Significance
Amyotrophic lateral sclerosis (ALS) is an invariably fatal neurodegenerative disease. Autosomal dominant mutations in the SOD1 gene are responsible for 12% of familial ALS cases and 1.5% of sporadic cases. However, it remains unknown whether the large fibrillar aggregates formed by misfolded SOD1 are a causative agent in disease progression. By designing mutations that specifically stabilize SOD1 fibrils or nonnative oligomers, we found that the assembly into insoluble fibrils mitigated the neurotoxic effects caused by aberrant conformation of trimers. We also demonstrated with electron microscopy that toxic SOD1 trimers displayed heterogeneous structures, in concert with computational studies. Our findings suggest a protective role of fibrils and a plausible pharmaceutical strategy, as promoting SOD1 fibrillogenesis reduces the population of neurotoxic species.
Keywords: ALS, SOD1, neurotoxicity, oligomer, fibril
Abstract
Aberrant accumulation of misfolded Cu, Zn superoxide dismutase (SOD1) is a hallmark of SOD1-associated amyotrophic lateral sclerosis (ALS), an invariably fatal neurodegenerative disease. While recent discovery of nonnative trimeric SOD1-associated neurotoxicity has suggested a potential pathway for motor neuron impairment, it is yet unknown whether large, insoluble aggregates are cytotoxic. Here we designed SOD1 mutations that specifically stabilize either the fibrillar form or the trimeric state of SOD1. The designed mutants display elevated populations of fibrils or trimers correspondingly, as demonstrated by gel filtration chromatography and electron microscopy. The trimer-stabilizing mutant, G147P, promoted cell death, even more potently in comparison with the aggressive ALS-associated mutants A4V and G93A. In contrast, the fibril-stabilizing mutants, N53I and D101I, positively impacted the survival of motor neuron-like cells. Hence, we conclude the SOD1 oligomer and not the mature form of aggregated fibril is critical for the neurotoxic effects in the model of ALS. The formation of large aggregates is in competition with trimer formation, suggesting that aggregation may be a protective mechanism against formation of toxic oligomeric intermediates.
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by progressive muscle weakness and eventual paralysis (1, 2). Misfolding and aggregation of the cytosolic antioxidant enzyme Cu, Zn superoxide dismutase (SOD1) represent common factors for both familial and sporadic cases of ALS (3–5). The formation of SOD1 oligomers and aggregates is critically dependent on the dissociation of the native dimer into monomers, followed by metal loss, reduction of intramolecular disulfide bond, and monomer misfolding (6, 7). By perturbing (stabilizing or destabilizing) a series of sparsely populated conformational states along the SOD1 misfolding pathway, researchers have gained valuable insight into the nonnative SOD1 conformers and subsequent cytotoxicity. For instance, a phosphomimetic mutation results in increased thermodynamic stability of the SOD1 native dimer and positively impacts motor neuron survival in a model of ALS (8). In contrast, Proctor et al. (9) identified trimer-stabilizing SOD1 mutants and demonstrated that the elevated population of SOD1 trimers promoted cell death. For the fibrillar form of SOD1 aggregates, studies revealed that the intrinsic rate of SOD1 amyloidogenesis was increased by certain ALS-linked mutations and that the fibrillization rate might explain the speed of disease progression (10). The perturbations introduced through various mutations, however, have an impact on the disparate nonnative states of SOD1 (monomers, oligomers, and fibrils). For example, inhibiting trimer formation may simultaneously increase the population of native dimers, misfolded monomers, or large aggregates. More importantly, we do not have complete knowledge of the impact of individual species appearing during the aggregation pathway on motor neuron viability. Uncovering roles of the dominant species appearing during aggregation is, therefore, key to untangling origins of motor neuron death, which is the key mystery in ALS.
It is debated whether the large, insoluble aggregates represent a cause of pathogenesis in neurodegenerative disorders, or whether the nonnative oligomers correlate with processes leading to cytotoxicity and neuron death (11, 12). The discovery of aberrant protein deposits, including amyloid-β (Aβ)1–40 and Aβ1–42 in Alzheimer’s disease (13), α-synuclein in Parkinson’s disease (14), and SOD1 in ALS (5), has been critical to the development of the amyloid cascade hypothesis, which emphasizes the aberrant accumulation of amyloid fibrils as a major cause of neuron impairment (15). The corresponding pharmaceutical strategy is to inhibit the formation of amyloid fibrils or remove the plaque in patients’ brain. However, recent clinical trials of Aβ-specific antibodies have failed to show improvements for Alzheimer’s disease patients’ dementia and may even worsen symptoms, despite the common success in reducing the Aβ-plaque load (16). These observations bring into question the pathological role of protein fibrils, and suggest a plausible role of transient oligomeric species in induced cytotoxicity. Indeed, studies on prion proteins revealed that the fibrils confer protection against ischemic damage in an acute stroke model (17). Similarly, propagating Aβ-fibrils exhibited antibiosis functions in mouse and worm models of Alzheimer’s disease (18). In the motor neuron disease ALS, recent studies suggest that nonnative SOD1 trimers are the cytotoxic species implicated in motor neuron death and disease progression (9, 19, 20). However, it is currently unknown whether large fibrillar aggregates are cytotoxic to motor neurons. This question is critical, as its answer has broad implications not only for understanding the etiology of ALS but also for that of many other neurodegenerative diseases where protein aggregation has been the key signature. If both the oligomers and large aggregates are cytotoxic, this fact would suggest multiple pathways to pathological development, potentially requiring untangling the roles of oligomers and fibrils independently. If the large aggregates are not cytotoxic, their formation would compete with the formation of small oligomers, suggesting they are protective to cells. Hence here we examined whether the large, matured form of SOD1 aggregates promoted cell death in a model of motor neurons.
We addressed this question through a combination of biophysical techniques. We designed mutations that specifically stabilize the fibrillar form or the trimeric state of SOD1, which promoted formation of longer fibrils or the population of oligomers, respectively. We applied size-exclusion chromatography (SEC) and electron microscopy (EM) approaches spanning a wide range of sample sizes and molecular weights to characterize the population distribution of native dimers, oligomers, and fibrils. The impact of the designed mutants on cellular viability was compared with that of wild-type protein (WT-SOD1) and two aggressive ALS-associated mutants (A4V- and G93A-SOD1). We showed that SOD1 mutants designed to promote SOD1 fibrils alleviated neurotoxicity. In contrast, the increasing population of trimers correlated with a higher death ratio in a cell model of ALS. These observations support an emerging hypothesis that the SOD1 trimer, and not the mature fibril, exerts the neurotoxic effects in motor neurons.
Results
We chose three mutants of SOD1 (N53I, D101I, and G147P) proposed by Proctor et al. (9) for their effect on trimer stability. To estimate the impact of these mutations on SOD1 fibrillar aggregates, we adopted the crystal structures (for the D101 and G147 positions) and computational model (for the G147 position) of SOD1 fibrils, recently determined by the Eisenberg group (21, 22), and calculated the change of free energies upon mutation using the molecular design suite Eris (SI Appendix, Fig. S1) (23, 24). Mutations N53I and D101I destabilize the trimer and promote the population of large aggregates, while G147P stabilizes the trimer. A4V and G93A, as the positive controls for ALS-relevant neurotoxicity, exacerbate SOD1 oligomerization as well as the exposure of a toxicity-associated epitope (20). In previous studies, the stability and morphology of the SOD1 trimer were examined in an acidic and demetalating condition (pH 3.5 and 10 mM EDTA), which accelerates the misfolding process (7, 9). We first examined our samples under the same condition with physiological concentration and temperature (30 μM and 37 °C). SOD1 trimers accumulated within the 2- to 24-h time window and then evolved to energetically favorable states of monomers and large aggregates (SI Appendix, Fig. S2). The population of native dimers significantly decreased as a consequence of low pH and loss of metals. Upon extended incubation, misfolded monomers and aggregates became the dominant species. In agreement with previous findings, our results suggest the monomeric species forms the available pool for further oligomerization and fibrillation of SOD1, and that the trimeric species represents an intermediate, metastable state (9, 25).
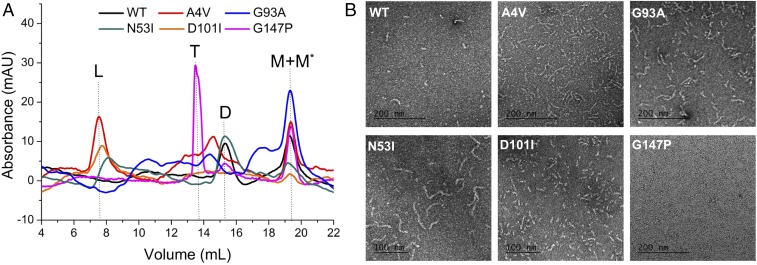
To gain a better understanding of SOD misfolding under physiological conditions, we separated the monomeric species of each mutant and reequilibrated the protein solution to neutral pH (20 mM Tris, 150 mM NaCl, pH 7.4). The misfolded SOD1 monomer (M* in Fig. 1), as reported previously, represents the precursor of nonnative SOD1 oligomers and fibrils. We used analytical SEC to resolve the distribution of each conformational state (Fig. 1A). The behavior of WT-SOD1 and five SOD1 mutants fell into four categories: (i) N53I-SOD1 formed large aggregates, as well as a population of dimers comparable to that of WT-SOD1; (ii) A4V- and G93A-SOD1 contained a significant amount of both large aggregates and trimers; (iii) the majority of D101I-SOD1 formed large aggregates, with little population of trimers and dimers; and (iv) the majority of G147P-SOD1 formed trimers. We quantified the amounts of each individual species by separating them through SEC (Fig. 2A). The size of SOD1 fibrils, however, fell well beyond the detection limitation of SEC. Thus, the presence or morphology of this insoluble form could not be readily detected.
Fig. 1.
SOD1 mutants designed to stabilize fibrils (N53I and D101I) and trimers (G147P) vary in the distribution of oligomeric states (D, dimers; L, large aggregates; M, monomers; M*, misfolded monomers; T, trimers). (A) Analytical size-exclusion chromatograms resolved the populations of each species. Samples were taken after incubation at physiological conditions (30 μM SOD1 in 20 mM Tris, 150 mM NaCl, pH 7.4) for 2 d. G147P-SOD1 exhibited an elevated population of trimers among all SOD1 mutants. (B) Electron microscopy negative-stain images presented the fibrillar form of SOD1. The samples were incubated at the same condition as in SEC. N53I- and D101I-SOD1 formed more fibrils in comparison with G147P-SOD1.
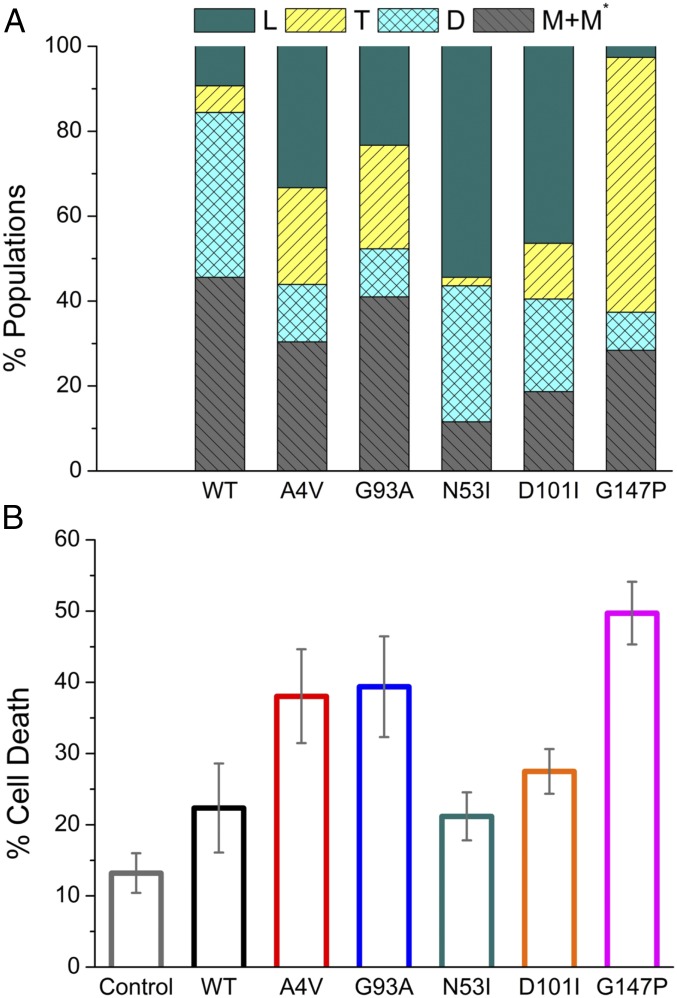
Fig. 2.
Elevated population of trimers is correlated with a higher death ratio in NSC-34 cells. (A) Distribution of native (D) and nonnative species (L, T, M, and M* as defined in Fig. 1) in WT-SOD1 and five mutants. Different oligomeric states were separated by size-exclusion chromatography, followed by measurement of their concentrations using UV absorption and BCA assays (Materials and Methods). (B) Average cell-death ratio measured by the percentage of SYTOX Green-stained cells within the SOD1-expressing cells (Fig. 4). Control: cells with mCherry expression only. Error bars represent the SEM; n = 3.
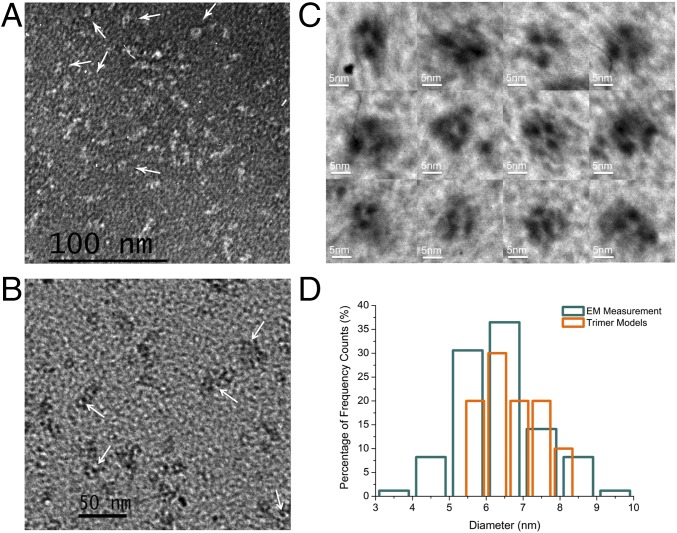
To overcome the barrier of SEC detection limits, we used EM to analyze the fibrils formed by each mutant (Fig. 1B and SI Appendix, Fig. S3). Among them, N53I-SOD1 formed the longest fibril [average length 87 ± 2.2 nm, number of fibrils or particles surveyed (n) = 108]. D101I-, A4V-, and G93A-SOD1 samples contained shorter fibrils, with average lengths of 23 ± 1.7, 43 ± 2.5, and 51 ± 2.8 nm, respectively (n = 156, 140, and 112). In comparison, G147P-SOD1 did not form significant amounts of fibrils or other forms of large aggregates. Interestingly, a careful examination of D101I- and G147P-SOD1 samples revealed ring-like structures in the background (Fig. 3 A and B, white arrows). The diameters of the rings (6.37 ± 1.23 nm, n = 15) are comparable to that of the SOD1 trimer (oligomer volume 71.7 nm3 and average diameter 5.2 nm) (9), suggesting they are possibly trimers detected by negative-stain EM. To better distinguish the trimer-like particles from the background, we applied metal-shadowing EM with the same samples (Fig. 3C). The EM images illustrated structural heterogeneity of possible SOD1 trimers with a mean diameter of 6.27 ± 1.11 nm (n = 85), which is close to a measurement on computationally derived trimer models (6.65 ± 0.70 nm, n = 10; Fig. 3D and SI Appendix, Fig. S4). The EM characterization of oligomeric SOD1 corroborates the previous study using molecular dynamics simulation and structural modeling (9), which together suggest that nonnative SOD1 trimers are a highly dynamic ensemble of diverse structures (SI Appendix, Fig. S5). We observed more of these rings in G147P-SOD1 in comparison with D101I-, N53I-, or WT-SOD1 samples (SI Appendix, Fig. S3). Hence, the majority of N53I-SOD1 formed long fibrils, while the majority of G147P-SOD1 formed oligomers (possibly trimers). Overall, the results obtained by EM are in full agreement with the quantitative measurement made by SEC.
Fig. 3.
Structures of nonnative SOD1 oligomers revealed by EM. (A) Negative-stain images of D101I-SOD1. (B) Negative-stain images of G147P-SOD1. The white arrows indicate the presence of rings with a diameter (6.3 nm) comparable to that of the SOD1 trimer (5.2 nm). (C) Metal-shadowing images of SOD1 trimer-like structures in D101I- and G147P-SOD1. (D) The distribution of SOD1-trimer diameters. Eighty-five trimer-like particles in the EM images and 10 in silico trimer models were included in the calculation of diameters. Cyan: trimer-like particle in EM (Fig. 3C); orange: trimer structural models (SI Appendix, Fig. S4).
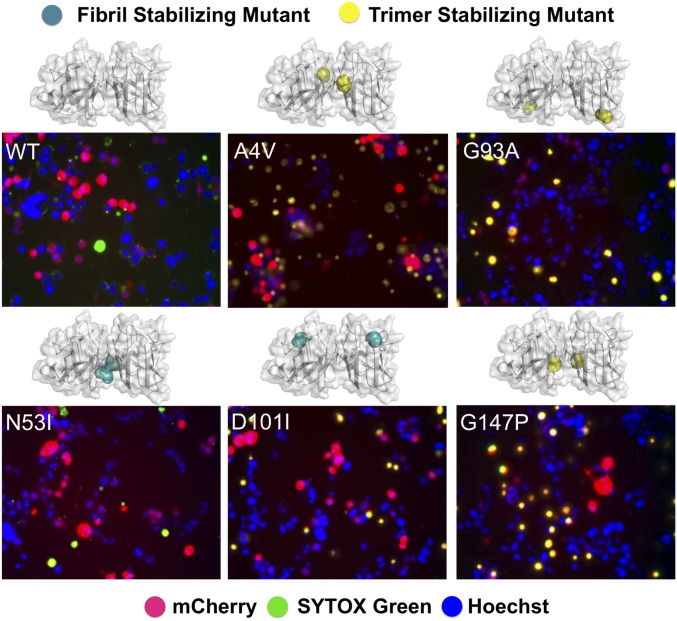
To gain insights into the physiological relevance of the designed SOD1 mutants in a cellular environment, we tested their toxicity in motor neuron-like cells (neuroblastoma spinal cord hybrid cell line; NSC-34) (26, 27). A4V- and G93A-SOD1 were used as positive controls for ALS-relevant neurotoxicity. First, we determined the suitable conditions for NSC-34 cell differentiation and the optimal time point for measurement of cell viability. Using immunofluorescence microscopy, we demonstrated that NSC-34 cells expressed the neuron-specific marker β-III-tubulin 2 d after differentiation (SI Appendix, Fig. S6). Expression of SOD1 proteins was accomplished by transfection of SOD1-mCherry expression constructs (SOD1 and mCherry genes expressed separately). The red fluorescence from mCherry proteins indicated successful transfection and expression of our vectors. The death ratio of differentiated NSC-34 cells was quantified by calibrating SYTOX Green-stained cells (green fluorescence) among mCherry-expressing cells (red fluorescence) (Fig. 4). N53I-, D101I-, and G147P-SOD1 exerted increasing neurotoxicity on NSC-34 cells (Fig. 2B; mean cell death: 21, 27, and 49%, respectively). Expression of G147P-SOD1 led to more cell death than the aggressive ALS mutants A4V and G93A (mean cell death 38 and 39%, respectively), possibly due to a larger portion of trimer populations seen in G147P-SOD1, while the presence of fibrils in A4V- and G93A-SOD1 reduced the available pool for formation of toxic species. In contrast, the expression of N53I-SOD1 resulted in reduced cell death of NSC-34 cells, likely because N53I-SOD1 formed the longest fibrils among all mutants characterized in this study (the reduced toxicity of N53I-SOD1 has also been reported by Proctor et al. in ref. 9). Importantly, we compared the cell death observed with the various SOD1 mutants with the propensity of these mutants to form the various oligomeric species. The correlation between cell death and increasing amount of SOD1 trimer (or decreasing amount of SOD1 fibrils) suggests that the SOD1 trimer, instead of the insoluble form of fibrillar aggregates, mediates the neurotoxic effects in the cell model of ALS.
Fig. 4.
Fibril-stabilizing mutants (N53I and D101I) exerted reduced neurotoxicity compared with the trimer-stabilizing mutant (G147P) in NSC-34 cells. A4V and G93A are positive controls for ALS-relevant cell death. The cell viabilities were measured 2 d after differentiation (blue: Hoechst stain; red: mCherry fluorescence; green: SYTOX Green stain; yellow: overlap between red and green staining, showing transfected cells that are undergoing death). The point mutations are indicated as spheres in the SOD1 structural model.
Discussion
SOD1 represents a major cause of familial ALS, and is involved in sporadic presentations (3, 28). However, a recent test for the presence of misfolded SOD1 deposits suggested they were below the detection limits of seven conformationally sensitive antibodies in spinal cord and cortex tissues from sporadic ALS patients (29). Hence, large aggregates of SOD1 may not be responsible for the neurotoxicity in the majority of ALS cases (sporadic ALS). Instead, our results suggested that the SOD1 trimer, which may escape immunohistochemistry detection due to its small size and transient nature, was a primary origin of motor neuron impairment. Furthermore, research with chimeric mouse embryos expressing SOD1-GFP fusion proteins unveiled that pathogenic cytosolic SOD1 could transfer between spinal cord motor neurons (30). We posit that in comparison with insoluble fibrils, SOD1 trimers can translocate between cells more readily. This translocation of misfolded oligomers could help the spreading of cytotoxicity and disease states. In accordance with previous work, our study indicates that the formation of nonnative oligomers leads to cell death.
Most familial SOD1 mutations decrease SOD1 stability (31). The oxidative posttranslational modification glutathionylation also results in dramatic impairment of SOD1 native structure via altering the rate of monomer association (20, 32), suggesting a critical role of oxidative stress in disease etiology. A comprehensive inspection of different structural species and their global effects on cellular viability could help resolve the toxic pathways leading to cell death (7). The current study suggests not only that dimer destabilization is associated with disease etiology but also that the stabilization of the nonnative trimeric SOD1 strongly affects motor neuron survival. Several viable pharmaceutical strategies can be potentially pursued based on our findings: (i) identifying small molecules that strengthen the dimer interface and prevent dimer dissociation, thus reducing the monomeric precursors of toxic species (8); (ii) identifying small molecules that convert toxic oligomers to nontoxic fibrils. As demonstrated in an Alzheimer’s disease model, acceleration of Aβ-fibrillogenesis could effectively reduce Aβ42-associated toxicity (33); and (iii) identifying small molecules that destabilize the SOD1 trimer directly.
The oligomeric form of SOD1 may consist of a series of sparsely presented and transient conformers, including trimers and higher-order oligomers. For instance, an 11-residue segment constituting antiparallel, out-of-register β-strands was identified as an oligomeric form of SOD1 aggregation (34). Based on its structure, the authors proposed a model of toxic SOD1 oligomers formed by 16 protomers. The study of NMR to probe the free-energy landscape of misfolded SOD1 indicated that different ALS-associated mutants could access significant varying excited states and that these conformational states were interconvertible (35, 36). In contrast, the ground, native state of SOD1 was structurally stable. In the current study, the biochemical assays (SEC and EM) revealed a clearly recognizable trimeric state among all oligomeric states, which correlated with the degeneration of motor neuron-like cells. Nonetheless, the high–molecular-weight region in size-exclusion chromatograms may contain lower populations of other misfolded forms. A plausible mechanism for SOD1-mediated ALS etiology is that different oligomers represent the divergent stages of misfolding and evolution of neurotoxic species, which present a synergy effect of interrupting the cellular machinery. To summarize our current findings, we conclude that the small oligomers of SOD1 exert neurotoxic effects that are mitigated by forming large, insoluble SOD1 assemblies.
Materials and Methods
Computation.
Designing mutations that perturb the stabilities of SOD1 aggregates was accomplished by Eris, an algorithm that performs automated side-chain repacking and backbone relaxation and calculates the changes in free energy upon mutation (eris.dokhlab.org) (23, 24). The input structural models were crystal structures [Protein Data Bank (PDB) ID code 4NIN, 101-DSVISLS-107; PDB ID code 4NIO, 147-GVTGIAQ-153] (21). For N53I, a model was built by the ZipperDB web server (https://services.mbi.ucla.edu/zipperdb/) using the SOD1 sequence as query input (22). The trimer models were described previously (9). The detailed procedures for Eris calculation were described previously (37). For mutations requiring significant backbone adjustment (e.g., small to large residues, Gly-to-Pro mutations), the flexible backbone mode was adopted in Eris, followed by molecular dynamic simulations as described previously (8).
Cloning, Expression, and Purification of SOD1.
The SOD1 mutant vectors (N53I-SOD1, D101I-SOD1, G147P-SOD1, A4V-SOD1, and G93A-SOD1 in the yeast expression vector YEp-351) were produced with a Q5 Site-Directed Mutagenesis Kit (NEB) following the provided protocol. Expression, purification, and separation of modified populations were carried out as described previously (19). Briefly, human SOD1 was expressed in the EG118 yeast strain at 30 °C. After treatment with ethanol/chloroform, the fractions containing SOD1 proteins were enriched by ammonium sulfate precipitation. Hydrophobic (phenyl Sepharose column; GE Healthcare) and ion-exchange (monoQ column; GE Healthcare) chromatography were then applied to isolate the SOD1 proteins.
Size-Exclusion Chromatography.
The SOD1 samples were dialyzed against a denaturing buffer (50 mM acetate/sodium acetate, 150 mM NaCl, 10 mM EDTA, pH 3.5) or physiological-condition buffer (20 mM Tris, 150 mM NaCl, pH 7.4). After equilibration overnight, the proteins solutions were diluted to 30 μM using the same dialysis buffer and then incubated at 37 °C. Typically the samples were incubated in denaturing buffer for 12 h and then in physiological-condition buffer for 2 d. After incubation, the samples were analyzed using a Superdex 75 (denaturing buffer) or 200 (for the physiological condition) 10/300 GL column (GE Healthcare).
Redler et al. (20) have previously performed dynamic light scattering and correlated the elution volume on a Superdex 200 10/300 GL column to the molecular weight of SOD1: 11.5 mL, hexamer; 12.2 mL, tetramer; 13.4 mL, trimer; 15.5 mL, dimer; and 17.6 mL, monomer (void volume 8 mL).
To determine the approximate amount of each species, the data curves of size-exclusion chromatograms were deconvoluted as described previously (19). The integrated area of each peak corresponds to the amount of a specific species (trimer, dimer, monomer, etc.). The concentration of each separated peak was also measured by bicinchoninic acid assay (BCA Assay Kit; Thermo Fisher Scientific). The large aggregates include the peak eluted at the void volume and the samples recovered from the syringe filter (0.22 μm).
Electron Microscopy.
Samples to be visualized by negative staining were adsorbed for 3 min to thin glow discharge-treated carbon foils suspended over 400-mesh copper grids. The samples were at a concentration of 10 μg/mL in a buffer of 50 mM acetate/sodium acetate, 150 mM NaCl, and 10 mM EDTA (pH 3.5). The grids were then stained with 2% uranyl acetate for 5 min and air-dried. The samples were imaged in an FEI Tecnai T12 instrument at 80 kV. Images were collected using a Gatan Orius real-time 2Kx2K CCD camera. Images for publication were arranged and contrast-adjusted using Adobe Photoshop. For tungsten metal shadow casting, the samples were applied to the thin carbon supports as above, and then washed in a water-ethanol series and air-dried followed by evaporation of tungsten metal with rotation at a vacuum of 1 × 10−6 torr. Imaging in the T12 instrument was at 40 kV.
SOD1 Plasmids for Cell Assays.
The mammalian expression vector (pCI-neo) contains mCherry genes for fluorescence measurements. The SOD1 gene and mCherry gene are insulated with an internal ribosome entry site and give red fluorescence when transfected.
Cell Culture, Transfection, Differentiation, and Cell-Viability Assay.
Neuroblastoma spinal cord cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (P/S). Cells were passaged every 2 to 3 d. To determine cell viability after introduction of the SOD1 mutant plasmids, NSC-34 cells were plated at a concentration of 3 × 105 cells per well in six-well plates. The following day, cells were transfected with 2.5 µg of the mCherry-SOD1 plasmids using Lipofectamine 2000 (Invitrogen). Five hours later, the media were replaced with differentiation media: DMEM, 1% FBS, 1% P/S, 1% nonessential amino acids, and 10 µM all-trans retinoic acid. Adequate differentiation was determined before cell-viability experiments by performing immunofluorescence with the neuronal specific antibody β-III-tubulin. Two days after transfection and differentiation, cells were stained with a 1:30,000 dilution of SYTOX Green (Thermo Fisher Scientific) to assess dead cells and a 1:10,000 dilution of Hoechst 33342 (Invitrogen) to visualize all cells present. Fluorescent images were captured in red (transfected SOD1 plasmid), blue (Hoechst 33342), and green (SYTOX Green) channels. Images were then overlaid in ImageJ (NIH) for analysis. The number of green cells (dead) was counted within a population of 100 red cells (transfected with the SOD1 plasmid) to determine the cell-death ratio among cells containing the SOD1 plasmids.
Supplementary Material
Acknowledgments
We thank Dr. Elizabeth A. Proctor, Dr. Yazhong Tao, Kasey Skinner, Moza Hamud, Aspen Gutgsell, Dr. M. Ashhar I. Khan, Dr. Konstantin I. Popov, and Edgar M. Faison for valuable discussion and outstanding support of the study. This work was supported by NIH Grants R01GM114015 and GM123247 (to N.V.D.), GM118331 (to M.D.), and GM31819 and ES013773 (to J.D.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800187115/-/DCSupplemental.
References
- 1.Boillée S, Vande Velde C, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 3.Cirulli ET, et al. FALS Sequencing Consortium Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng HX, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 5.Redler RL, Dokholyan NV. The complex molecular biology of amyotrophic lateral sclerosis (ALS) Prog Mol Biol Transl Sci. 2012;107:215–262. doi: 10.1016/B978-0-12-385883-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay M, et al. The disulfide bond, but not zinc or dimerization, controls initiation and seeded growth in amyotrophic lateral sclerosis-linked Cu,Zn superoxide dismutase (SOD1) fibrillation. J Biol Chem. 2015;290:30624–30636. doi: 10.1074/jbc.M115.666503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fay JM, et al. A phosphomimetic mutation stabilizes SOD1 and rescues cell viability in the context of an ALS-associated mutation. Structure. 2016;24:1898–1906. doi: 10.1016/j.str.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proctor EA, et al. Nonnative SOD1 trimer is toxic to motor neurons in a model of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2016;113:614–619. doi: 10.1073/pnas.1516725113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdolvahabi A, et al. Kaplan-Meier meets chemical kinetics: Intrinsic rate of SOD1 amyloidogenesis decreased by subset of ALS mutations and cannot fully explain age of disease onset. ACS Chem Neurosci. 2017;8:1378–1389. doi: 10.1021/acschemneuro.7b00029. [DOI] [PubMed] [Google Scholar]
- 11.Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 12.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 13.De Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 14.Wong YC, Krainc D. α-Synuclein toxicity in neurodegeneration: Mechanism and therapeutic strategies. Nat Med. 2017;23:1–13. doi: 10.1038/nm.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrup K. The case for rejecting the amyloid cascade hypothesis. Nat Neurosci. 2015;18:794–799. doi: 10.1038/nn.4017. [DOI] [PubMed] [Google Scholar]
- 16.Holmes C, et al. Long-term effects of Abeta42 immunisation in Alzheimer’s disease: Follow-up of a randomised, placebo-controlled phase I trial. Lancet. 2008;372:216–223. doi: 10.1016/S0140-6736(08)61075-2. [DOI] [PubMed] [Google Scholar]
- 17.Steele AD, et al. Context dependent neuroprotective properties of prion protein (PrP) Prion. 2009;3:240–249. doi: 10.4161/pri.3.4.10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar DK, et al. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci Transl Med. 2016;8:340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redler RL, et al. Glutathionylation at Cys-111 induces dissociation of wild type and FALS mutant SOD1 dimers. Biochemistry. 2011;50:7057–7066. doi: 10.1021/bi200614y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redler RL, Fee L, Fay JM, Caplow M, Dokholyan NV. Non-native soluble oligomers of Cu/Zn superoxide dismutase (SOD1) contain a conformational epitope linked to cytotoxicity in amyotrophic lateral sclerosis (ALS) Biochemistry. 2014;53:2423–2432. doi: 10.1021/bi500158w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanova MI, et al. Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc Natl Acad Sci USA. 2014;111:197–201. doi: 10.1073/pnas.1320786110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin S, Ding F, Dokholyan NV. Eris: An automated estimator of protein stability. Nat Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 24.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 25.Khan MAI, et al. Cu/Zn superoxide dismutase forms amyloid fibrils under near-physiological quiescent conditions: The roles of disulfide bonds and effects of denaturant. ACS Chem Neurosci. 2017;8:2019–2026. doi: 10.1021/acschemneuro.7b00162. [DOI] [PubMed] [Google Scholar]
- 26.Cashman NR, et al. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- 27.Sabitha KR, Sanjay D, Savita B, Raju TR, Laxmi TR. Electrophysiological characterization of Nsc-34 cell line using microelectrode array. J Neurol Sci. 2016;370:134–139. doi: 10.1016/j.jns.2016.09.038. [DOI] [PubMed] [Google Scholar]
- 28.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 29.Da Cruz S, et al. Misfolded SOD1 is not a primary component of sporadic ALS. Acta Neuropathol. 2017;134:97–111. doi: 10.1007/s00401-017-1688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas EV, Fenton WA, McGrath J, Horwich AL. Transfer of pathogenic and nonpathogenic cytosolic proteins between spinal cord motor neurons in vivo in chimeric mice. Proc Natl Acad Sci USA. 2017;114:E3139–E3148. doi: 10.1073/pnas.1701465114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci USA. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilcox KC, et al. Modifications of superoxide dismutase (SOD1) in human erythrocytes: A possible role in amyotrophic lateral sclerosis. J Biol Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bieschke J, et al. Small-molecule conversion of toxic oligomers to nontoxic β-sheet-rich amyloid fibrils. Nat Chem Biol. 2011;8:93–101. doi: 10.1038/nchembio.719. [DOI] [PubMed] [Google Scholar]
- 34.Sangwan S, et al. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS) Proc Natl Acad Sci USA. 2017;114:8770–8775. doi: 10.1073/pnas.1705091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luchinat E, et al. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat Commun. 2014;5:5502. doi: 10.1038/ncomms6502. [DOI] [PubMed] [Google Scholar]
- 36.Sekhar A, et al. Probing the free energy landscapes of ALS disease mutants of SOD1 by NMR spectroscopy. Proc Natl Acad Sci USA. 2016;113:E6939–E6945. doi: 10.1073/pnas.1611418113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu C, Mowrey DD, Dokholyan NV. Computational protein design through grafting and stabilization. Methods Mol Biol. 2017;1529:227–241. doi: 10.1007/978-1-4939-6637-0_11. [DOI] [PubMed] [Google Scholar]