Significance
The physical basis for the regulation of protein MAP kinases is key to understanding their role in cellular signaling in human biology and disease. However, the biochemical events that lead to MAPK activation are not well understood. Here, using biomolecular NMR dynamics measurements, we show that kinase activation is not due simply to phosphorylation of the activation loop. Rather, we show that phosphorylation and substrate binding cause the dynamics of p38 to change from a state in which they are uncoordinated to one that is uniform across the entire molecule. That is, synchronized dynamics coordinates enzyme activity, which leads to a shift in the energy landscape and ultimately drives enzyme function. This makes essential elements of allostery and activation dynamically accessible.
Keywords: MAP kinase, NMR spectroscopy, NMR dynamics, kinase activation, signaling
Abstract
Mitogen-activated protein kinases, which include p38, are essential for cell differentiation and autophagy. The current model for p38 activation involves activation-loop phosphorylation with subsequent substrate binding leading to substrate phosphorylation. Despite extensive efforts, the molecular mechanism of activation remains unclear. Here, using NMR spectroscopy, we show how the modulation of protein dynamics across timescales activates p38. We find that activation-loop phosphorylation does not change the average conformation of p38; rather it quenches the loop ps-ns dynamics. We then show that substrate binding to nonphosphorylated and phosphorylated p38 results in uniform µs-ms backbone dynamics at catalytically essential regions and across the entire molecule, respectively. Together, these results show that phosphorylation and substrate binding flatten the energy landscape of the protein, making essential elements of allostery and activation dynamically accessible. The high degree of structural conservation among ser/thr kinases suggests that elements of this mechanism may be conserved across the kinase family.
Mitogen-activated protein kinases have evolved to transduce environmental and developmental signals into adaptive and programmed responses such as differentiation, inflammation, and apoptosis. The importance of these processes is well-illustrated by the many inherited or acquired human diseases that stem from abnormalities in MAPK-signaling pathways, including Parkinson’s disease, inflammatory disorders, and cancer. p38α (MAPK14; α-isoform; 360 aa; 41.3 kDa; hereafter referred to as p38) is one of the best structurally and biologically characterized MAPKs (Fig. 1A) (1, 2). It adopts a typical kinase fold, including N- (residues 1–105) and C-terminal lobes (residues 114–316) that are connected via a hinge (residues 106–113). The N-terminal lobe mediates ATP binding, while the C-terminal lobe both facilitates effector/substrate binding at the kinase interaction motif (KIM- or d-motif) binding site and has a known allosteric ligand-binding pocket in the MAPK-specific insert. Based on hundreds of p38 crystal structures (>200 PDB entries), the widely held view is that p38 activation is achieved by phosphorylation of its activation loop, an event that results in a rigid-body rotation of the N- and C-terminal lobes by 5–10° and is argued to facilitate phosphoryl transfer (3, 4). However, structures of nonphosphorylated and phosphorylated p38 in complex with a peptide from the mitogen-activated protein kinase MKK3b, an enzyme which activates p38 by phosphorylating its activation loop, is not consistent with this view, and it has become clear that additional factors are likely necessary to activate p38 (5–7).
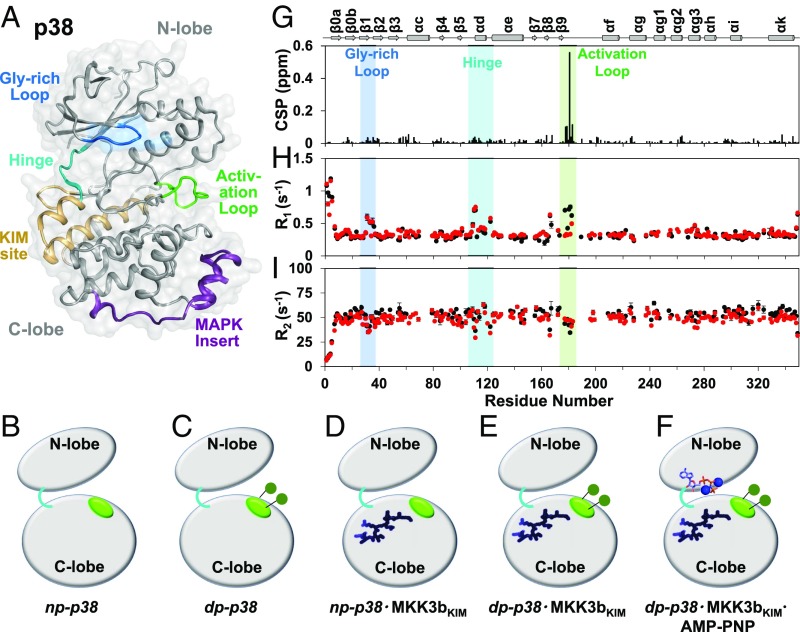
Fig. 1.
Phosphorylation affects fast timescale dynamics at the activation loop. (A) p38 adopts a typical kinase fold with an N- and a C-terminal lobe. Gly-rich loop, KIM-binding site, hinge, activation loop, and the MAPK-specific insert are highlighted. (B–F) Cartoon representation of the p38 states investigated in this work: (B) np-p38, (C) dp-p38, (D) np-p38⋅MKK3bKIM, (E) dp-p38⋅MKK3bKIM, and (F) np-p38⋅MKK3bKIM⋅AMP-PNP. Activation loop (green), KIM-peptide (dark blue), and AMP-Mg+2–binding sites (sticks) are highlighted. (G) Chemical-shift perturbations (CSP) upon activation-loop phosphorylation vs. residue number. (H) The 15N longitudinal relaxation rates (R1) and (I) transverse relaxation rates (R2) for np-p38 (black) and dp-p38 (red) vs. residue number. The Gly-rich loop (blue), hinge (cyan), and activation loop (green) are highlighted.
The emerging view is that enzymes, including kinases, require not only conformational changes but also dynamic events to perform their functions (8–13). Some of these events are required for catalysis and some, for a subset of enzymes, are required for the allosteric control of the catalytic cycle. However, the “rules” for motion (conformational entropy) and the dynamical transmission of allostery in enzymes are only beginning to be established. Initial studies on p38 and related kinases by us and others have suggested that dynamic conformational rearrangements are critical to the basic function and regulation of this important class of enzymes (8, 14). However, a detailed understanding of how p38 dynamics are correlated with activity and regulation is unknown.
Because the physical basis for MAPK activation is key to understanding its role in cellular signaling in human biology and disease, we initiated a comprehensive effort to determine how the 15N backbone fast timescale (ps-ns) and intermediate timescale motions (µs-ms) of p38 control its regulation and activity. Our NMR dynamics data reveal that phosphorylation of the p38 activation loop leads to increased rigidity of the this loop in the fast timescale (ps-ns), but, unexpectedly, does not lead to a change in the relative position of the N- and C-terminal lobes in solution; i.e., they do not close about the active site. This demonstrates that the molecular basis for the activation of p38 by phosphorylation occurs by a different mechanism. Using constant-time Carr–Purcell–Meiboom–Gill (ct-CPMG) NMR dynamics measurements, we then showed that µs-ms motions are coordinated by substrate binding to the KIM-motif–binding site, which induces allostery between the KIM and the hinge region that connect the N- and C-lobes. However, it is only when both KIM-binding and activation-loop phosphorylation are present that uniform µs-ms dynamics in p38—allowing for coordinated recruitment of ATP—is achieved. Thus, the data show that the changes in the dynamics of p38 shift the energy landscape to orchestrate a synchronized activation of p38 and, in turn, make essential elements of allostery and activation dynamically possible. Together, these data not only provide fundamental insights into the mechanism of MAPK activation, but can also now be leveraged to develop therapeutics that target p38 kinase dynamics.
Results
Activation-Loop Phosphorylation Does Not Change the Average Conformation of p38.
To better understand the dynamical and structural basis for the activation of p38 by phosphorylation we studied five primary states of the p38 protein by an array of biophysical methods: (i) np-p38, nonphosphorylated p38; (ii) dp-p38, doubly phosphorylated p38; (iii) np-p38⋅MKK3bKIM, the binary complex between a canonical KIM peptide and nonphosphorylated p38; (iv) dp-p38⋅MKK3bKIM, the doubly phosphorylated counterpart; and (v) dp-p38⋅MKK3bKIM⋅AMP-PNP, AMP-PNP–bound dp-p38⋅MKK3bKIM (Fig. 1 B–F). The canonical KIM peptide (MKK3bKIM) corresponds to the KIM-binding sequence of the MKK3b kinase and was chosen because the corresponding crystal structures are available.
Two-dimensional [1H,15N] transverse relaxation optimized spectroscopy (TROSY) spectra of p38 in these multiple functional states are of excellent quality, allowing for the comprehensive analysis (334 amide H-N reporters) of the structural and dynamical aspects of p38 activation and regulation (15) (SI Appendix, Fig. S1). MKK3bKIM binds np- and dp-p38 with similar dissociation constants (KD ∼ 5 µM) (SI Appendix, Figs. S2 and S3 and Table S1). Phosphorylation of Thr180 and Tyr182 was achieved in vitro using MKK6. The 2D [1H,15N] TROSY spectrum of dp-p38 showed only local chemical-shift perturbations (CSPs) near the phosphorylated residues (Fig. 1G). This indicates that p38 phosphorylation does not result in a rigid-body rotation around the hinge region in solution, an observation consistent with data reported for methyl-labeled p38 (additional information in SI Appendix, Fig. S4) (16).
Dual Phosphorylation Quenches Activation-Loop ps-ns Dynamics.
It is now recognized that protein dynamics forms an integral part of enzyme function (8–13). To gain further insight in the role of dynamics in p38 activation, we characterized fast ps-ns motion on the backbone in various states using NMR relaxation measurements. The 15N-backbone dynamics of np-p38 are uniform throughout p38, with three regions showing increased flexibility: the glycine-rich loop (residues 30–40), the hinge region (residues 100–120, connecting the N- and C-lobe), and the activation loop (residues 172–183). Phosphorylation of p38 leads to a change in the ps-ns dynamics (Fig. 1 H and I and SI Appendix, Fig. S5), where the activation loop becomes as rigid as to become indistinguishable from the rest of the p38 backbone, while the glycine-rich loop and the hinge region are unchanged; i.e., they stay flexible in dp-p38. Furthermore, a plot of R2/R1 vs. p38 residue for np-p38 and dp-p38 confirms that phosphorylation leads to a reduction of fast timescale dynamics (SI Appendix, Fig. S6). The addition of the MKK3bKIM peptide to np- and dp-p38 does not alter p38’s fast dynamics; i.e., they mirror those of np- and dp-p38, respectively (SI Appendix, Figs. S7 and S8). This shows that while phosphorylation of the activation-loop residues influences the fast timescale backbone dynamics of p38, KIM binding does not.
Nonphosphorylated p38 Has Heterogeneous Dynamics in the µs-ms Time Regime.
Enzymatic reactions can have catalytic turnover rates that correlate with protein dynamics in the μs-ms timescale (17–19). This correlation has been previously used to unravel how enzymes function (14, 20, 21). We thus examined the nature of slower motions in the major states of p38 using 15N constant-time Carr–Purcell–Meiboom–Gill (ct-CPMG) relaxation dispersion experiments (22), which probe motions in the μs-ms timescale. Residues undergoing μs-ms conformational exchange dynamics show changes in the effective relaxation rate R2 (R2,eff), which was measured as a function of the repetition frequency (νCPMG). Plots of R2,eff vs. νCPMG show a curvature for residues exhibiting μs-ms timescale dynamics. Fitting these curves to a two-state model (Carver–Richards) allows the populations (pA and pB) and the exchange rates (kex) to be extracted. To determine the kinetic and thermodynamic parameters defining the interconversion of states connected by motion on the µs-ms timescale, 15N ct-CPMG dispersion profiles were obtained at two magnetic fields (11.7 and 19.96 T).
All states of p38 displayed numerous residues having conformational exchange dynamics on the µs-ms timescale. Importantly, while individual sites can generally be fitted with a two-site exchange model, it was impossible to globally fit the observed dispersion profiles to a single two-state model. This simply indicates the presence of a heterogeneous dynamic profile. Next we considered the possibility that p38 has multiple clusters of residues that follow similar exchange dynamics. To avoid overfitting, we clustered using the Bayesian Information Criterion (BIC, ref. 23). Applying this analysis to np-p38 identified multiple clusters with exchange motions >2,500 s−1 (Fig. 2A and SI Appendix, Fig. S9 and Table S2). Most of these clusters did not form a contiguous group within np-p38, i.e., residues that are either in proximity based on p38 primary or tertiary structure, or based on functionally important regions. Furthermore, many residues showed intermediate exchange dynamics that did not conform to one of these dynamic groups of residues. This shows np-p38 has heterogeneous dynamics in the µs-ms time regime. To establish the robustness of the simultaneous cluster selection and Richard–Carver equation fitting, we calculated reduced χ2 (χ2ν) surfaces (SI Appendix, Fig. S10, Table S3, and SI Methods). The χ2ν showed that this approach identifies the statistically relevant groups in a statistically robust manner.
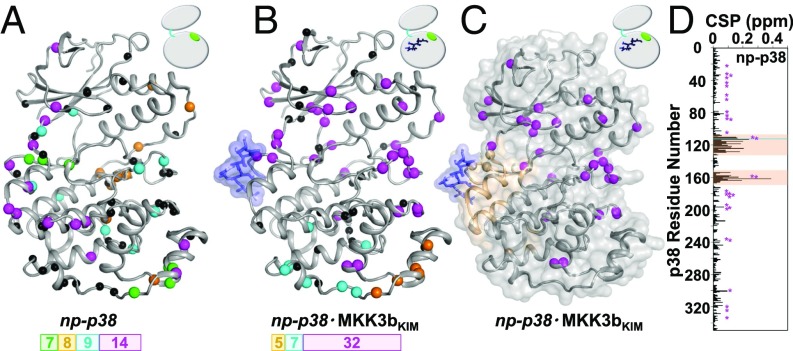
Fig. 2.
KIM binding induces exchange dynamics beyond the peptide-binding site. Clusters of residues with uniform exchange dynamics (rates and population), color coded on the basis of kex, are shown for (A) np-p38 (magenta, >2,500 s−1; cyan, >2,500 s−1; orange, 1,687 ± 134 s−1; and green, 1,452 ± 187 s−1) and (B) np-p38⋅MKK3bKIM (magenta, >2,500 s−1; cyan, 1,251 ± 68 s−1; and orange, 2,403 ± 332 s−1) (blue sticks/surface). Residues that cannot be fit within a cluster in a statistically meaningful manner are shown as black spheres. Number of residues in the largest clusters is shown. (C) MKK3bKIM peptide binding (blue sticks/surface) leads to CSPs (beige) only in the KIM-binding pocket, whereas residues beyond the KIM-binding pocket cluster with uniform exchange dynamics (magenta spheres; includes the KIM-binding pocket, the hinge, the N-terminal lobe, and the activation loop). (D) Residues with µs-ms dynamics are highlighted (magenta) on the histogram showing the 1H/15N CSPs of np-p38 upon MKK3bKIM binding vs. residue number.
Allostery Between the Activation Loop and the Hinge Region Is Mediated by Intermediate Exchange Dynamics.
Strikingly different results are obtained from the BIC analysis for np-p38⋅MKK3bKIM (Fig. 2B and SI Appendix, Fig. S11 and Table S2). A single large cluster of p38 residues was identified that includes residues from the p38 activation loop, the hinge region, and the N-terminal lobe, as well as p38 residues near the MAPK-specific insert motif. A number of additional, though smaller, clusters of residues with statistically similar exchange behaviors were also identified. Most importantly, the binding of MKK3bKIM dynamically links the activation loop with the hinge region and the N-terminal lobe. The CSP analysis of MKK3bKIM binding to np-p38 showed excellent agreement with the binding interface detected in the np-p38:MKK3bKIM crystal structure (5). Notably, no CSPs were identified in the N-terminal loop, the hinge regions, and the activation loop, indicating that the MKK3bKIM peptide-binding allostery between the activation loop and the hinge region is largely mediated by intermediate exchange dynamics and not conformational changes in p38 (Fig. 2 C and D).
Unlike ERK2, Phosphorylation of the p38 Activation Loop Does Not Lead to Global Changes in Conformational Exchange Dynamics.
Phosphorylation of p38 also changes the dynamics in the µs-ms timescale (Fig. 3A and SI Appendix, Fig. S12 and Table S2). Four clusters of residues in dp-p38 exhibited a more coherent structural and functional proximity than those of np-p38, but much less than the np-p38⋅MKK3bKIM complex. Strikingly, the cluster that connects the p38 activation loop, the hinge region, and the N-terminal lobe in np-p38⋅MKK3bKIM was lost in dp-p38. This contrasts with the dynamics observed for the MAPK ERK2, where activation-loop phosphorylation leads to global changes in conformational exchange dynamics (8).
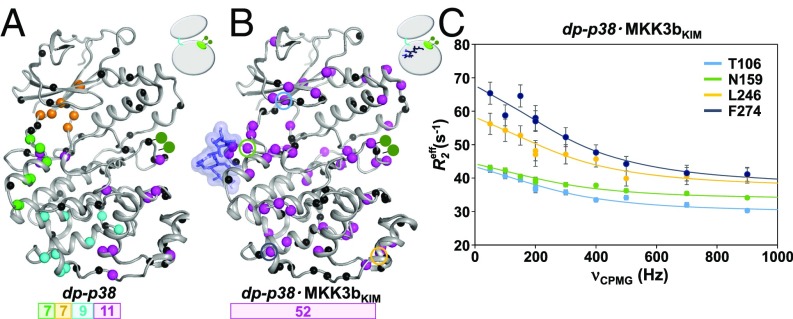
Fig. 3.
Phosphorylation and KIM-peptide binding induces uniform exchange dynamics throughout p38. Clusters with uniform exchange dynamics, residues color coded on the basis of on kex, in (A) dp-p38 (magenta, 1,304 ± 58 s−1; cyan, 1,649 ± 106 s−1; orange, 962 ± 98 s−1; and green, >2,500 s−1) and (B) dp-p38⋅MKK3bKIM (magenta, 1,826 ± 80 s−1). Residues that could not be fit within a cluster in a statically meaningful manner are shown as black spheres. (C) Representative 15N ct-CPMG dispersion data (dp-p38⋅MKK3bKIM) for residues Thr106 (cyan), Asn159 (green), Leu246 (orange), and Phe274 (blue); corresponding residues are highlighted in B. The lines show cluster-fits to the Richard–Carver equation, and error bars are derived from duplicate measurements.
Activation-Loop Phosphorylation Coupled with KIM Binding Synchronizes p38 Intermediate Exchange Dynamics.
We then studied the exchange dynamics of dp-p38⋅MKK3bKIM. Remarkably, it was possible to fit a large group of residues to a single exchange rate and population using a global fit (kex = 1,826 ± 120 s−1; pB = 0.06 ± 0.02) (Fig. 3 B and C and SI Appendix, Fig. S13 and Table S2). This cluster includes all regions of p38 that have a functional, biological role, including the N-terminal lobe, the hinge, the activation loop, and the C-terminal lobe including the MAPK-specific insert. Hence, MKK3bKIM-binding and activation-loop phosphorylation are necessary to achieve uniform µs-ms dynamics throughout p38, in principle, enabling communication throughout the enzyme. Based on our data, it is likely that MKK3bKIM binding leads to a dynamic communication between the hinge and the activation loop (as displayed in np-p38⋅MKK3bKIM), whereas activation-loop phosphorylation, which reduces the ps-ns fast timescale backbone motion of the activation loop, enables a dynamic communication between the lobes. These changes likely correspond to a refining of the local energy landscape, as large-scale motions that are exemplified by slow timescale dynamics reshape (flatten) the energy landscape to include additional functional elements of p38. This flattening reduces the heterogeneity of motion and provides access to optimally functional states. This ultimately allows for the synchronization of the dynamics in p38 and the dynamic enzymatic activation of p38. This view can be extended to p38 substrates beyond the particular KIM peptide sequence used in this study, since they share highly conserved KIM-binding motifs (SI Appendix, Fig. S14).
Synchronized µs-ms Dynamics of dp-p38⋅MKK3bKIM Is Lost upon AMP-PNP Binding.
A recent report showed that dp-p38⋅MK2peptide complex has enhanced ATP binding (16), indicative of the allosteric nature of the system. To explore this, we examined the dp-p38⋅MKK3bKIM⋅AMP-PNP ternary complex under saturating conditions of the ATP analog (1:17 ratio). Many residues show increased CSPs compared with CSPs in dp-p38 (SI Appendix, Figs. S15 and S16), without a change in fast timescale backbone dynamics (SI Appendix, Fig. S17). Furthermore, more than 12 residues are broadened beyond detectability in the 2D [1H,15N] TROSY spectrum. We repeated the 15N ct-CPMG measurements for dp-p38⋅MKK3bKIM⋅AMP-PNP. Unfortunately, severe exchange broadening lowered the quality of the obtained data for dp-p38⋅MKK3bKIM⋅AMP-PNP, and fewer residues were quantitatively assessed. Nevertheless, the remaining sites indicate that AMP-PNP binding disrupts the coherent dynamics that were present in dp-p38⋅MKK3bKIM (SI Appendix, Figs. S18 and S19 and Table S2). This shows that both phosphorylation and substrate binding are necessary to induce uniform exchange dynamics in p38, which subsequently allows for the observed increased binding affinity for AMP-PNP.
Discussion
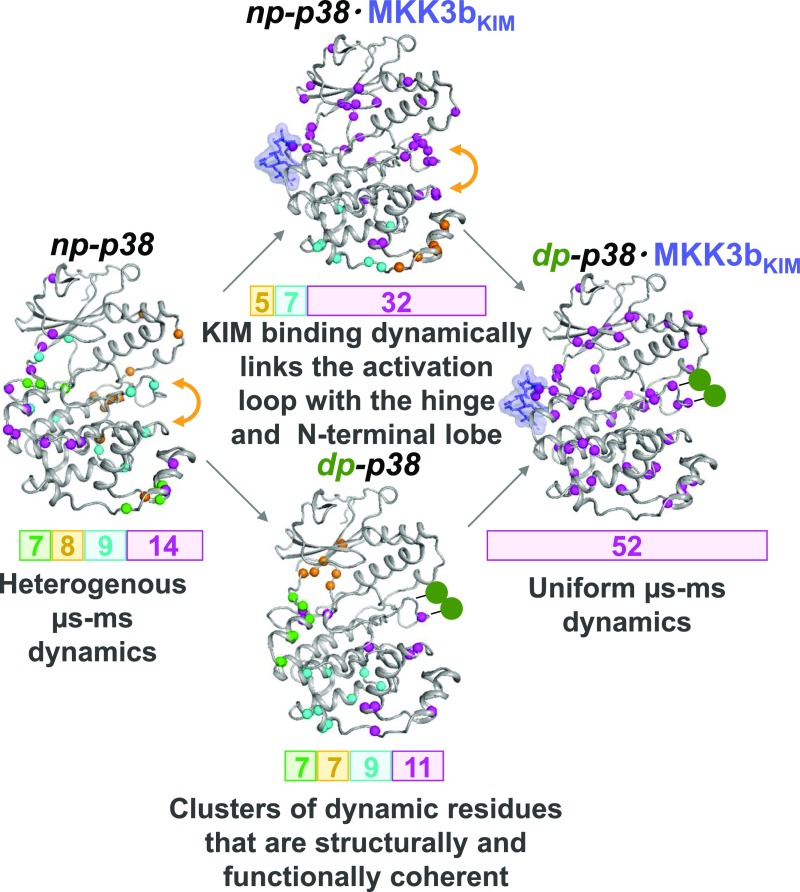
Our study shows that activation of p38 is accompanied by changes in fast and intermediate timescale dynamics that dominate the lack of apparent structural changes. Further, we also show that both p38 phosphorylation and KIM binding are necessary to coordinate this dynamic activation (Fig. 4), which reflects a manipulation of motions that control the energy landscape of the p38 backbone upon a change in a functional state. Thus, in contrast to previous reports suggesting that dynamics is necessary for enzymatic activity, here motions are necessary for the control of enzymatic activity through the step-wise assembly of an enzymatic competent state of p38.
Fig. 4.
Dynamic activation of p38. KIM-peptide binding (blue sticks/surface) leads to uniform µs-ms dynamics between the hinge and activation loop, while the activation loop stays dynamic in the ps-ns timescale (yellow arrow). Phosphorylation (green balls) rigidifies the activation loop and, together with KIM-peptide binding, manipulates the motions that regulate the energy landscape of p38.
One outstanding question is how conserved this process is for kinases generally. Here, we show that the motions of the unphosphorylated state of p38 (np-p38) are uncorrelated and not organized for catalysis. This observation is similar to that reported for both np-ERK2 (8), a second MAP kinase, and PKA (14), another ser/thr kinase (the dynamics of ERK2 and PKA were studied using 13C methyl (Ile, Val, and Leu [ILV]) side-chain dynamics experiments); namely, the dynamics of np-ERK2 and PKA are also uncorrelated. However, while all three kinases exhibit a similar lack of correlated motions in their inactive states, the dynamics of p38 and ERK2 diverge upon phosphorylation. Specifically, when ERK2 is dually phosphorylated in its activation loop (dp-ERK2), two independent clusters of correlated motions on the µs-ms timescale are observed: one cluster that connects the activation loop to the hinge region and a second cluster within the MAP kinase insert region. In contrast, the µs-ms motions of phosphorylated p38 (dp-p38) are still not correlated; rather, the only change observed upon p38 phosphorylation is a rigidification of the ps-ns motions in the p38 activation loop. In the case of p38, it is only the addition of a ligand (regulator or substrate) to either np-p38 or dp-p38 that leads to correlated µs-ms motions. A further difference is that 13C ILV methyl side-chain chemical-shift data show that phosphorylation of ERK2 leads to significant chemical-shift changes throughout ERK2 and especially in the N-terminal lobe. This is not observed in p38. Rather, 15N backbone chemical-shift data show that phosphorylation of p38 results in only local chemical-shift changes, namely, within the activation loop. Taken together, these dynamics data show that the two MAPKs, ERK2 and p38, are differentially activated.
In contrast, a comparison of the dynamics of PKA with that of p38 shows that PKA is more similar to that of p38. Namely, converting PKA into a binary (PKA:ATPɣ-C–bound) or to a ternary state (PKA:ATPɣ-C:substrate–bound) not only coordinated its dynamics, but did so with a global exchange rate (kex = 2,500 ± 300 s−1) comparable to that reported here for the dp-p38⋅MKK3bKIM complex.
Reports have shown that protein dynamics can play a critical role in modulating allostery (24). A report showed that cAMP binding to the catabolite activator protein (CAP) induces dynamic allostery in the absence of any conformational change (25). In a different report, it was shown that the truncation of the C-terminal α-helix of a PDZ domain leads to reduced-affinity peptide ligand binding, despite the fact that the helix is not abutting the peptide-binding site (26). Further investigation showed that truncation of the α-helix increased the flexibility of the PDZ domain, leading to unfavorable conformational entropy that led to a decrease in the binding affinity to the peptide. Lisi et al. (27) showed that indole–3-glycerol–phosphate synthase is dependent on millisecond conformational motions for efficient catalysis. Using the example of calmodulin in complex with different peptides it was shown that conformational entropy is correlated to the overall binding entropy and hence the binding energetics of protein–ligand interaction (28). Last, we have shown that PTP1B uses dynamics to allow for allostery (13). Here, we have shown that synchronized dynamics directly correlates with the p38 activity by changing the energy landscape.
In summary, in contrast to previous reports suggesting that dynamics is necessary for enzymatic activity, for kinases it is clear that dynamics control enzymatic activity through the step-wise assembly of an enzymatic competent state. The high degree of structural conservation among kinases suggests that elements of this assembly may be conserved within the entire kinase family. Furthermore, the concept that motions control the energy landscape of an enzyme to systematically control its activity may be a general strategy exploited by enzymes that require multiple coordinated events (phosphorylation and substrate docking, among others) in order for catalysis to occur.
Materials and Methods
Extended materials and methods are included in SI Appendix.
NMR Spectroscopy.
All NMR spectra were recorded at 293 K on either a Bruker Avance II 500 MHz or a Bruker Avance IIIHD 850 MHz (1H Larmor) spectrometer both equipped with a TCI HCN z-gradient cryoprobe. NMR samples were prepared in NMR buffer containing 10% (vol/vol) D2O. MKK3bKIM was titrated with [2H,15N]-labeled np-p38 and [2H,15N]-labeled dp-p38 at increasing molar ratios (one, two, four, and six) to generate a saturated complex of p38 with MKK3bKIM, a ratio which was used for all subsequent NMR relaxation measurements. Similarly, saturated complex of AMP-PNP with various states of p38 (np-p38, dp-p38, np-p38⋅MKK3bKIM, and dp-p38⋅MKK3bKIM) were generated by titrating increasing molar ratios of AMP-PNP (5, 10, 12, 15, and 17) with the respective p38 states. Backbone amide chemical-shift deviations were calculated using the formula: Δδav = √(0.5((δHN,bound − δHN,free)2 + 0.04 (δN,bound − δN,free)2)). The sequence-specific backbone resonance assignments of dp-p38 and dp-p38⋅MKK3bKIM states were obtained using the combination of 2D [1H,15N] TROSY, 3D TROSY-HNCA, 3D TROSY-HN(CO)CA, and 3D TROSY-HNCACB.
The 15N longitudinal (R1) and transverse (R2) relaxation rates and 15N[1H]-NOE (hetNOE) measurements were acquired in an interleaved manner as follows (29). T1 and T2 experiments were acquired at 850 MHz with a recycle delay of 3 s between experiments and the following relaxation delays for T1: 101, 401, 401, 801, 1,201, 1,601, 2,001, 3,001, 3,001, and 4,001 ms; and T2: 3.6, 7.2, 14.4, 14.4, 18, 21.6, 25.2, 32.4, 32.4, 43.2, 54, and 72 ms. Relaxation delays for data collected at 500 MHz are as follows: T1: 100, 500, 500, 800, 1,100, 1,400, 1,700, 2,000, 2,500, 3,000, and 3,000 ms; and T2: 3.6, 14.4, 14.4, 18, 21.6, 25.2, 28.8, 32.4, 32.4, 43.2, 54, and 72 ms. Two relaxation experiments were repeated to determine an experimental error of the measurements. The 15N{H}-NOE measurements were determined from a pair of interleaved spectra acquired with or without presaturation and a recycle delay of 5 s at 500 MHz.
Ct–Carr–Purcell–Meiboom–Gill (ct-CPMG) relaxation dispersion experiments (22) were performed at 293 K using two magnetic field strengths (500 and 850 MHz). The experiment was arrayed with different delays (2τ) between 15N refocusing pulses, with a total time of 20 ms (Trelax). The experiment was acquired with 12 different τ times corresponding to CPMG frequencies of 0, 50, 100, 150, 200, 200, 300, 400, 500, 700, and 900 Hz. Two τ times were collected in duplicate to determine the experimental measurement error. The effective decay rate (R2,eff) was calculated by the equation R2,eff = −1/T ln[I(νCPMG)/I0], where νCPMG = 1/4τ, 2τ being the interval between successive 180 degree 15N refocusing pulses, and IνCPMG and I0 are the intensities of peaks recorded with and without CPMG period, respectively. All NMR data were processed and analyzed using NMRPipe (30) and Sparky (31, 32). The p38 concentration for all NMR measurements was 0.4 ± 0.1 mM. Within experimental parameters, the concentration of all samples was kept constant between different measurements to reduce possible effects on R2,eff due to changes in solution viscosity or possible nonspecific aggregation. All relaxation experiments were recorded using freshly prepared p38, and MKK3bKIM peptide was carefully titrated to achieve full saturation. When saturation was achieved (chemical shift of the peaks stopped changing), additional MKK3bKIM peptide was added to ensure that all experiments were performed under fully saturated conditions and thus that the observed CPMG dispersions do not reflect the ligand on/off. The same approach was used for AMP-PNP titrations.
CPMG Analysis.
Initially, all p38 residues were fit individually to the Carver–Richards equation for a system in two-state exchange using a Levenberg–Marquardt algorithm. No particular clustering was evident from this approach. Therefore, in an initial pass, residues adjacent to each other in sequence or space were clustered together for the purpose of a mutual fit. The quality of a group fit was assessed using the Bayesian Information Criterion (BIC) to compare the group fit to the results of the individual fits using ΔBIC = BICgroup − BICindividual. In this case, ΔBIC favors the group fit when it is more negative. Residues were kept in groups that satisfied the following criteria: (i) the cumulative ΔBIC for the group was negative and (ii) the ΔBIC for the individual residue within the group was no greater than +1. Groups were subsequently expanded by adding adjacent residues, residues with individual fit parameters similar to the group parameters, and by unifying small groups with similar dynamic parameters, so long as conditions i and ii were met.
No detailed rates were calculated for clusters with kex > 2,500 s−1. If pB = 0.5, it was classified as N.D., as this condition results from a set of residues in fast exchange, where the population cannot reliably be separated from (pApB)Δω2. No group of residues with BIC/residue values ≥ −6 was considered statistically meaningful.
Supplementary Material
Acknowledgments
This work was supported by an NIH-R01GM100910 grant (to A.J.W. and W.P.). This research is based in part on data obtained at the Brown University Structural Biology Core Facility, which is supported by the Division of Biology and Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 27273 and 27274).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721441115/-/DCSupplemental.
References
- 1.Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson KP, et al. Crystal structure of p38 mitogen-activated protein kinase. J Biol Chem. 1996;271:27696–27700. doi: 10.1074/jbc.271.44.27696. [DOI] [PubMed] [Google Scholar]
- 3.Bellon S, Fitzgibbon MJ, Fox T, Hsiao HM, Wilson KP. The structure of phosphorylated p38gamma is monomeric and reveals a conserved activation-loop conformation. Structure. 1999;7:1057–1065. doi: 10.1016/s0969-2126(99)80173-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y-Y, Wu J-W, Wang Z-X. Mitogen-activated protein kinase (MAPK) phosphatase 3-mediated cross-talk between MAPKs ERK2 and p38alpha. J Biol Chem. 2011;286:16150–16162. doi: 10.1074/jbc.M110.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 6.Akella R, Min X, Wu Q, Gardner KH, Goldsmith EJ. The third conformation of p38α MAP kinase observed in phosphorylated p38α and in solution. Structure. 2010;18:1571–1578. doi: 10.1016/j.str.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 8.Xiao Y, et al. Phosphorylation releases constraints to domain motion in ERK2. Proc Natl Acad Sci USA. 2014;111:2506–2511. doi: 10.1073/pnas.1318899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogtherr M, et al. NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew Chem Int Ed Engl. 2006;45:993–997. doi: 10.1002/anie.200502770. [DOI] [PubMed] [Google Scholar]
- 10.Masterson LR, et al. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci USA. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc Natl Acad Sci USA. 2013;110:E4437–E4445. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy MS, et al. Conformational rigidity and protein dynamics at distinct timescales regulate PTP1B activity and allostery. Mol Cell. 2017;65:644–658.e5. doi: 10.1016/j.molcel.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, et al. A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci Adv. 2017;3:e1600663. doi: 10.1126/sciadv.1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis DM, et al. Structural basis of p38α regulation by hematopoietic tyrosine phosphatase. Nat Chem Biol. 2011;7:916–924. doi: 10.1038/nchembio.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokunaga Y, Takeuchi K, Takahashi H, Shimada I. Allosteric enhancement of MAP kinase p38α’s activity and substrate selectivity by docking interactions. Nat Struct Mol Biol. 2014;21:704–711. doi: 10.1038/nsmb.2861. [DOI] [PubMed] [Google Scholar]
- 17.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 18.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 20.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 21.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: Application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 23.d’Auvergne EJ, Gooley PR. The use of model selection in the model-free analysis of protein dynamics. J Biomol NMR. 2003;25:25–39. doi: 10.1023/a:1021902006114. [DOI] [PubMed] [Google Scholar]
- 24.Tzeng SR, Kalodimos CG. Protein dynamics and allostery: An NMR view. Curr Opin Struct Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisi GP, East KW, Batista VS, Loria JP. Altering the allosteric pathway in IGPS suppresses millisecond motions and catalytic activity. Proc Natl Acad Sci USA. 2017;114:E3414–E3423. doi: 10.1073/pnas.1700448114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 30.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Goddard TD, Kneller DG. 2004 Sparky–NMR Assignment Program. Available at https://www.cgl.ucsf.edu/home/sparky/. Accessed April 3, 2018.
- 32.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]