
We then tested the recombination efficiency between linear DNA and circular DNA (LCHR) of different recombinase combinations under the control of inducible Rha promoter in E. coli and DSM 7029 using plasmid and genome modification assay, respectively (SI Appendix, Fig. S2 A and B). Redαβ7029 functions both in E. coli, with colony number per milliliter (cnpm) of ∼400 (Fig. 2B), and in DSM 7029, with cnpm of ∼40 (Fig. 2C). The hypothetical protein H7029 had no positive effect on recombination (SI Appendix, Fig. S2C). The addition of Redγ into Redαβ7029 remarkably enhanced the recombination efficiency in both E. coli (Fig. 2B) and DSM 7029 (Fig. 2C) as it synergizes in function with E. coli Redαβ (37), whereas, Pluγ or HBDU did not. The ETh-BDU recombinase pair and the TEpsy system from Pseudomonas syringae (25) were also functional, but at a low efficiency, with both cnpm less than 10. The Redαβγ from E. coli or Redγ alone did not exhibit recombination activity in DSM 7029 (Fig. 2C). The Redαβ7029 operon was more efficient in the LCHR assay (SI Appendix, Fig. S3), and the recombination efficiency increased significantly with increase in homology arm length (SI Appendix, Fig. S2D). Redγ-Redαβ7029 is the most efficient recombinase combination in DSM 7029 (cnpm was ∼160), prompting its use for further optimization and genome engineering.
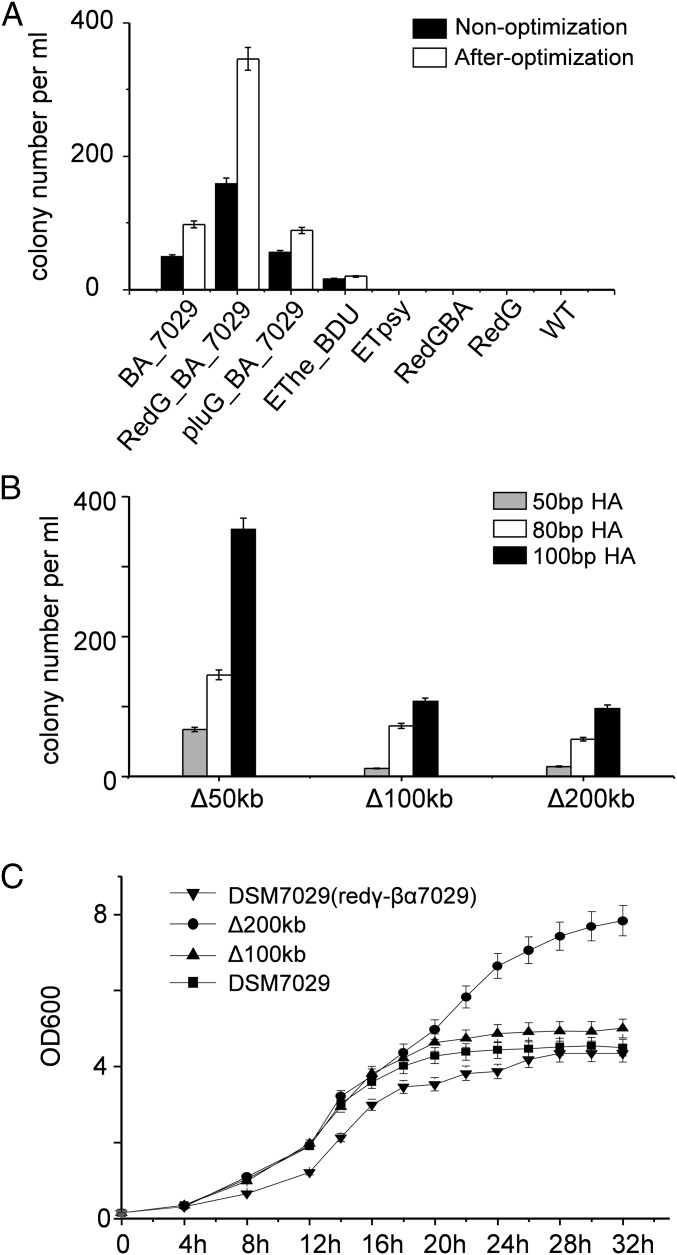
Because competent cell preparation and electroporation are critical for efficient recombineering in bacteria, we established optimal conditions for recombineering in DSM 7029 (SI Appendix, Fig. S4 A–E). By doing so, the recombination efficiency was enhanced by threefold compared with initial conditions (Fig. 3A). A detailed protocol for recombineering in DSM 7029 is included in SI Appendix. We then performed replacements of large DNA regions in the DSM 7029 genome. A 200-kb genome sequence (3054675–3254511) encoding two BGCs was successfully replaced using 50-bp homology arms (Fig. 3B and SI Appendix, Fig. S4F). Furthermore, the biomass and growth rate of the 100-kb and 200-kb deletion mutants were improved compared with the wild type (Fig. 3C). These results established a technical platform for precise DNA engineering to promote cryptic BGC mining and future genome reduction of DSM 7029 as a chassis for the expression of microbial natural products (43).
Fig. 3.
Optimization of the Redγ-Redαβ7029 recombineering procedure. (A) Comparison of different recombinase combinations as indicated in Fig. 2 in genome modification of DSM 7029 between the initial and optimized procedures. (B) Replacement of large genome sequences (50 kb to 200 kb) with an apramycin resistance gene containing varying homologous arms (50 bp to 100 bp) in DSM 7029 using optimized conditions. Error bars, SD; n = 3. (C) Growth curves of DSM 7029 and 100-kb and 200-kb deletion mutants.
Redβ7029 Catalyzes Highly Efficient Single-Stranded DNA Recombination.
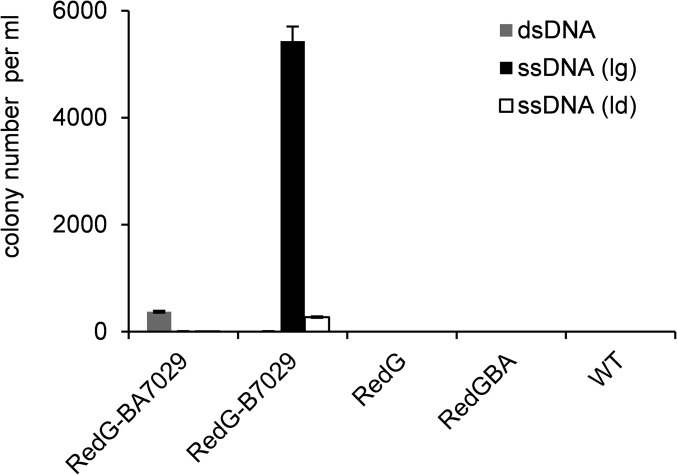
Phage recombinases mediating single-stranded oligonucleotide recombination is also efficient for genome engineering in E. coli (11, 44). We investigated the recombination efficiency of the Redγ-Redαβ7029 and Redγ-Redβ7029 mediated genome modification in DSM 7029 using a linear dsDNA and two ssDNA (leading strand and lagging strand) as substrates with the optimal procedure (SI Appendix, Figs. S2B and S4G). Redγ-Redαβ7029 also enabled the use of linear ssDNA as a substrate to modify the genome, but at extremely low efficiency, with cnpm less than 10. Redγ-Redβ7029 was competent at ssDNA recombination; especially when using the lagging ssDNA, the efficiency is substantially improved (cnpm was more than 5,000), 13 times higher than the Redγ-Redαβ7029 mediated dsDNA recombination (cnpm was ∼300) (Fig. 4). Thus, Redβ7029 catalyzes high-efficiency ssDNA recombination for genome engineering, greatly increasing the utility of this recombinase pair. Because the generation of long ssDNA requires in vitro exonuclease digestion (45, 46), we instead utilized the dsDNA recombination directly using a PCR product as a substrate for genome engineering.
Fig. 4.
Comparison of Redαβ7029 recombinase combinations using dsDNA and ssDNA (ld, leading strand; lg, lagging strand) as substrates to modify the genome of DSM 7029 as indicated in SI Appendix, Fig. S2B. Error bars, SD; n = 3.
Strategy for Deciphering Cryptic BGCs in DSM 7029.
The precise insertion of a functional promoter is a proven method for activating silent BGCs and enabling production (47). Thus, we employed this genetic tool to perform systematic interrogation of cryptic NRPS-PKS BGCs at low transcriptional levels (SI Appendix, Table S3) by comparative metabolite analysis between an inactivated mutant and a promoter-inserted activated mutant. The DNA sequence of the first NRPS-PKS gene in the target BGC was replaced with an antibiotic selection marker to generate a completely inactivated mutant. The same antibiotic marker along with its constitutive promoter was precisely inserted upstream of the first biosynthetic gene, substituting the promoter region of the main biosynthetic operon and generating an activated mutant in which the antibiotic resistance gene-driven target BGC is within a functional operon (Fig. 1). Using this approach, natural products encoded by cryptic BGCs were predicted, isolated, and characterized on the basis of bioinformatic, mass spectrometry, and NMR analysis.
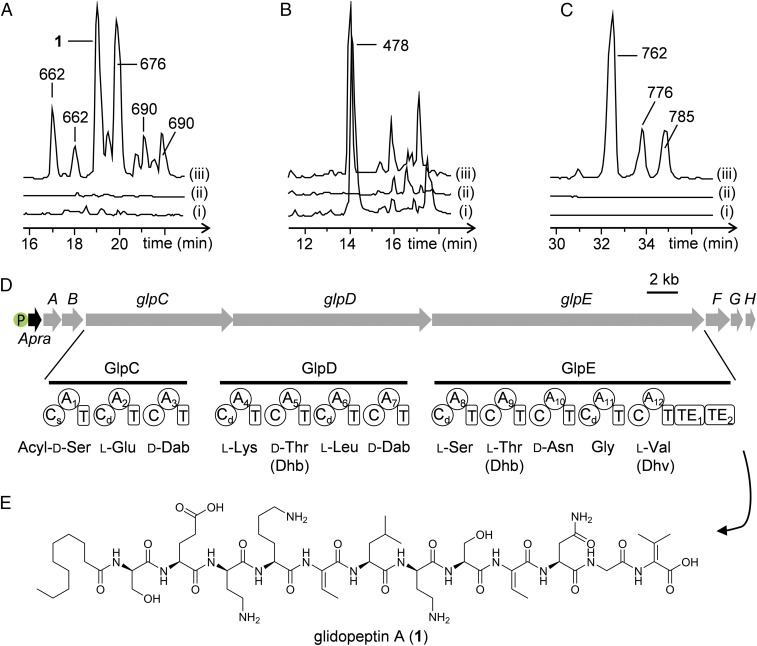
To eliminate the interference of the cytotoxic glidobactins, the predominant natural products in DSM 7029, we completely deleted the glidobactin biosynthetic genes glbB-glbG (48) using Redγ-Redαβ7029 recombineering without addition of an antibiotic marker (SI Appendix, Fig. S5). We then constructed inactivated and promoter knock-in mutants of three cryptic BGCs in this glb deletion mutant. The products of three cryptic BGCs, 6A, 7, and 11, were detected by comparing their metabolite profiles using LC-MS (Fig. 5 and SI Appendix, Figs. S6–S8). BGCs 6A and 11 from activated mutants produced several extra peaks at m/z 662, 676 (1), and 690 [M + 2H]2+, and at m/z 762, 776, and 785 [M + 2H]2+, respectively, compared with that of the inactivated mutants or the wild-type strain (Fig. 5). These results demonstrated that BGCs 6A and 11 remained silent under the laboratory fermentation conditions, but were effectively activated by promoter exchange using this system. An extra peak at m/z 478 [M + 2H]2+ was identified in the BGC 7 activated mutant and the wild-type strain but was absent in the inactivated mutant as well.
Fig. 5.
Mining of three cryptic gene clusters from DSM 7029. (A) HPLC-HRMS analysis (BPC+ 650 to 700) of extracts from (i) DSM 7029-Δglb, (ii) inactivated mutant DSM 7029-Δglb-ΔBGC6A, and (iii) activated mutant DSM 7029-Δglb-PApra-BGC6A; the m/z of each peak is indicated, except 1 (m/z = 676). (B) HPLC-HRMS analysis (BPC+ 450 to 500) of extracts from (i) DSM 7029-Δglb, (ii) DSM 7029-Δglb-ΔBGC7, and (iii) DSM 7029-Δglb-PApra-BGC7. (C) HPLC-HRMS analysis (BPC+ 750 to 800) of extracts from (i) DSM 7029-Δglb, (ii) DSM 7029-Δglb-ΔBGC11, and (iii) DSM 7029-Δglb-PApra-BGC11. (D) BGC6A (glpA-H) encoding the glidopeptins with promoter knock-in (P-Apra), and predicted NRPS domain and A domain selectivity for GlpCDE. Parentheses indicate amino acids observed in the NMR-determined final glidopeptin structure that differ from the bioinformatics prediction. A, adenylation domain; C, condensation domain; Cd, dual condensation/epimerization domain; CS, starter condensation domain; Dab, 2,4-diaminobutyric acid; Dhb, 2,3-dehydrobutyric acid; Dhv, α,β-dehydrovaline; T, peptidyl carrier domain. (E) Complete structure of glidopeptin A (1).
Identification of the Lipopeptide Glidopeptin A from DSM 7029.
One compound, namely glidopeptin A (1), was isolated from BGC 6A activated mutant. The glidopeptin gene cluster (glp, BGC 6A) contains eight unidirectional genes (glpA-H; Fig. 5D and SI Appendix, Table S5). Three large genes, glpCDE, encode NRPSs predicted to form a 12-amino acid lipopeptide because the first condensation (C) domain belongs to a starter type C domain that incorporates an acyl group as a starter unit (49). The additional genes are predicted to be responsible for product transport and for generating unnatural amino acids for use by the adenylation (A) domains of NRPS. The glpA gene encodes a 2,4-diaminobutyrate-4-aminotransferase that is commonly used in 2,4-diaminobutyric acid (Dab) biosynthesis, which is supported by the predicted Dab substrate binding specificity of two A domains, A3 and A7 (Fig. 5D). The glpF, glpB, glpG, and glpH genes are predicted to encode for a hydroxylase, a major facilitator transporter in charge of product transport, a histidinol phosphatase, and a membrane protein, respectively (SI Appendix, Table S5).
Biosynthesis was activated by promoter knock-in, and the main compound glidopeptin A (1) was isolated as a colorless oil with a molecular formula established as C60H102N16O19 by the high-resolution electrospray ionization mass spectroscopy (HR ESIMS) ion at m/z 676.3833 [M + 2H]2+ (calculated 676.3826). Its structure was determined on the basis of analysis of 1H and 13C NMR spectroscopy as well as 2D correlation spectroscopies including 1H-1H correlation spectroscopy, heteronuclear multiple-bond correlation (HMBC), and heteronuclear single-quantum coherence. Interpretation of the NMR data established seven proteinogenic amino acid residues including Glu, Lys, Leu, Asn, Gly, and two Ser; five nonproteinogenic amino acid residues including two Dab, two 2,3-dehydrobutyric acids (Dhb), and one α,β-dehydrovaline (Dhv); and a decanoic acid residue (Fig. 5 and SI Appendix, Tables S6 and S7). These residues were connected to form a linear 12-amino acid lipopeptide on the basis of HMBC correlations (SI Appendix, Fig. S15). The configurations of the amino acid residues were assigned as d-Ser, l-Glu, d-Dab, l-Lys, l-Leu, d-Dab, l-Ser, and l-Asn, using Marfey’s method combined with the presence of dual condensation/epimerization domains (Fig. 5D and SI Appendix, Fig. S13). Therefore, glidopeptin A was characterized to be a linear 12-amino acid lipopeptide containing a rare C-terminal Dhv that was only found in the myxobacterial nonribosomal peptide myxovalargin (50, 51).
The glidopeptin amino acid sequence is consistent with the prediction from glpCDE NRPS (Fig. 5) except that two Dhb and one Dhv are incorporated at the positions predicted to contain Thr (positions 5 and 9) and Val (position 12), respectively. The tandem thioesterases (TE1-TE2) observed at the C terminus of the NRPS are commonly found in large lipopeptide BGCs and generally predicted to catalyze both intramolecular cyclization and proofreading to guarantee fidelity of the biosynthetic assembly line (52, 53). However, our data showed that glidopeptin is a linear lipopeptide, indicating that the glp cluster indeed encodes a family of structural lipopeptides.
Extended Applications of Redαβ7029 Recombinases to Other Burkholderiales Strains for Genome Mining.
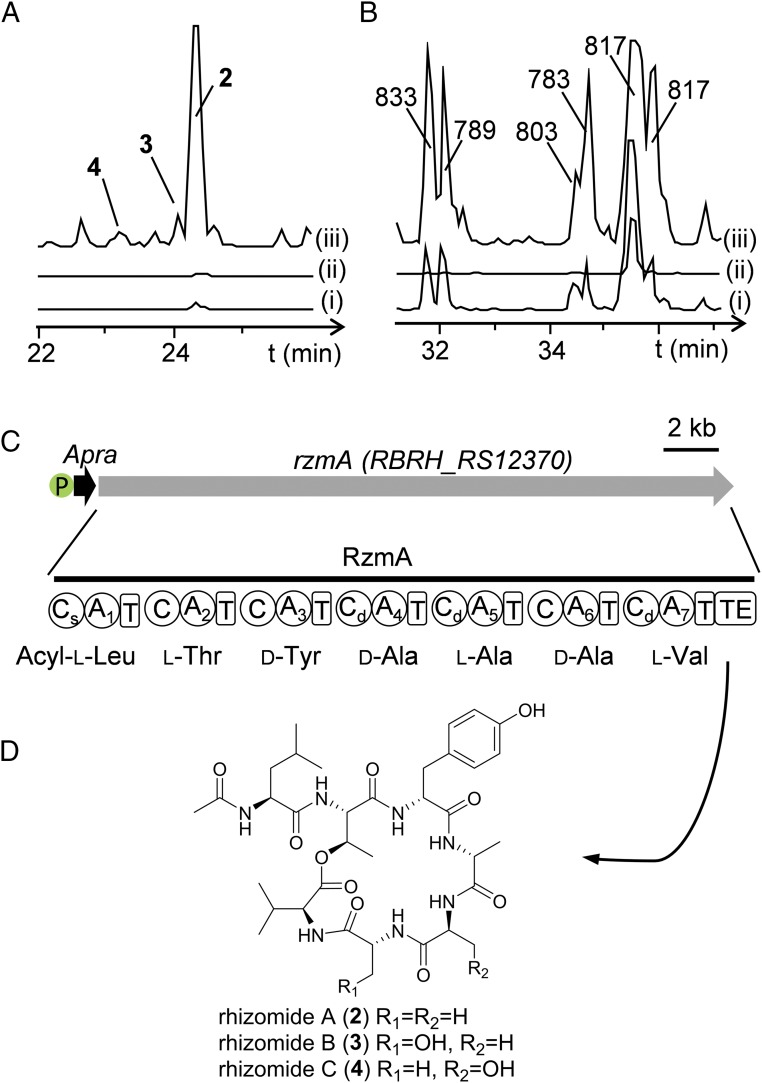
This recombineering and BGC mining strategy was not only functional in the native host but was also applied to other Burkholderiales strains that lack native Redαβ recombinase homologs, including P. rhizoxinica HKI 454, an endosymbiont of Rhizopus microspores (36), and Paraburkholderia phytofirmans strain PsJN, a plant growth-promoting endophyte (54). P. rhizoxinica HKI 454 produces antimitotic NRPS-PKS macrolide rhizoxins and harbors 14 additional NRPS gene clusters. However, no peptides corresponding to the NRPSs have been previously isolated (55). The rhizoxins account for a very large proportion of metabolites (56), and their biosynthetic pathway was successfully inactivated by gene replacement using Redγ-Redαβ7029 recombineering with high efficiency, greatly diminishing the metabolite content (SI Appendix, Fig. S9). Afterward, inactivated and promoter knock-in mutants for selected cryptic NRPS BGCs were generated via the above strategy (SI Appendix, Figs. S10 and S11). The products of BGC P1 rhizomides A-C [m/z 732 (2), 748 (3, 4)] and BGC P7 (m/z 833, 789, 803, 783, 817) from plasmid pBRH01were identified (Fig. 6). The yield of rhizomides in promoter knock-in mutants was 70-fold higher than that in the wild-type strain, indicating that the strong promoter remarkably enhanced product yields. Redγ-Redαβ7029 recombineering was also effective in P. phytofirmans strain PsJN for BGC manipulation, but no products were detected (SI Appendix, Fig. S12).
Fig. 6.
Mining of two cryptic gene clusters from P. rhizoxinica HKI 454. (A) HPLC-HRMS analysis (BPC+ 700 to 750) of extracts from (i) HKI 454-Δrhi, (ii) HKI 454-Δrhi-ΔBGC P1, and (iii) HKI 454-Δrhi-PApra-BGC P1. (B) HPLC-HRMS analysis (BPC+ 750 to 850) of extracts from (i) HKI 454-Δrhi, (ii) HKI 454-Δrhi -ΔBGC P7, and (iii) HKI 454-Δrhi-PApra-BGC P7; the m/z of each peak is indicated. (C) BGC P1 encoding the rhizomides (gene RBRH_01504), with promoter insertion (P-Apra), predicted NRPS domain, and A domain selectivity for RzmA. (D) Complete structure of rhizomides A−C (2 to 4).
Identification of Peptide Rhizomides from P. rhizoxinica HKI 454.
The single rhizomide (rzm, BGC P1) biosynthetic gene RBRH01504 encodes a seven-module NRPS with a starter C domain, indicating that its product may be a class of lipopeptides (Fig. 6C). Compounds 2 to 4, namely rhizomides A to C, were purified as white amorphous powder and analyzed by HR ESIMS and NMR. The molecular formula of the dominant product 2 was determined to be C35H53N7O10 according to the HR ESIMS ion at m/z 732.3930 [M + H]+ (calculated 732.3927). The NMR data indicated the presence of seven amino acids (SI Appendix, Fig. S15 and Table S8). HMBC correlations between amide protons and adjacent carbonyl groups defined the order of the seven amino acids, an acetyl group attached to the Leu N terminus, and intramolecular cyclization through the Thr side chain (SI Appendix, Fig. S15). The absolute configurations of the amino acids were confirmed by Marfey’s method and bioinformatics analysis of the C domains (Fig. 6C and SI Appendix, Fig. S14). The two minor products 3 and 4 differ from 2 with one more oxygen atom as indicated by the HR ESIMS-determined formula C35H53N7O11 (m/z 748.3873 [M + H]+, calculated 748.3876), which was supported by the NMR comparison of 3 or 4 with that of 2, indicating that a Ser residue in 3 and 4 replaced the third Ala or the second Ala in 2 (SI Appendix, Fig. S15 and Tables S9 and S10), respectively. Marfey’s analysis of 3 and 4 demonstrated the presence of d- and l-Ser in 3 and 4 (SI Appendix, Fig. S14), respectively, supporting the proposed structures (Fig. 6D). Therefore, rhizomides A−C (2 to 4) were determined to be seven-amino acid cyclic lipopeptides with an N-acetyl-Leu linking to a rare 19-membered lactone. These structures are consistent with the bioinformatic analysis of NRPS RzmA. The starter C domain condenses an unusual short fatty acid chain acetyl-CoA and Leu, and chain extension continues with the incorporation of l-Thr, d-Tyr, d-Ala, l-Ala/Ser, d-Ala/Ser, and l-Val by Rzm modules 2 to 7. The peptide chain is then passed to the TE domain, and is catalyzed into a 19-membered ring. The broad substrate binding specificity of the A5 and A6 domains allowed loading and incorporation of Ser into the extended chain to form the minor rhizomides C and B, respectively.
Bioactivity of Glidopeptin A and Rhizomide A.
Glidopeptin A and rhizomide A showed weak antitumor activity against several human tumor cells lines (IC50 34 μM to 96 μM) (SI Appendix, Table S12). Glidopeptin A and rhizomide A also showed weak protective activity against cucumber downy mildew (Pseudoperonospora cubenis) (SI Appendix, Table S11) and weak in vitro inhibition activities against Staphylococcus aureus and Bacillus subtilis (SI Appendix, Fig. S50), respectively.
Discussion
The chance of a successful mining of cryptic BGCs in a native host is generally higher than that in a heterologous host. However, it is usually inefficient due to a low homologous recombinant efficiency, especially in nonmodel organisms. To address this challenge, we used genome-guided discovery of host-specific phage recombinases and a transient expression system to develop an efficient recombineering method in a natural product producer. This method greatly expedites the construction of an inactive mutant by gene replacement and the activation of the cryptic BGCs by precise promoter insertion upstream of the main biosynthetic gene. This method is superior to the classical but laborious single crossover mutagenesis, especially considering the scalable manipulation of abundant BGCs. The transient expression of the recombinase ensures high-level expression for efficient DNA recombination, and diminishes the toxicity of recombinases and undesirable genome rearrangements. In addition, we used the same antibiotic selection for BGC inactivation and activation, which minimized disturbances during metabolite analysis. The use of an antibiotic promoter driving a BGC bypassed the complex screening of a feasible promoter for each producer. In addition to precise promoter insertion, more complex engineering such as domain/module swapping and seamless mutagenesis can be accomplished using the recombineering method in native producers. Once a potentially new compound is detected, biosynthesis dissection by in-frame gene deletion, structural optimization by combinatorial biosynthesis, and yield increase by host improvement or manipulation of regulators could easily be performed in the original host.
Recombineering based on the host-specific recombinases Redαβ7029, pair of native bacteriophage recombinases in Burkholderiales, allowed us to investigate cryptic BGCs efficiently not only in the native DSM 7029 but also in other Burkholderiales species that lack effective genetic tools or native Redαβ recombinase homologs. A total of 15 metabolites from five BGCs was discovered in this study, and four metabolites were isolated and elucidated as compounds. Intriguingly, of five activated BGCs, two did not produce detectable products in the native producers under the same growth conditions, indicating that the promoter insertion strategy activates the expression of silent clusters. One cluster also showed a remarkably increased yield in the promoter knock-in mutant. Admittedly, some cryptic BGC products remain undiscovered, partially because they are present in very low abundance despite the improved expression level of biosynthetic genes using the promoter insertion strategy. It is possible that the small quantity of products escaped detection, due to the limits of the analytical methods or statistical data evaluation. Our strategy, combined with comprehensive metabolome mining, could further improve the likelihood of discovering specialized metabolites using genome mining (57, 58).
In conclusion, we have demonstrated that a recombinase transient expression strategy was able to systematically activate cryptic gene clusters from Burkholderiales species. The metabolites can be detected by LC-MS and characterized by NMR. This strategy can be applied to more natural product producers for cryptic BGC investigation, and, when combined with medium optimization or metabolome mining, can mine the biosynthetic potential of a strain for the discovery of new molecules. Recombineering of the BGCs in native producers greatly facilitates genome engineering for host amelioration, speeds up the discovery of new compounds with attractive bioactivities, and optimizes metabolite production through combinatorial biosynthesis for pharmaceutical and agricultural applications.
Materials and Methods
Methods describing plasmid construction, promoter screening, detailed recombineering protocol, gene cluster inactivation, promoter insertion, fermentation and extraction of strains, HPLC-ESI-HRMS analyses, isolation and characterization of compounds, and bioactivity test are described in detail in SI Appendix, SI Materials and Methods.
SI Appendix tables include strains, plasmids, mutants in this work (SI Appendix, Table S2), recombinase pairs (SI Appendix, Table S1), gene annotation of involved gene clusters (SI Appendix, Table S5), primers used in this study (SI Appendix, Table S13), transcript level of selected genes (SI Appendix, Table S3), promoter sequences (SI Appendix, Table S4) and NMR data of compounds (SI Appendix, Tables S6–S10), and the protective activity of glidopeptin A and rhizomide A against six plant diseases (SI Appendix, Table S11). SI Appendix figures include inducible promoter screening (SI Appendix, Fig. S1), recombination efficiency of different recombinases in E. coli and DSM 7029 (SI Appendix, Fig. S2), the recombination efficiency of the LCHR and LLHR mediated by Redγ-Redαβ7029 in E. coli (SI Appendix, Fig. S3), optimization of the work condition of Redγ-Redαβ7029 in DSM 7029 (SI Appendix, Fig. S4), seamless deletion of glidobactin gene cluster (SI Appendix, Fig. S5), inactivation and activation of BGC 6A, BGC 7, and BGC 11 in DSM 7029 (SI Appendix, Figs. S6–S8), inactivation of rhizoxin gene cluster in P. rhizoxinica HKI 454 (SI Appendix, Fig. S9), inactivation and activation of BGC P1 and BGC P7 in P. rhizoxinica HKI 454 (SI Appendix, Figs. S10 and S11), inactivation and activation of BGC 2 in P. phytofirmans PsJN (SI Appendix, Fig. S12), and NMR spectra of compounds 1 to 4 (SI Appendix, Figs. S15–S49). Cytotoxic activity and bioactivity of glidopeptin A and rhizomide A are described (SI Appendix, Fig. S50 and Table S12).
Supplementary Material
Acknowledgments
We thank Prof. Du-Qiang Luo (Hebei University) and Prof. Naiguo Si (Shenyang Sinochem Agrochemicals R&D Co. Ltd) for bioactivity assessment, and Prof. Blaine Pfeifer and Prof. Liangcheng Du for constructive proofreading of this manuscript. This work was supported by grants from the National Key R&D Program of China (2017YFD0201405), the National Natural Science Foundation of China (31670098, 31500033, and 31670097), the Major Basic Program of the Natural Science Foundation of Shandong Province, China (ZR2017ZB0212), and the Higher Education Discipline Innovation Introduction Program (B16030). This work was also supported by the Recruitment Program of Global Experts (Y.Z.) and the Qilu Youth Scholar Startup Funding of Shandong University (to X.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720941115/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Cantrell CL, Dayan FE, Duke SO. Natural products as sources for new pesticides. J Nat Prod. 2012;75:1231–1242. doi: 10.1021/np300024u. [DOI] [PubMed] [Google Scholar]
- 3.Bode HB, Müller R. The impact of bacterial genomics on natural product research. Angew Chem Int Ed Engl. 2005;44:6828–6846. doi: 10.1002/anie.200501080. [DOI] [PubMed] [Google Scholar]
- 4.Cimermancic P, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee S, et al. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat Biotechnol. 2017;35:676–683. doi: 10.1038/nbt.3886. [DOI] [PubMed] [Google Scholar]
- 6.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J Ind Microbiol Biotechnol. 2014;41:175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montiel D, Kang HS, Chang FY, Charlop-Powers Z, Brady SF. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc Natl Acad Sci USA. 2015;112:8953–8958. doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang MM, et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol. 2017;13:607–609. doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 13.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: A homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu J, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 15.Muyrers JPP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering–New options for cloning and manipulating DNA. Trends Biochem Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 16.Bunny K, Liu J, Roth J. Phenotypes of lexA mutations in Salmonella enterica: Evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J Bacteriol. 2002;184:6235–6249. doi: 10.1128/JB.184.22.6235-6249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei D, Wang M, Shi J, Hao J. Red recombinase assisted gene replacement in Klebsiella pneumoniae. J Ind Microbiol Biotechnol. 2012;39:1219–1226. doi: 10.1007/s10295-012-1117-x. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, et al. Genome engineering of Agrobacterium tumefaciens using the lambda Red recombination system. Appl Microbiol Biotechnol. 2014;98:2165–2172. doi: 10.1007/s00253-013-5412-x. [DOI] [PubMed] [Google Scholar]
- 19.Lesic B, Rahme LG. Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol Biol. 2008;9:20. doi: 10.1186/1471-2199-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett AR, et al. Genetic tools for allelic replacement in Burkholderia species. Appl Environ Microbiol. 2008;74:4498–4508. doi: 10.1128/AEM.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, et al. Knockout and pullout recombineering for naturally transformable Burkholderia thailandensis and Burkholderia pseudomallei. Nat Protoc. 2011;6:1085–1104. doi: 10.1038/nprot.2011.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol. 2003;38:113–116. doi: 10.1016/S0928-8244(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 23.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 24.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: Characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–1107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 25.Swingle B, Bao Z, Markel E, Chambers A, Cartinhour S. Recombineering using RecTE from Pseudomonas syringae. Appl Environ Microbiol. 2010;76:4960–4968. doi: 10.1128/AEM.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Pijkeren JP, Britton RA. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012;40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xin Y, Guo T, Mu Y, Kong J. Identification and functional analysis of potential prophage-derived recombinases for genome editing in Lactobacillus casei. FEMS Microbiol Lett. 2017;364:fnx243. doi: 10.1093/femsle/fnx243. [DOI] [PubMed] [Google Scholar]
- 28.Yin J, et al. A new recombineering system for Photorhabdus and Xenorhabdus. Nucleic Acids Res. 2015;43:e36. doi: 10.1093/nar/gku1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H, Tao W, Gong F, Li Y, Zhang Y. A functional recT gene for recombineering of Clostridium. J Biotechnol. 2014;173:65–67. doi: 10.1016/j.jbiotec.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Cheng YQ. Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biotechnol. 2014;41:275–284. doi: 10.1007/s10295-013-1376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esmaeel Q, et al. Burkholderia genome mining for nonribosomal peptide synthetases reveals a great potential for novel siderophores and lipopeptides synthesis. MicrobiologyOpen. 2016;5:512–526. doi: 10.1002/mbo3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minowa Y, Araki M, Kanehisa M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J Mol Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- 33.Esmaeel Q, Pupin M, Jacques P, Leclère V. Nonribosomal peptides and polyketides of Burkholderia: New compounds potentially implicated in biocontrol and pharmaceuticals. Environ Sci Pollut Res Int. 2017 doi: 10.1007/s11356-017-9166-3. [DOI] [PubMed] [Google Scholar]
- 34.Depoorter E, et al. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol. 2016;100:5215–5229. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 35.Tang B, Yu Y, Zhang Y, Zhao G, Ding X. Complete genome sequence of the glidobactin producing strain [Polyangium] brachysporum DSM 7029. J Biotechnol. 2015;210:83–84. doi: 10.1016/j.jbiotec.2015.06.417. [DOI] [PubMed] [Google Scholar]
- 36.Lackner G, Moebius N, Partida-Martinez L, Hertweck C. Complete genome sequence of Burkholderia rhizoxinica, an Endosymbiont of Rhizopus microsporus. J Bacteriol. 2011;193:783–784. doi: 10.1128/JB.01318-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, et al. An improved recombineering approach by adding RecA to lambda Red recombination. Mol Biotechnol. 2006;32:43–53. doi: 10.1385/mb:32:1:043. [DOI] [PubMed] [Google Scholar]
- 38.Rang A, Will H. The tetracycline-responsive promoter contains functional interferon-inducible response elements. Nucleic Acids Res. 2000;28:1120–1125. doi: 10.1093/nar/28.5.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman JR, Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 40.Binder D, et al. Light-controlled cell factories: Employing photocaged isopropyl-β-d-thiogalactopyranoside for light-mediated optimization of lac promoter-based gene expression and (+)-valencene biosynthesis in Corynebacterium glutamicum. Appl Environ Microbiol. 2016;82:6141–6149. doi: 10.1128/AEM.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egan SM, Schleif RF. A regulatory cascade in the induction of rhaBAD. J Mol Biol. 1993;234:87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat Protoc. 2016;11:1175–1190. doi: 10.1038/nprot.2016.054. [DOI] [PubMed] [Google Scholar]
- 43.Bian X, et al. Heterologous production and yield improvement of epothilones in Burkholderiales strain DSM 7029. ACS Chem Biol. 2017;12:1805–1812. doi: 10.1021/acschembio.7b00097. [DOI] [PubMed] [Google Scholar]
- 44.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avci-Adali M, Paul A, Wilhelm N, Ziemer G, Wendel HP. Upgrading SELEX technology by using lambda exonuclease digestion for single-stranded DNA generation. Molecules. 2009;15:1–11. doi: 10.3390/molecules15010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maresca M, et al. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myronovskyi M, Luzhetskyy A. Native and engineered promoters in natural product discovery. Nat Prod Rep. 2016;33:1006–1019. doi: 10.1039/c6np00002a. [DOI] [PubMed] [Google Scholar]
- 48.Schellenberg B, Bigler L, Dudler R. Identification of genes involved in the biosynthesis of the cytotoxic compound glidobactin from a soil bacterium. Environ Microbiol. 2007;9:1640–1650. doi: 10.1111/j.1462-2920.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- 49.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol. 2007;7:78–92. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irschik H, Gerth K, Kemmer T, Steinmetz H, Reichenbach H. The myxovalargins, new peptide antibiotics from Myxococcus fulvus (Myxobacterales). I. Cultivation, isolation, and some chemical and biological properties. J Antibiot (Tokyo) 1983;36:6–12. doi: 10.7164/antibiotics.36.6. [DOI] [PubMed] [Google Scholar]
- 51.Gille F, Kirschning A. Studies on the synthesis of peptides containing dehydrovaline and dehydroisoleucine based on copper-mediated enamide formation. Beilstein J Org Chem. 2016;12:564–570. doi: 10.3762/bjoc.12.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hou J, Robbel L, Marahiel MA. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem Biol. 2011;18:655–664. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 53.Roongsawang N, Washio K, Morikawa M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int J Mol Sci. 2010;12:141–172. doi: 10.3390/ijms12010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weilharter A, et al. Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J Bacteriol. 2011;193:3383–3384. doi: 10.1128/JB.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. Evolution of an endofungal lifestyle: Deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011;12:210. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scherlach K, Partida-Martinez LP, Dahse HM, Hertweck C. Antimitotic rhizoxin derivatives from a cultured bacterial endosymbiont of the rice pathogenic fungus Rhizopus microsporus. J Am Chem Soc. 2006;128:11529–11536. doi: 10.1021/ja062953o. [DOI] [PubMed] [Google Scholar]
- 57.Krug D, Müller R. Secondary metabolomics: The impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31:768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- 58.Hoffmann T, Krug D, Hüttel S, Müller R. Improving natural products identification through targeted LC-MS/MS in an untargeted secondary metabolomics workflow. Anal Chem. 2014;86:10780–10788. doi: 10.1021/ac502805w. [DOI] [PubMed] [Google Scholar]