Significance
This study identifies a mechanistic basis for the enigmatic, long-observed interaction between the rheumatoid arthritis shared epitope (SE)—the most significant genetic risk factor in this disease—and exposure to environmental pollutants, such as cigarette smoke. Specifically, we show that the SE, acting as a signal transduction ligand, cooperates with the aryl hydrocarbon receptor-activated pathway, and together facilitate cellular events that culminate in inflammation and bone destruction in experimental autoimmune arthritis. The cross-talk between the two pathways is mediated by nuclear factor kappa B.
Keywords: shared epitope, aryl hydrocarbon receptor, signaling, gene–environment interaction, nuclear factor kappa B
Abstract
The susceptibility to autoimmune diseases is affected by genetic and environmental factors. In rheumatoid arthritis (RA), the shared epitope (SE), a five-amino acid sequence motif encoded by RA-associated HLA-DRB1 alleles, is the single most significant genetic risk factor. The risk conferred by the SE is increased in a multiplicative way by exposure to various environmental pollutants, such as cigarette smoke. The mechanism of this synergistic interaction is unknown. It is worth noting that the SE has recently been found to act as a signal transduction ligand that facilitates differentiation of Th17 cells and osteoclasts in vitro and in vivo. Intriguingly, the aryl hydrocarbon receptor (AhR), a transcription factor that mediates the xenobiotic effects of many pollutants, including tobacco combustion products, has been found to activate similar biologic effects. Prompted by these similarities, we sought to determine whether the SE and AhR signaling pathways interact in autoimmune arthritis. Here we uncovered a nuclear factor kappa B-mediated synergistic interaction between the SE and AhR pathways that leads to markedly enhanced osteoclast differentiation and Th17 polarization in vitro. Administration of AhR pathway agonists to transgenic mice carrying human SE-coding alleles resulted in a robust increase in arthritis severity, bone destruction, overabundance of osteoclasts, and IL17-expressing cells in the inflamed joints and draining lymph nodes of arthritic mice. Thus, this study identifies a previously unrecognized mechanism of gene–environment interaction that could provide insights into the well-described but poorly understood amplification of the genetic risk for RA upon exposure to environmental pollutants.
Millions of people are afflicted with autoimmune diseases, and many more are at risk for developing such disorders. The etiology of autoimmunity is incompletely understood, but there is strong evidence that both genetic and environmental factors play a role (1–3). Over the years, there has been growing realization that genes and environment may interact in autoimmunity in an additive or multiplicative fashion. One condition in which such interaction is apparent is rheumatoid arthritis (RA). Recent evidence indicates that the risk of developing this common disease is significantly amplified in genetically susceptible individuals who have been exposed to various environmental pollutants (4–6).
The mechanisms governing the genetic and environmental risks for RA, let alone the synergism between these two factors, are presently unknown. Two-thirds of RA risk is attributed to genetic factors, primarily the HLA-DRB1 locus. The vast majority of RA patients carry particular HLA-DRB1 alleles that encode a five-amino acid sequence motif called the shared epitope (SE) in the third allelic hypervariable region of the HLA-DRβ chain (7). The SE is the strongest genetic risk factor for severe RA known to date, but its mechanism of action in RA is unknown.
Over the past few years we have gained important new insights into the functional role of the SE in RA (8–23). Our findings indicate that the SE, located near the tip of a prominent fold in the DRβ chain (15), acts as a ligand that interacts with cell surface calreticulin (CRT) (11, 13, 14). SE-activated signaling depends also on CRT transmembrane coreceptor, CD91 (also known as LRP1) (11). Activation of this pathway leads to Th17 polarization and increased osteoclast (OC) differentiation and activity, both in vitro and in vivo (12, 17, 20). Importantly, the SE signaling effects are independent of its role in antigen presentation, do not involve interaction with T-cell receptors, and can be seen with natural, cell surface-expressed HLA-DR molecules, or physiologically folded cell-free HLA-DR tetramers, as well as soluble synthetic peptides or peptidomimetics expressing the SE sequence motif. When administered to mice with collagen-induced arthritis (CIA), synthetic SE ligands enhanced arthritis severity and bone damage (17, 20).
In addition to genetic predisposition, environmental factors significantly influence RA susceptibility as well. For example, the prevalence of RA is higher in urban populations, and the disease is associated with exposure to environmental pollutants, such as dioxin-like compounds (24) and tobacco smoke (25). Furthermore, cigarette smoke increases the disease risk of SE-positive individuals in a multiplicative, dose-dependent fashion (26, 27). The mechanistic basis of this gene–environment interaction is unknown.
One of the theories concerning the impact of environmental pollutants on autoimmunity is focused on the aryl hydrocarbon receptor (AhR) (24), an intracellular ligand-activated transcription factor that mediates the effects of various polycyclic aromatic hydrocarbons, including dioxin and tobacco combustion products. Intriguingly, reminiscent of the SE ligand, AhR agonists have been shown to perturb the balance between regulatory T (Treg) cells and Th17 cells (28) and activate OCs (29). Moreover, the AhR pathway has been implicated in the pathogenesis of autoimmune diseases (30–32).
Prompted by these parallels, here we sought to determine whether the SE and AhR pathways interact functionally in inflammatory arthritis. Our findings show that the SE ligand and AhR agonists operate synergistically and together markedly enhance OC and Th17 cell differentiation and exacerbate disease severity and bone destruction in CIA. The cross-talk between the two pathways is mediated by nuclear factor kappa B (NF-κB).
Results
Synergistic Proosteoclastogenic Interaction Between the SE and AhR Pathways in Vitro.
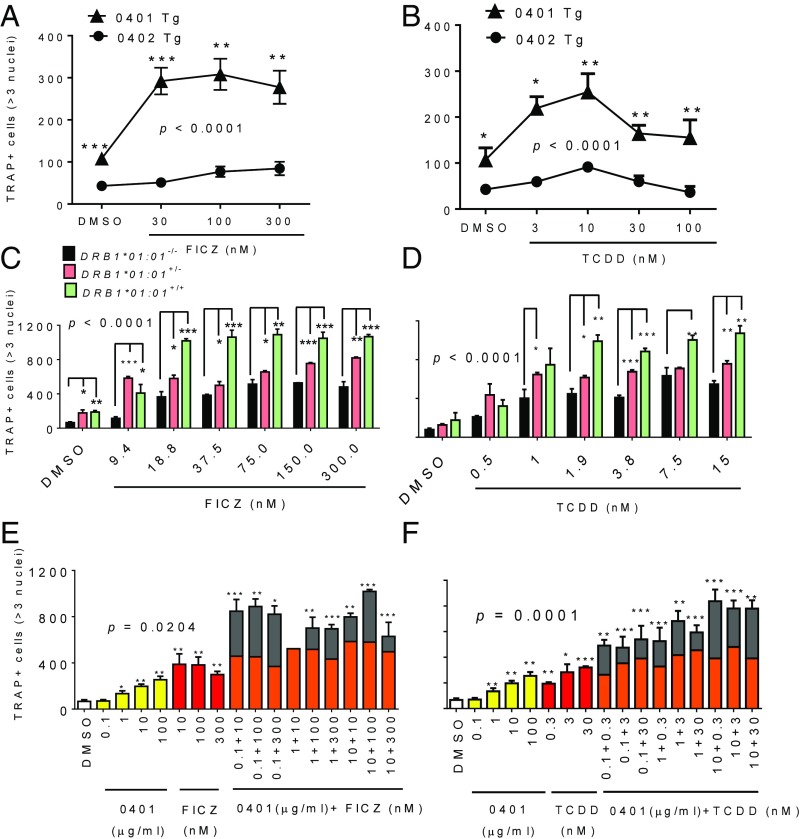
OC-mediated bone destruction is a major disease severity factor in RA (33). Since the SE and AhR agonists have been independently shown to facilitate OC differentiation, we first undertook to determine whether the two pathways cooperate during OC differentiation in vitro. To this end we studied transgenic (Tg) mice that express on their cell surface SE-positive human HLA-DR molecules with a 70-QKRAA-74 SE sequence in the DRβ chain, coded by the HLA-DRB1*04:01 allele (34) (referred to herein as “0401 Tg”). As control, we studied similar background mice expressing SE-negative (70-DERAA-74) HLA-DR molecules, whose β-chain is coded by the allele HLA-DRB1*04:02 (“0402 Tg”). When exposed to the AhR agonists 6-formylindolo[3,2-b]carbazole (FICZ) (Fig. 1A) or 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Fig. 1B), bone marrow cells (BMCs) from SE-positive 0401 Tg mice, but not the SE-negative 0402 Tg, displayed significantly (P < 0.0001 by two-way ANOVA) facilitated OC differentiation [F(1, 8) = 92.01 for FICZ; F(1, 10) = 70.91 for TCDD]. A similar pattern was seen in another HLA-DRB1*04:01 Tg mouse line on a B10.M background (35) (Fig. S1). Thus, an in vivo-expressed SE specifically augments the proosteoclastogenic effects of AhR agonists.
Fig. 1.
Synergistic SE–AhR interaction in osteoclastogenesis. BMCs from SE-positive (0401 Tg, triangles) or SE-negative (0402 Tg, circles) mice were cultured for 6 d in OC-differentiating medium in the absence or presence of various concentrations of AhR agonists FICZ (A) or TCDD (B) and the number of OC (TRAP-positive multinucleated cells) was determined. (C and D) BMCs from SE homozygous (SE+/+, green), heterozygous (SE+/−, pink), or null (SE−/−, black) B10.M-HLA-DR1 (DRB1*01:01) Tg mice were cultured with various doses of FICZ (C) or TCDD (D) and OCs were quantified as above. (E and F) RAW 264.7 cells cultured in OC-differentiating medium in the absence or presence of various concentrations of the SE ligand 65–79*0401 (0401, yellow) and in the presence of various doses of FICZ (E) or TCDD (F) alone (red) or in combination with AhR agonist plus SE ligand (orange/gray). Osteoclastogenic effects of SE–AhR agonist combinations (gray) which exceed the arithmetic sum (orange) of the effects by the respective agents alone denote synergism. Multiple comparison analysis concerning the significance of the genotype (A and B) and synergism (C–F) was performed using two-way ANOVA, and shown as P values in each figure. Results in all experiments represent mean and SEM of biologic replicates (n = 3). In all figures: *P < 0.05; **P < 0.01; ***P < 0.001.
To better quantify the interaction, we next addressed the effect of SE “dose” by analyzing the impact of SE-coding gene zygosity. To this end, BMCs from HLA-DR1 Tg mice on a B10.M background (36) with homozygous (SE+/+), heterozygous (SE+/−), or null (SE−/−) genotype of a SE (70-QRRAA-74)-coding allele HLA-DRB1*01:01 were cultured under OC differentiation conditions in the presence of different concentrations of FICZ (Fig. 1C) or TCDD (Fig. 1D). A two-way ANOVA analysis indicted a statistically significant synergistic interaction between AhR agonist and the SE-coding gene zygosity [F(12, 42) = 10.13] (P < 0.0001) for FICZ treatment and [F(12, 42) = 2.84] (P < 0.0001) for TCDD treatment.
As discussed above, the SE signaling effects are independent of SE’s role in antigen presentation, and can be seen with naturally folded HLA-DR molecules, as well as cell-free synthetic peptides expressing the SE sequence motif (8, 9, 11, 12, 17, 20). Consistent with these previous findings, as shown in Fig. 1 E and F, SE–AhR synergism could be seen in combinatorial dose–response experiments in which a soluble synthetic SE ligand 65–79*0401 (8) was used in OC differentiation assays in SE-negative mouse RAW 264.7 cells. The effect of SE ligand–AhR agonist combinations was greater than the arithmetic sum of the effects of the different agents individually. A two-way ANOVA analysis revealed significant synergistic interaction between SE and FICZ [F(9, 32) = 2.65] (P = 0.0204), and between the SE ligand and TCDD [F(9, 32) = 5.51] (P = 0.0001). Thus, taken together, Fig. 1 demonstrates that the SE ligand, either in its physiological cell surface conformation, or as a soluble synthetic ligand, interacts synergistically with AhR agonists to facilitate osteoclastogenesis.
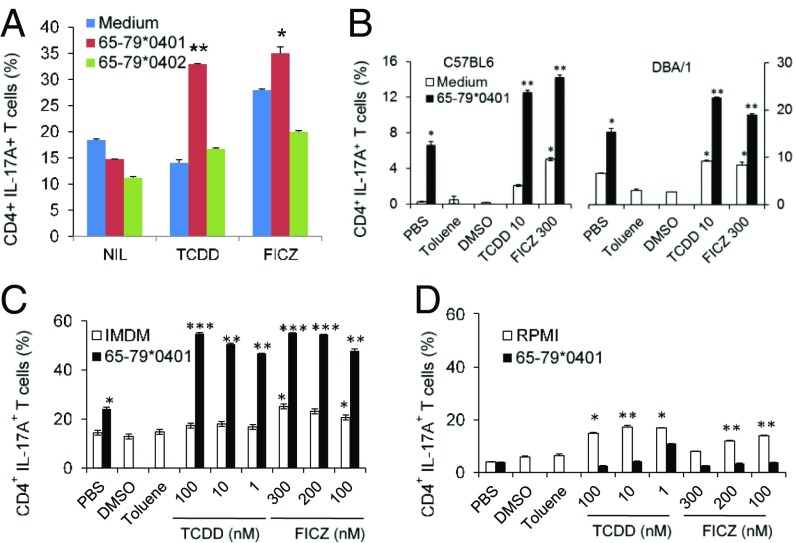
In addition to the effect on osteoclastogenesis, AhR–SE interaction strongly promoted Th17 cell polarization (Fig. 2A). This interaction was mouse strain independent as it could be seen in genetically disparate C57BL6 (H2b) and DBA/1 (H2q) mice (Fig. 2B). Additionally, consistent with the known enhancing effect of tryptophan on AhR-activated Th17 polarization (37), Fig. 2C and D show that SE–AhR interaction effect on Th17 polarization was tryptophan dependent. The reason for the paradoxical effect in tryptophan-deficient media in the presence of the SE ligand (Fig. 2D) is unclear, but could be due to the fact that the SE inhibits the tryptophan-catabolizing enzyme indoleamine 2,3 dioxygenase (12). One scenario to consider is that under these “double hit” conditions TCDD produced a paradoxical Th17 differentiation effect. This question, as well as the in vivo significance of tryptophan and its catabolites in Th17 polarization under SE–AhR interaction conditions require further study.
Fig. 2.
SE–AhR interaction promotes Th17 polarization in vitro. (A) In vitro Th17 differentiation of T cells derived from SE-negative DBA/1 mice in the presence or absence of 100 μg/mL of SE ligand 65–79*0401 (red) or SE-negative 65–79*0402 control peptide (green), in the presence or absence of TCDD (10 nM) or FICZ (200 nM). Asterisks denote significant increase relative to medium. (B) Interaction between the SE ligand and AhR agonists is not strain restricted, as it could be seen in both C57BL6 (H2b) and DBA/1 (H2q) mice. (C and D) DBA/1 naïve T cells were polarized in vitro in the presence or absence of 100 μg/mL of SE ligand 65–79*0401 and TCDD or FICZ at various concentrations. As can be seen, SE–AhR interaction effect could be demonstrated in tryptophan-rich Iscove’s modified Dulbecco’s medium (IMDM) (C), but not in tryptophan-poor Roswell Park Memorial Institute medium (RPMI)-1640 (D). Asterisks denote statistical significance relative to the respective vehicle (peptide versus PBS; TCDD versus toluene; FICZ versus DMSO). Mean and SD of biological replicates (n = 2) in a representative, one of two or more independent experiments. In all figures, statistical analysis was performed using a two-tailed t test. *P < 0.05; **P < 0.01; ***P < 0.001.
SE–AhR interaction led to synergistic facilitation of inflammatory arthritis-related factors. For example, coculturing RAW 264.7 cells with TCDD and the SE ligand 65–79*0401 led to markedly enhanced mRNA expression levels of the bone-destroying cathepsin K (Ctsk) gene (Fig. S2) and to increased joint tissue protein expression levels of the chemokine (C-C motif) ligand 2 (Ccl2) (Fig. S3).
A Role for NF-κB.
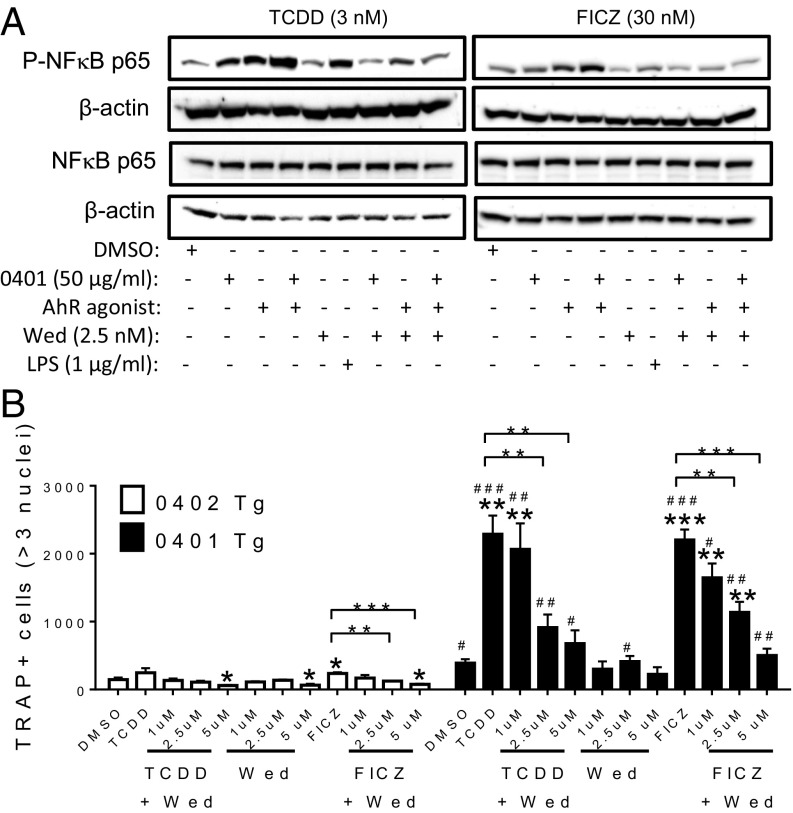
It is worth noting that Ctsk and Ccl2, which are established markers of OC differentiation (38, 39), are both up-regulated by NF-κB (40, 41), a pathway which has known roles in inflammatory arthritis, osteoclastogenesis, and AhR-activated pathways (40–43). We therefore undertook to examine the involvement of NF-κB in SE–AhR interaction. As can be seen in Fig. 3A, the combination of the SE ligand plus TCDD or FICZ facilitated NF-κB signaling, as evidenced by increased p65 nuclear translocation. The NF-κB pathway inhibitor wedelolactone efficiently blocked the activation. To validate these findings, RAW 264.7 cells were transiently transfected with an NF-κB–inducible secreted alkaline phosphatase (SEAP) construct. When these cells were cotreated with the SE ligand and FICZ, enhanced NF-κB–induced transcriptional activation was observed. The interaction was completely blocked by cotransfecting cells with a dominant negative IκBαDN construct (Fig. S4). Together, these findings confirm the role of NF-κB in the interaction.
Fig. 3.
SE–AhR interaction is mediated by NF-κB. (A) Western blot analysis of NF-κB nuclear translocation. RAW 264.7 cells were treated for 30 min with or without 50 μg/mL of SE ligand 65–79*0401 (0401) in the presence or absence of 3 nM of TCDD (Left) or 30 nM of FICZ (Right). LPS was used as a positive control and wedelolactone (Wed) as an NF-κB pathway inhibitor. Immunoblots of nuclear (Top two panels) and total (Bottom two panels) proteins are shown. A representative, one of five independent experiments is shown. (B) BMCs harvested from SE-positive 0401 Tg (black bars) or SE-negative 0402 Tg (white bars) mice were cultured for 6 d in OC-differentiating medium with or without 3 nM TCDD or 100 nM FICZ in the presence or absence of various concentrations of the NF-κB pathway inhibitor, Wed. OCs were counted as above. Data (mean and SEM) represent biologic replicates (n = 3). Asterisks denote significance within the respective Tg mouse lines (*P < 0.05; **P < 0.01; ***P < 0.001); # symbols denote statistical significance between 0401 Tg versus 0402 Tg mice (#P < 0.05; ##P < 0.01; ###P < 0.001).
We next determined the involvement of NF-κB in SE–AhR interaction during osteoclastogenesis in BMCs isolated from SE-positive 0401 Tg and SE-negative 0402 Tg mice (Fig. 3B). There was only minor effect of AhR agonists on OC differentiation in BMCs from SE-negative 0402 Tg mice. In contrast, a markedly enhanced OC differentiation was seen in the SE-positive 0401 Tg mice. The NF-κB inhibitor wedelolactone blocked AhR–SE synergism in a dose-dependent fashion (Fig. 3B). Similar trends were seen in another Tg mouse line expressing DRB1*04:01-coded SE-positive DR4 molecules on a B10.M background (Fig. S5). Thus, we conclude that the interaction between SE and AhR agonists during osteoclastogenesis is mediated by the NF-κB signaling pathway.
Synergistic Interaction Between the SE and AhR Pathways in Vivo.
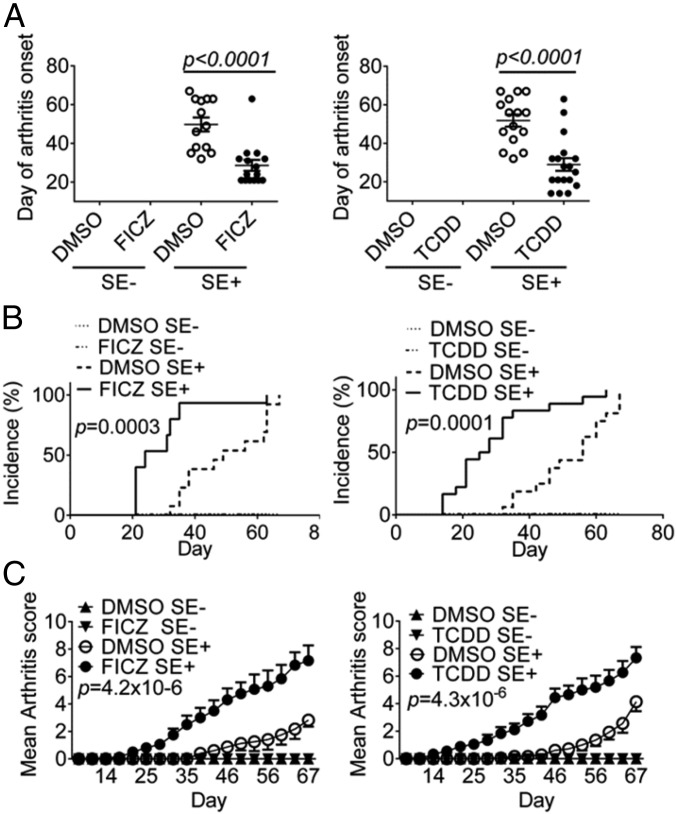
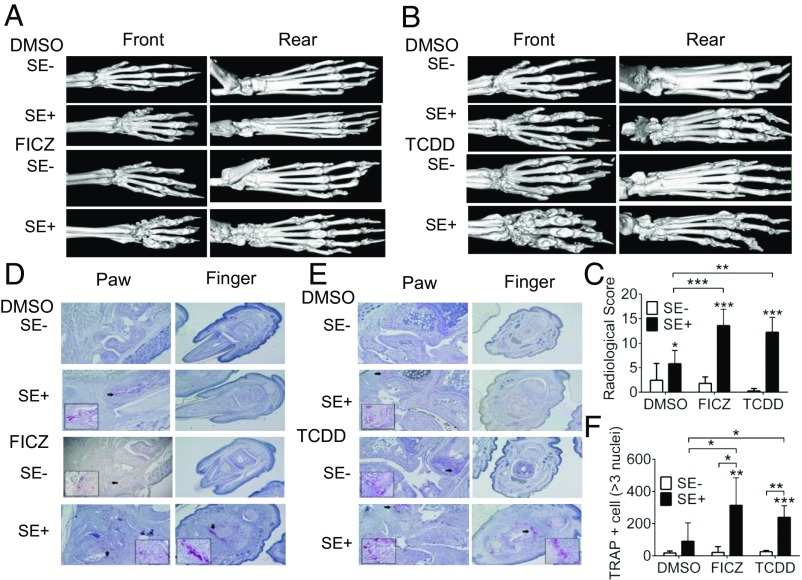
Given the important roles that OC and Th17 cells play in RA and its experimental models, we proceeded to determine the arthritogenic effect of SE–AhR interaction in vivo in HLA-DRB1*04:01 Tg mice on a B10.M background (35). In this model, the SE+/+ genotype confers embryonal lethality; SE−/− mice are resistant to CIA, while SE+/− mice develop moderately severe erosive arthritis, thereby offering a suitable system for studying disease-worsening interventional protocols. As can be seen in Fig. 4, both FICZ and TCDD significantly facilitated the day of onset (Fig. 4A) and disease progression (Fig. 4B), and worsened joint swelling (Fig. 4C). Microcomputerized tomography (CT) imaging (Fig. 5 A and B) and radiologic scores (Fig. 5C) demonstrated significantly worse bone erosions in SE-positive CIA mice that were treated with AhR agonists FICZ or TCDD.
Fig. 4.
Interaction between the SE and AhR pathways in vivo. CIA was induced in B10.M Tg mice carrying either a DRB1*04:01−/+ (SE+), or DRB1*04:01−/− (SE−) genotype. Mice were injected i.p. weekly with FICZ, 28 ng/gm body weight, or TCDD, 0.3 ng/gm body weight, or DMSO. In A, P values were calculated using a two-tailed t test. In B, P values were calculated using a log-ranked (Mantel–Cox) test. In C, P values were calculated using a paired t test for the entire disease course. Results are compiled data from two experiments (n = 16 per group in FICZ treatment experiments, and n = 18 per group in TCDD treatment experiments). In all figures, error bars represent SEM.
Fig. 5.
AhR–SE interaction facilitates bone damage. (A and B) Representative micro-CT images of CIA mice treated with FICZ (A), or TCDD (B), or DMSO. (C) Radiologic scores. Mean and SEM; n = 5 per treatment group. *P < 0.05; **P < 0.01; ***P < 0.001. (D and E) Representative histologic images of TRAP-stained paw and finger tissue sections of CIA mice treated with FICZ (D), TCDD (E), or DMSO. Original magnification is 10×. Areas of high OC abundance are marked by arrows and shown in a 40× magnification in the boxed image in the Lower corner. (F) Quantification of the total number of TRAP-positive cells in front and rear paws of CIA mice treated with FICZ, TCDD, or DMSO (n = 3 in the FICZ treatment group and n = 4 in the TCDD treatment group). *P < 0.05; **P < 0.01; ***P < 0.001.
Tartrate-resistant acid phosphatase (TRAP) staining of joint tissues showed increased OC infiltration in AhR agonist-treated SE-positive mice (Fig. 5 D and E). When cultured ex vivo in OC-differentiating conditions, BMCs isolated from SE-positive CIA mice that were treated in vivo with AhR agonists showed a significantly more robust osteoclastogenesis (Fig. 5F). Additionally, synovial tissues from SE-positive, AhR agonists-treated mice showed significantly higher in situ abundance of IL17 (Fig. S6), and their spleens and regional lymph nodes had a higher percentage of Th17 cells, with a reciprocal decline of Treg cells compared with SE+-untreated, and SE-treated mice (Fig. S7).
Discussion
This study uncovers a previously unknown mechanistic basis for gene–environment interaction in autoimmune arthritis. Case studies have long demonstrated a multiplicative interaction between SE-coding HLA-DRB1 alleles and cigarette smoking as risk factors for seropositive RA patients, with dose–effect relationships for both the SE zygosity and the extent of tobacco exposure (26, 27). The mechanistic basis of this interaction is unknown. Prevailing hypothesis for SE–smoking interaction in RA postulate that cigarette smoke may activate protein citrullination in the lungs, which in turn can become antigenic in SE-positive individuals with resultant anticitrullinated protein antibodies (44, 45). One caveat of the lung-centered hypotheses is the fact that RA risk has been shown to associate with in utero exposure in nonsmoker offspring of smoking mothers (46) despite the fact that the exposure had happened during a period when lungs were not yet exposed to air.
The findings presented here uncover an NF-κB–mediated SE–AhR pathways cross-talk that leads to enhancement of cell differentiation and functional activation of proarthritogenic mechanisms in vitro, as well as increased disease severity in experimental arthritis in vivo. To the best of our knowledge, this study provides a unique experimentally validated mechanism of SE interaction with an environmental factor.
This study offers several notable insights. First, SE–AhR interaction could be documented in OC and Th17 cells, which have both been previously independently found to play important roles in the pathogenesis of autoimmune arthritis (4). The interaction was seen in both primary and long-term cultured cells, derived from several immunogenetically distinct mouse strains. The effect could be found both in vitro and in vivo. Furthermore, the interactions could be seen with the SE in its cell surface-expressed, physiologic conformation, and as a soluble cell-free ligand. This versatility provides internal corroboration and attests to the biologic significance of the discovered mechanism.
Second, the parallels between the interactions found here and previously reported epidemiologic observations in human RA are compelling. Similar to the experimental data shown here, both SE-coding HLA-DRB1 allele dose and the extent of environmental exposure have been found to positively impact disease risk (26, 27). Furthermore, we validated the interaction in two distinct human HLA-DRB1 alleles, *04:01 and *01:01 (coding for the two most common SE motif sequences QKRAA and QRRAA, respectively).
Third, the SE ligand interacted with both exogenous, synthetic (TCDD) and endogenous, physiological (FICZ) AhR agonists. These findings are consistent with other studies showing Th17-polarizing and autoimmune effects by FICZ in experimental autoimmune encephalomyelitis (EAE) (30) but appear to differ with some reports concerning TCDD effect (31). The apparent inconsistency may be due to inherent differences between the EAE and CIA disease models, in the latter of which the AhR pathway has been previously implicated as a facilitator of bone damage (47). Additionally, the apparent inconsistency could be due to experimental protocol or mouse age and strain differences. Noteworthy, TCDD dose differences could well explain the apparent inconsistency. While the in vivo FICZ doses previously reported in EAE (30) were equivalent to those used here, and produced similar autoimmune enhancing effects, the study reporting anti-EAE effects by TCDD (31) used a 166-fold higher dose than the TCDD dose administered here.
AhR and its agonists have been previously proposed to play a pathogenic role in RA by activating synovial fibroblasts (48), dendritic cells (49), macrophages (50), and osteoclasts (51), and inhibiting osteoblasts (47), thereby facilitating inflammation and bone destruction in RA (52). Our previous studies (8–23) have demonstrated that the SE ligand activates similar cellular and pathologic effects, suggesting that the two pathways operate in a similar fashion. It is therefore intriguing that the NF-κB pathway, an important activator of several pathogenic mechanisms in RA (42, 43, 53), and a downstream effector mechanism of AhR-activated RA-relevant effects (24, 54), was identified here as the pathway that mediates SE–AhR interaction. Thus, the data presented here provide a molecular basis for cross-talk between the two pathways. Based on these findings, we propose a mechanistic model of gene–environment interaction in inflammatory arthritis (Fig. S8).
Methods
Reagents, Cells, and Mice.
Reagents, cells, and mice in this study were as previously described (8, 20, 34–36). For details see SI Materials and Methods.
Mice were housed at the University of Michigan’s Unit for Laboratory Animal Medicine facility. All experiments were performed in accordance with protocols approved by the Committee on Use and Care of Animals.
Quantification of OC Differentiation in Vitro.
The quantification of OC differentiatin in vitro was performed as previously described (8, 17, 20). For details see SI Materials and Methods.
T-Cell Differentiation Assays and Flow Cytometry.
T-cell differentiation assays and flow cytometry were performed as we previously described (12). For details see SI Materials and Methods.
Western Blots.
NF-κB p65 nuclear translocation was determined in 70% confluent RAW 264.7 cells pretreated for 1 h with or without the NF-κB inhibitor wedelolactone (7-methoxy-5, 11, 12-trihydroxycoumestan; Sigma-Aldrich) before stimulating them with various agents in DMSO, or the vehicle alone. After treatment, cells were rinsed twice with ice-cold PBS, harvested, and resuspended in 200 μL of buffer A (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl and 0.1% Nonidet P-40) containing 1× protease/phosphatase inhibitor mixture (Cell Signaling Technology). After spin down for 30 s at 14,000 rpm, the cytosolic portion of the protein was transferred to a new tube. Cell pellets were resuspended in 50 μL of buffer B (20 mM Hepes, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA and 25% V/V glycerol) containing 1× protease/phosphatase inhibitor mixture (Cell Signaling Technology). After incubation on ice for 30 min with constant vortex, the samples were spun down at 14,000 rpm for 15 min in 4 °C, and nuclear proteins were then transferred to a new tube. Proteins were quantified using a BCA Kit (Thermo Fisher Scientific). Equal quantities of nuclear or cytosolic protein were separated by 10% Tris-glycine SDS/PAGE gels (Life Technologies) and transferred onto a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% nonfat milk in Tris-phosphate buffer containing 0.1% Tween 20 (TBS-T) for 1 h, the membranes were further incubated overnight at 4 °C with primary antibodies including anti–NF-κB p65, or antiphospho–NF-κB (Ser536) (both from Cell Signaling Technology). As a loading control, the blots were also probed for β-actin (Thermo Fisher Scientific). The next day, horseradish peroxidase-conjugated secondary antibodies (GE Healthcare Life Sciences) were applied. Peroxidase-conjugated streptavidin and substrate were used for detection. The blot images were captured using a quantitative Western imaging system (Aplegen’s Omega Lum C).
CIA Induction and in Vivo AhR Agonist Administration.
To induce autoimmune arthritis in the HLA-DR mouse strains (B10.M background), 8- to 10-wk-old mice were immunized s.c. at the base of the tail with 100 µg of bovine CII emulsified in complete Freund’s adjuvant (CFA) containing 4 mg/mL of heat-killed Mycobacterium (H37Ra, Difco). Starting at day 18 postimmunization, mice were examined three times per week and the presence of arthritic limbs, the number of affected limbs, and the severity of the arthritis were assessed. Severity of disease was evaluated by visual inspection and each limb was assigned a score using a scale of 0–4 as described (55). TCDD (0.3 ng/gm body weight) or FICZ (28 ng/gm body weight) were administered in 50 µL of DMSO once per week i.p. Control mice received equal volume of the DMSO vehicle only. Arthritis severity was determined as previously described (20).
Joint Tissue Studies.
Joint tissue studies were performed as previously described (20, 23). For details see SI Materials and Methods.
Radiological Imaging.
Radiological imaging was performed as previously described (20, 23). For details see SI Materials and Methods.
Statistical Analysis.
Student’s t test, log-ranked (Mantel–Cox) test, or two-way ANOVA test using Prism 6.0 (GraphPad) software was used. In Fig. 1 C–F, statistical analyses of synergism were performed using a two-way ANOVA by SAS software (version 9.4). A fixed-effect, general linear model was created in SAS for the interactions of the independent variables of AhR agonist (FICZ or TCDD) concentrations and either SE genotype or soluble SE-ligand concentration and for their effect on the number of TRAP+ cells. An F test was conducted for each model to determine the presence of a statistically significant synergistic effect as indicated by the probability of observing an F value greater than or equal to the f statistic of the interaction term under the null hypothesis of an F(DFinteraction, DF error) distribution. Under the null hypothesis, there is no interaction between the independent variables. An f statistic is defined as . The mean squares error is the variance of data not explained by the model and is defined as . An α = 0.05 was selected for statistical significance. P values <0.05 were considered significant; P values >0.05 were considered nonsignificant (NS). All flow cytometry data were analyzed with FlowJo (TreeStar). No statistical methods were used to predetermine sample size.
Supplementary Material
Acknowledgments
We thank Prashasinka Gehlot, Emily Henson, and Ying Liu for technical assistance. This work was supported by the National Institute of Environmental Health Sciences (Grant R21ES024428), the National Institute of General Medical Sciences (Grant R01GM088560), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (Grants R01AR059085, R61AR073014, and T32AR07080). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest statement: J.H. and S.L. are named inventors on technology owned by the Regents of The University of Michigan, which is licensed to Cadila Healthcare Limited.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722124115/-/DCSupplemental.
References
- 1.Selmi C, et al. Mechanisms of environmental influence on human autoimmunity: A National Institute of Environmental Health Sciences expert panel workshop. J Autoimmun. 2012;39:272–284. doi: 10.1016/j.jaut.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Miller FW, et al. Epidemiology of environmental exposures and human autoimmune diseases: Findings from a National Institute of Environmental Health Sciences expert panel workshop. J Autoimmun. 2012;39:259–271. doi: 10.1016/j.jaut.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eringsmark Regnéll S, Lernmark A. The environment and the origins of islet autoimmunity and type 1 diabetes. Diabet Med. 2013;30:155–160. doi: 10.1111/dme.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 5.Karlson EW, Deane K. Environmental and gene-environment interactions and risk of rheumatoid arthritis. Rheum Dis Clin North Am. 2012;38:405–426. doi: 10.1016/j.rdc.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svendsen AJ, et al. On the origin of rheumatoid arthritis: The impact of environment and genes–A population based twin study. PLoS One. 2013;8:e57304. doi: 10.1371/journal.pone.0057304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 8.Ling S, Lai A, Borschukova O, Pumpens P, Holoshitz J. Activation of nitric oxide signaling by the rheumatoid arthritis shared epitope. Arthritis Rheum. 2006;54:3423–3432. doi: 10.1002/art.22178. [DOI] [PubMed] [Google Scholar]
- 9.Ling S, et al. The rheumatoid arthritis shared epitope increases cellular susceptibility to oxidative stress by antagonizing an adenosine-mediated anti-oxidative pathway. Arthritis Res Ther. 2007;9:R5. doi: 10.1186/ar2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holoshitz J, Ling S. Nitric oxide signaling triggered by the rheumatoid arthritis shared epitope: A new paradigm for MHC disease association? Ann N Y Acad Sci. 2007;1110:73–83. doi: 10.1196/annals.1423.009. [DOI] [PubMed] [Google Scholar]
- 11.Ling S, Pi X, Holoshitz J. The rheumatoid arthritis shared epitope triggers innate immune signaling via cell surface calreticulin. J Immunol. 2007;179:6359–6367. doi: 10.4049/jimmunol.179.9.6359. [DOI] [PubMed] [Google Scholar]
- 12.De Almeida DE, et al. Immune dysregulation by the rheumatoid arthritis shared epitope. J Immunol. 2010;185:1927–1934. doi: 10.4049/jimmunol.0904002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holoshitz J, De Almeida DE, Ling S. A role for calreticulin in the pathogenesis of rheumatoid arthritis. Ann N Y Acad Sci. 2010;1209:91–98. doi: 10.1111/j.1749-6632.2010.05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling S, Cheng A, Pumpens P, Michalak M, Holoshitz J. Identification of the rheumatoid arthritis shared epitope binding site on calreticulin. PLoS One. 2010;5:e11703. doi: 10.1371/journal.pone.0011703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Almeida DE, Ling S, Holoshitz J. New insights into the functional role of the rheumatoid arthritis shared epitope. FEBS Lett. 2011;585:3619–3626. doi: 10.1016/j.febslet.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Almeida DE, Holoshitz J. MHC molecules in health and disease: At the cusp of a paradigm shift. Self Nonself. 2011;2:43–48. doi: 10.4161/self.2.1.15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holoshitz J, et al. An HLA-DRB1-coded signal transduction ligand facilitates inflammatory arthritis: A new mechanism of autoimmunity. J Immunol. 2013;190:48–57. doi: 10.4049/jimmunol.1202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling S, Cline EN, Haug TS, Fox DA, Holoshitz J. Citrullinated calreticulin potentiates rheumatoid arthritis shared epitope signaling. Arthritis Rheum. 2013;65:618–626. doi: 10.1002/art.37814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naveh S, et al. Developing potent backbone cyclic peptides bearing the shared epitope sequence as rheumatoid arthritis drug-leads. Bioorg Med Chem Lett. 2012;22:493–496. doi: 10.1016/j.bmcl.2011.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu J, et al. A small shared epitope-mimetic compound potently accelerates osteoclast-mediated bone damage in autoimmune arthritis. J Immunol. 2013;191:2096–2103. doi: 10.4049/jimmunol.1203231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco LP, Plegue M, Fung-Leung W-P, Holoshitz J. Gender-biased regulation of human IL-17-producing cells in vitro by peptides corresponding to distinct HLA-DRB1 allele-coded sequences. J Immune Based Ther Vaccines Antimicrob. 2013;2:29–38. doi: 10.4236/jibtva.2013.23004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holoshitz J. The quest for better understanding of HLA-disease association: Scenes from a road less travelled by. Discov Med. 2013;16:93–101. [PMC free article] [PubMed] [Google Scholar]
- 23.Ling S, et al. Shared epitope-antagonistic ligands: A new therapeutic strategy in mice with erosive arthritis. Arthritis Rheumatol. 2015;67:2061–2070. doi: 10.1002/art.39158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi S, et al. A role for the aryl hydrocarbon receptor and the dioxin TCDD in rheumatoid arthritis. Rheumatology (Oxford) 2008;47:1317–1322. doi: 10.1093/rheumatology/ken259. [DOI] [PubMed] [Google Scholar]
- 25.Heliövaara M, Aho K, Aromaa A, Knekt P, Reunanen A. Smoking and risk of rheumatoid arthritis. J Rheumatol. 1993;20:1830–1835. [PubMed] [Google Scholar]
- 26.Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 2004;50:3085–3092. doi: 10.1002/art.20553. [DOI] [PubMed] [Google Scholar]
- 27.Karlson EW, et al. Gene-environment interaction between HLA-DRB1 shared epitope and heavy cigarette smoking in predicting incident rheumatoid arthritis. Ann Rheum Dis. 2010;69:54–60. doi: 10.1136/ard.2008.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Iqbal J, et al. Smoke carcinogens cause bone loss through the aryl hydrocarbon receptor and induction of Cyp1 enzymes. Proc Natl Acad Sci USA. 2013;110:11115–11120. doi: 10.1073/pnas.1220919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 31.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 32.Ishimaru N, et al. Neonatal exposure to low-dose 2,3,7,8-tetrachlorodibenzo-p-dioxin causes autoimmunity due to the disruption of T cell tolerance. J Immunol. 2009;182:6576–6586. doi: 10.4049/jimmunol.0802289. [DOI] [PubMed] [Google Scholar]
- 33.de Punder YM, van Riel PL. Rheumatoid arthritis: Understanding joint damage and physical disability in RA. Nat Rev Rheumatol. 2011;7:260–261. doi: 10.1038/nrrheum.2011.49. [DOI] [PubMed] [Google Scholar]
- 34.Taneja V, et al. Delineating the role of the HLA-DR4 “shared epitope” in susceptibility versus resistance to develop arthritis. J Immunol. 2008;181:2869–2877. doi: 10.4049/jimmunol.181.4.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosloniec EF, et al. Induction of autoimmune arthritis in HLA-DR4 (DRB1*0401) transgenic mice by immunization with human and bovine type II collagen. J Immunol. 1998;160:2573–2578. [PubMed] [Google Scholar]
- 36.Rosloniec EF, et al. An HLA-DR1 transgene confers susceptibility to collagen-induced arthritis elicited with human type II collagen. J Exp Med. 1997;185:1113–1122. doi: 10.1084/jem.185.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W, et al. C/EBPα regulates osteoclast lineage commitment. Proc Natl Acad Sci USA. 2013;110:7294–7299. doi: 10.1073/pnas.1211383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison NA, Day CJ, Nicholson GC. Dominant negative MCP-1 blocks human osteoclast differentiation. J Cell Biochem. 2014;115:303–312. doi: 10.1002/jcb.24663. [DOI] [PubMed] [Google Scholar]
- 40.Zheng W, et al. Role of osteopontin in induction of monocyte chemoattractant protein 1 and macrophage inflammatory protein 1beta through the NF-kappaB and MAPK pathways in rheumatoid arthritis. Arthritis Rheum. 2009;60:1957–1965. doi: 10.1002/art.24625. [DOI] [PubMed] [Google Scholar]
- 41.Otero JE, Chen T, Zhang K, Abu-Amer Y. Constitutively active canonical NF-κB pathway induces severe bone loss in mice. PLoS One. 2012;7:e38694. doi: 10.1371/journal.pone.0038694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Z, Boyle DL, Manning AM, Firestein GS. AP-1 and NF-kappaB regulation in rheumatoid arthritis and murine collagen-induced arthritis. Autoimmunity. 1998;28:197–208. doi: 10.3109/08916939808995367. [DOI] [PubMed] [Google Scholar]
- 43.Criswell LA. Gene discovery in rheumatoid arthritis highlights the CD40/NF-kappaB signaling pathway in disease pathogenesis. Immunol Rev. 2010;233:55–61. doi: 10.1111/j.0105-2896.2009.00862.x. [DOI] [PubMed] [Google Scholar]
- 44.Makrygiannakis D, et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann Rheum Dis. 2008;67:1488–1492. doi: 10.1136/ard.2007.075192. [DOI] [PubMed] [Google Scholar]
- 45.Bidkar M, et al. Cigarette smoke induces immune responses to Vimentin in both, arthritis-susceptible and -resistant humanized mice. PLoS One. 2016;11:e0162341. doi: 10.1371/journal.pone.0162341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaakkola JJ, Gissler M. Maternal smoking in pregnancy as a determinant of rheumatoid arthritis and other inflammatory polyarthropathies during the first 7 years of life. Int J Epidemiol. 2005;34:664–671. doi: 10.1093/ije/dyi006. [DOI] [PubMed] [Google Scholar]
- 47.Yu H, et al. The aryl hydrocarbon receptor suppresses osteoblast proliferation and differentiation through the activation of the ERK signaling pathway. Toxicol Appl Pharmacol. 2014;280:502–510. doi: 10.1016/j.taap.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 48.Lahoti TS, et al. Aryl hydrocarbon receptor antagonism attenuates growth factor expression, proliferation, and migration in fibroblast-like synoviocytes from patients with rheumatoid arthritis. J Pharmacol Exp Ther. 2014;348:236–245. doi: 10.1124/jpet.113.209726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazantseva MG, Highton J, Stamp LK, Hessian PA. Dendritic cells provide a potential link between smoking and inflammation in rheumatoid arthritis. Arthritis Res Ther. 2012;14:R208. doi: 10.1186/ar4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogando J, et al. Notch-regulated miR-223 targets the aryl hydrocarbon receptor pathway and increases cytokine production in macrophages from rheumatoid arthritis patients. Sci Rep. 2016;6:20223. doi: 10.1038/srep20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu TY, Pang WJ, Yang GS. Aryl hydrocarbon receptors in osteoclast lineage cells are a negative regulator of bone mass. PLoS One. 2015;10:e0117112. doi: 10.1371/journal.pone.0117112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen NT, et al. Aryl hydrocarbon receptor antagonism and its role in rheumatoid arthritis. J Exp Pharmacol. 2015;7:29–35. doi: 10.2147/JEP.S63549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyce BF, Xiu Y, Li J, Xing L, Yao Z. NF-κB-mediated regulation of osteoclastogenesis. Endocrinol Metab (Seoul) 2015;30:35–44. doi: 10.3803/EnM.2015.30.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel CF, et al. Cross-talk between aryl hydrocarbon receptor and the inflammatory response: A role for nuclear factor-κB. J Biol Chem. 2014;289:1866–1875. doi: 10.1074/jbc.M113.505578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosloniec EF, Cremer M, Kang AH, Myers LK, Brand DD. Collagen-induced arthritis. Curr Protoc Immunol. 2010;89:15.5.1–15.5.25. doi: 10.1002/0471142735.im1505s89. [DOI] [PubMed] [Google Scholar]