Significance
We present flySAM, a potent system for Cas9-based transcriptional activation (CRISPRa) in Drosophila. flySAM greatly improves on existing in vivo CRISPRa techniques in terms of potency, scalability, and ease of use, and provides a simple and general method for conducting overexpression experiments and screens. flySAM will now serve as the basis for our growing collection of publicly available CRISPRa transgenic fly lines.
Keywords: CRISPR/Cas9, CRISPRa, Cas9 activators, gain of function
Abstract
CRISPR/Cas9-based transcriptional activation (CRISPRa) has recently emerged as a powerful and scalable technique for systematic overexpression genetic analysis in Drosophila melanogaster. We present flySAM, a potent tool for in vivo CRISPRa, which offers major improvements over existing strategies in terms of effectiveness, scalability, and ease of use. flySAM outperforms existing in vivo CRISPRa strategies and approximates phenotypes obtained using traditional Gal4-UAS overexpression. Moreover, because flySAM typically requires only a single sgRNA, it dramatically improves scalability. We use flySAM to demonstrate multiplexed CRISPRa, which has not been previously shown in vivo. In addition, we have simplified the experimental use of flySAM by creating a single vector encoding both the UAS:Cas9-activator and the sgRNA, allowing for inducible CRISPRa in a single genetic cross. flySAM will replace previous CRISPRa strategies as the basis of our growing genome-wide transgenic overexpression resource, TRiP-OE.
A number of techniques have recently been developed for systematic gene overexpression studies based on CRISPR/Cas9 transcriptional activators (CRISPRa) (1). In CRISPRa, nuclease-dead Cas9 (dCas9) is used to guide a transcriptional activator complex to a target gene’s transcriptional start site via a short guide RNA (sgRNA). CRISPRa offers several advantages over traditional techniques for overexpression (reviewed in ref. 2) and represents an important complement to existing genome-wide resources for loss-of-function studies (3). A central challenge now is to adapt CRISPRa for in vivo use to allow for robust, systematic overexpression studies in an organismal context (2, 4–7).
We have recently shown that dCas9-VPR is an effective tool for CRISPRa in vivo in Drosophila (2, 7). In VPR, dCas9 is fused to a tripartite transcriptional activator domain (VP64-p65-Rta) (8). While VPR successfully activates target genes and generates phenotypes in vivo, this system has three important limitations. First, VPR typically requires two sgRNAs per target gene to reliably achieve consistent transcriptional activation (2). This greatly increases the cost and complexity of creating a large-scale resource for in vivo CRISPRa, and also doubles the chance of off-target effects. Second, because CRISPRa requires three independent transgenes in a single fly (Gal4, UAS:dCas9-VPR, and sgRNA), the use of this system is not as straightforward as standard Gal4-UAS–based tools, which require only a single genetic cross. Third, previous experiments in Drosophila cell culture suggest that an alternative CRISPRa technique, synergistic activation mediator (SAM), outperforms VPR in direct comparisons (1). However, previous attempts to express SAM components in vivo have failed due to toxicity (2).
In contrast to VPR, SAM involves two separate protein components—dCas9-VP64 and MCP:p65-HSF1—as well as a modified sgRNA containing two MS2 hairpin structures that recruit MCP:p65-HSF1 (9). To date, it has not been possible to apply SAM in vivo in flies because ubiquitous expression of UAS:MCP-p65-HSF1 is lethal in the absence of any sgRNA (2). In addition, attempts to express “SAM-like” components, including alternative transcriptional activation domains fused to MCP, failed to outperform VPR in vivo (2). For these reasons, the first generation of transgenic lines for in vivo CRISPRa (the “TRiP-OE” collection) was based on VPR (2).
Here we present flySAM, a robust, scalable, and simplified strategy for in vivo CRISPRa that overcomes previous toxicity issues and unambiguously outperforms VPR in direct comparisons. We show that flySAM using a single sgRNA typically outperforms VPR using two sgRNAs, and that the severity of flySAM phenotypes is comparable to traditional Gal4-UAS overexpression. We use flySAM to demonstrate multiplexed CRISPRa of multiple genes in vivo. Finally, we have greatly simplified the use of this system by combining flySAM with the sgRNA in a single transgenic vector, allowing for tissue-specific CRISPRa with a single genetic cross. flySAM thus represents a major improvement in the strength, scalability, and ease of use over existing CRISPRa strategies, and will replace VPR as the basis of our growing genome-wide transgenic CRISPRa resource, TRiP-OE.
Results and Discussion
Establishing in Vivo flySAM Using the T2A Self-Cleaving Peptide.
Previous attempts to express the SAM component UAS:MCP-p65-HSF1 in vivo used the pJFRC7 vector, which is designed for very high protein expression levels and includes 20XUAS-binding sites and a 5′ intervening sequence (IVS) to boost translational efficiency (2). We hypothesized that expressing MCP:p65-HSF1 at lower levels may overcome the lethality observed in previous reports (2); therefore, we designed a vector, flySAM1.0, expressing the SAM components dCas9-VP64 and MCP-p65-HSF1 separated by a T2A self-cleaving peptide, under 10XUAS control (Fig. 1A), reasoning that proteins in the second position of a T2A-containing bicistronic transcript are typically expressed at lower levels than proteins in the first position (10). We also constructed SAM-compatible sgRNA expression backbone vectors for expressing either a single sgRNA (U6:2-sRNA2.0) or multiple sgRNAs (Methods and Tables S1 and S2).
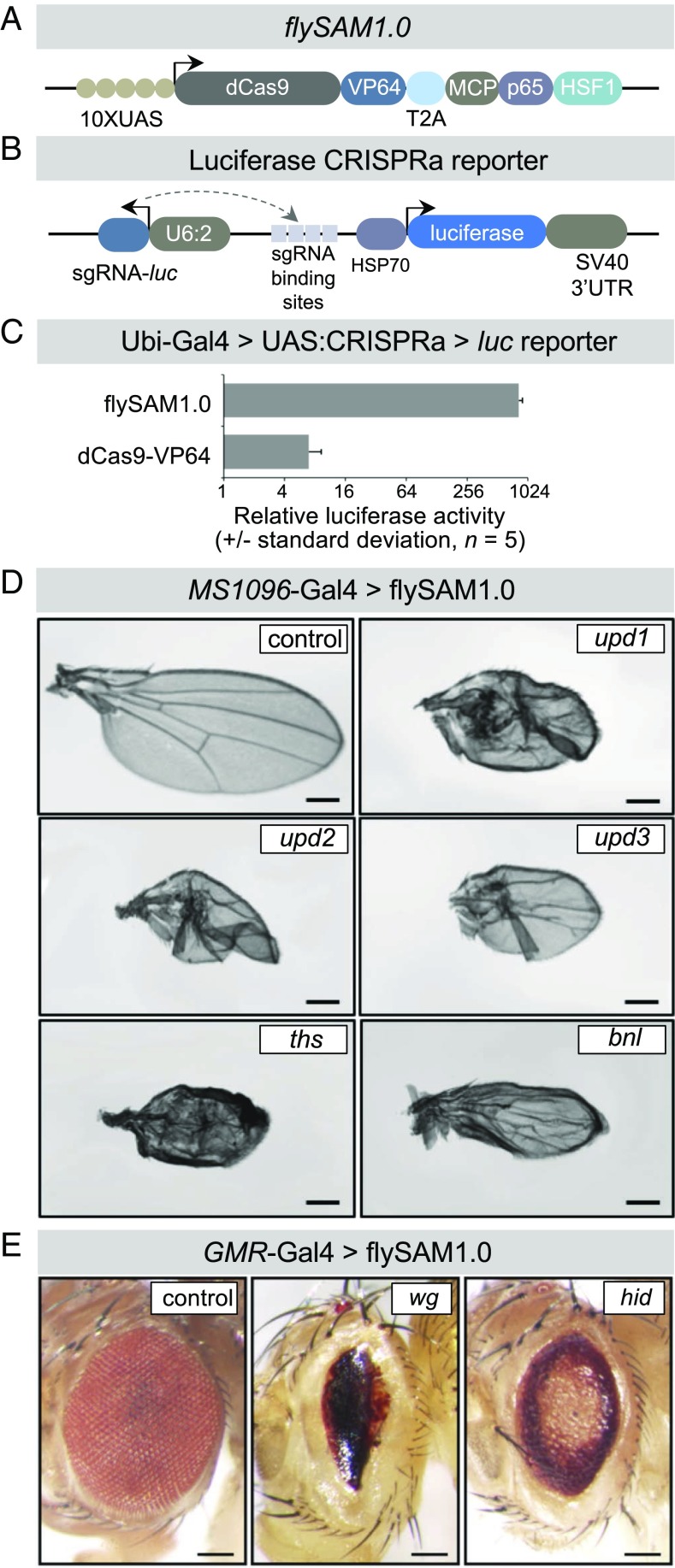
Fig. 1.
flySAM1.0 is a potent technique for in vivo CRISPRa. (A) Schematic of the flySAM1.0 construct, showing the two SAM components separated by a T2A peptide. Not to scale. (B) Schematic of the luciferase CRISPRa reporter containing the luciferase coding sequence downstream of four tandem binding sites for an sgRNA that is encoded as a separate transcript. (C) In vivo CRISPRa luciferase assay of flySAM1.0 compared with dCas9-VP64, driven by Ubi-Gal4. (D and E) flySAM1.0 activates a range of endogenous genes in vivo in the wing (MS1096-Gal4) (D) and eye (GMR-Gal4) (E), using a single sgRNA. (Scale bars: 250 µm.)
We tested whether flySAM1.0 can be expressed in vivo without toxicity by crossing this line to a ubiquitous Gal4 (actin-Gal4) in the absence of any sgRNA. In contrast to the 100% lethality observed for 20XUAS-IVS-UAS:MCP-p65-HSF1 (2), we observed normal survival rates and no visible phenotypes for flySAM1.0 (n = 125; Fig. 1). Similar results were obtained for a second ubiquitous driver, Ubi-Gal4. To further confirm that flySAM1.0 is not toxic in vivo, we crossed this line to a panel of tissue-specific Gal4 drivers (hh-Gal4, MS1096-Gal4, dMef2-Gal4, Lpp-Gal4, and nub-Gal4). In all cases, we observed normal survival rates and no visible morphological phenotypes in any of the targeted tissues (n = 30–135 offspring per genotype). We did observe, however, that expression of flySAM1.0 using tubulin-Gal4, an additional ubiquitous driver, was lethal (100% lethal; n = 110 siblings), indicating that this construct may be toxic when expressed ubiquitously at high levels.
To test whether flySAM successfully activates target genes in vivo, we constructed a luciferase reporter line containing the luciferase coding sequence downstream of four tandem sgRNA-binding sites, as well as an sgRNA targeting these binding sites, together in one vector (Fig. 1B). Using this reporter line, we measured the activity of UAS:flySAM1.0 relative to a UAS:dCas9-VP64 control using Ubi-Gal4, and observed a dramatic increase in luciferase activity, indicating that flySAM1.0 is effective in vivo (Fig. 1C). We also constructed two additional flySAM-like vectors containing alternative activation domains in place of MCP-p65-HSF1 (flySAM1.1: UAS:dCas9-VP64-T2A-MCP-p65-Rta and flySAM1.2: UAS:dCas9-VP64-T2A-MCP-Dorsal-dHSF). However, both lines had reduced survival or full lethality when expressed ubiquitously using actin-Gal4 and, furthermore, failed to outperform flySAM using our luciferase reporter assay with Ubi-Gal4 (Fig. S1).
We next tested whether flySAM1.0 is capable of activating endogenous genes. We generated U6:2-sgRNA2.0 lines targeting a number of genes: the secreted ligands upd1, upd2, upd3, ths, bnl, and wg and the proapoptotic gene hid. We drove flySAM in the wing (using MS1096-Gal4) or the eye (GMR-Gal4) and observed specific phenotypes for each sgRNA, indicating that flySAM1.0 can trigger physiologically relevant levels of transcriptional activation in vivo (Fig. 1 D and E). We note that these phenotypes were observed using a single sgRNA, while VPR typically requires two sgRNAs for effective CRISPRa (2, 7).
flySAM Outperforms VPR in Direct Comparisons.
To directly compare flySAM1.0 to VPR in vivo, we first used our luciferase reporter system, either containing MS2 hairpins (for flySAM1.0) or a standard sgRNA tail (for VPR). flySAM1.0 led to far higher levels of luciferase activity than VPR (Fig. 2A). The superior performance of flySAM is not due solely to MS2 loops in the sgRNA, as the presence of MS2 loops in fact decreased the performance of dCas9-VPR in cell culture (Fig. S2).
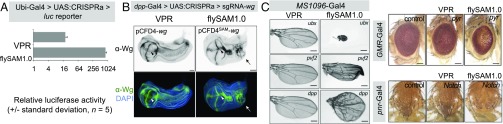
Fig. 2.
flySAM1.0 outperforms VPR. (A) In vivo CRISPRa luciferase assay of flySAM1.0 compared with dCas9-VPR, using Ubi-Gal4. (B) dpp-Gal4–driven ectopic activation of wg in the larval wing disk using flySAM1.0 vs. VPR using identical protospacer sequences. The arrowhead indicates a duplicated wing pouch, and arrows indicate ectopic Wg expression. (Scale bars: 50 µm.) (C) Comparison of flySAM1.0 and VPR in the wing, eye, and notum, using identical protospacer sequences, differentiated only by the presence of MS2 loops in the flySAM experiments. (Scale bars: 250 µm.)
To compare the ability of VPR and SAM to activate endogenous genes in vivo, we used an established system for comparing CRISPRa activity in which dpp-Gal4 is used to activate wg in an ectopic domain of the larval wing imaginal disk (2, 7). In these experiments, two sgRNAs were used that were identical in protospacer sequence and differed only in the presence of MS2 loops in the sgRNA tail. While both systems activated ectopic Wg expression, flySAM1.0 induced a duplication of the wing pouch, complete with a secondary dorsoventral axis, and visibly higher levels of ectopic Wg expression (Fig. 2B). A fully duplicated wing pouch is reminiscent of the phenotype observed using dpp-Gal4 > UAS:wg (11), and is never observed using VPR (this study and refs. 2 and 7). The phenotype observed with flySAM1.0 is thus unambiguously stronger than VPR.
Previous experiments comparing VPR and SAM in cell culture have shown that across multiple individual tests of different genes are some cases in which VPR can outperform SAM (1). Therefore, we tested an additional five genes (ubx, Pvf2, dpp, pyr, and N, all using single sgRNAs) in different tissues (wing, MS1096-Gal4; eye, GMR-Gal4; notum, pnr-Gal4), and found that flySAM generated stronger phenotypes than VPR for all five genes (Fig. 2C). Thus, for a given sgRNA sequence, flySAM outperforms VPR in vivo in all cases we have studied.
flySAM Phenotypes Resemble UAS:cDNA Overexpression Phenotypes.
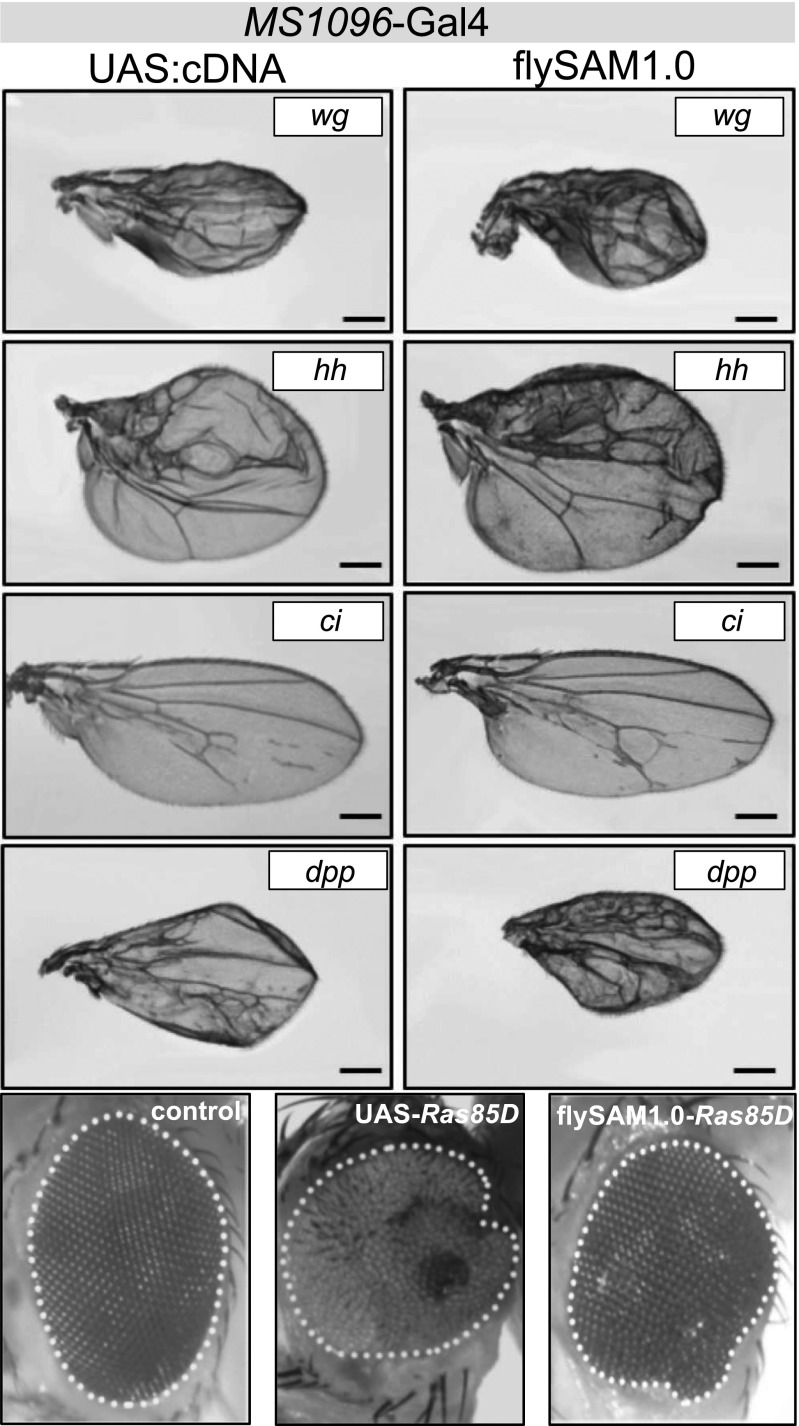
Traditionally, the most widely used technique for in vivo overexpression in Drosophila is cDNA overexpression using the Gal4-UAS system (12, 13). To directly compare flySAM to cDNA overexpression, we used U6:2-sgRNA2.0 lines for five genes for which UAS overexpression lines are available (wg, hh, ci, dpp, and Ras85D). Driving these constructs in the wing (MS1096-Gal4) or eye (eyeless-Gal4), we observed comparable phenotypes in all cases (Fig. 3). These results indicate that flySAM1.0 can recapitulate overexpression phenotypes comparable in strength to Gal4-UAS overexpression, using only a single sgRNA.
Fig. 3.
flySAM1.0 phenotypes recapitulate Gal4-UAS overexpression phenotypes. The phenotypes observed using flySAM1.0 (with one sgRNA) resemble those obtained using Gal4 > UAS-cDNA. Wing phenotype, MS1096-Gal4; eye phenotype, eyeless-Gal4. (Scale bars: 250 µm.)
flySAM Allows for Multiplexed CRISPRa.
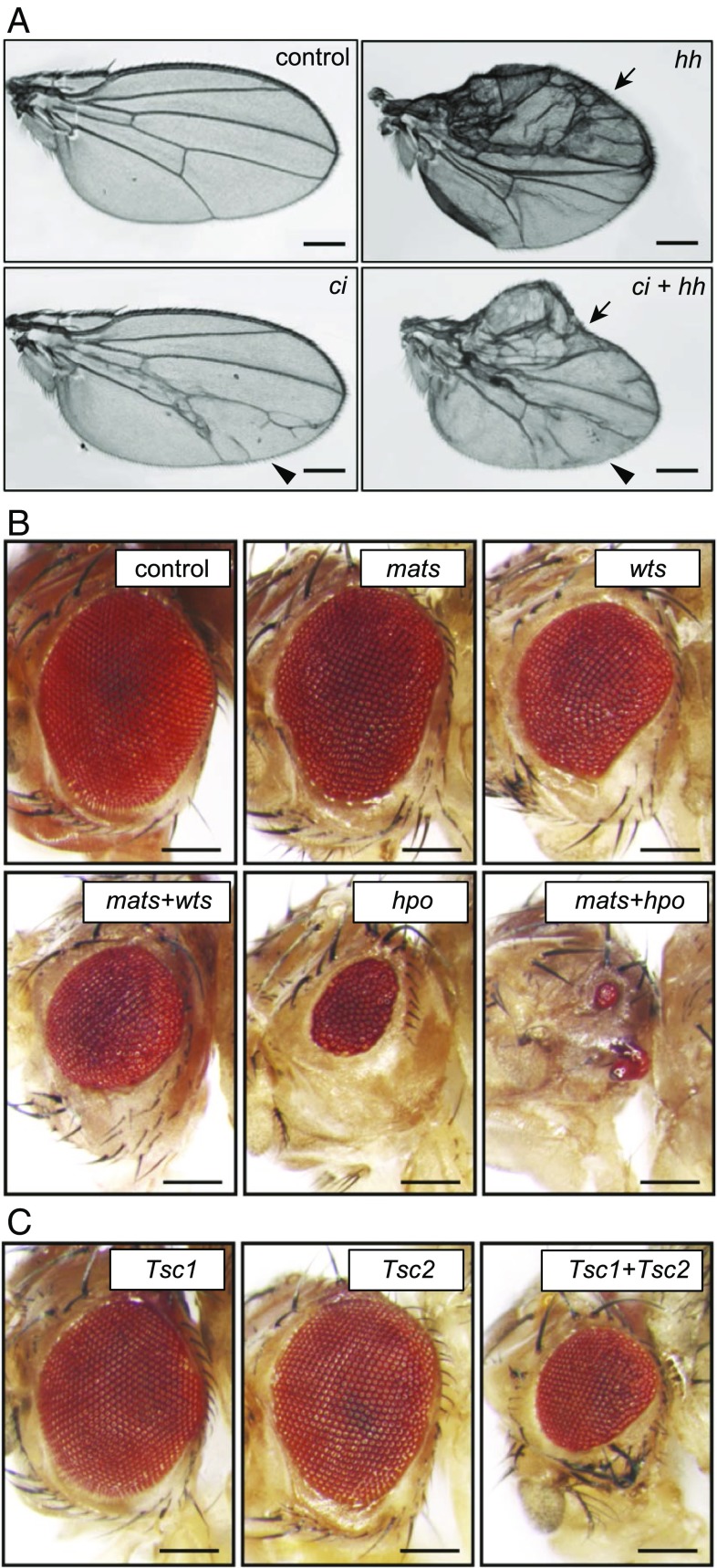
We tested whether it is possible to activate multiple genes simultaneously by coexpressing multiple sgRNAs in a single fly. We first activated ci and hh, both singly and together, in the wing using MS1096-Gal4, using a single sgRNA vector that expresses either one or both sgRNAs. Single gene activation of hh using MS1096-Gal4 results in overgrowth and patterning defects primarily in the anterior wing compartment, while ci activation results in vein defects primarily in the posterior wing compartment (Figs. 3 and 4A). When ci and hh were activated simultaneously, both phenotypes were observed in the wing (Fig. 4A), indicating successful multiplexed CRISPRa. We next tested eye-specific CRISPRa of genes from the Hippo pathway using eyeless-Gal4. Individual overexpression of wts, mats, or hpo (two or three sgRNAs per gene) produced relatively mild small-eye phenotypes, while multiplexed CRISPRa for wts+mats or mats+hpo showed strong enhancement of this phenotype (Fig. 4B). Similarly, while individual CRISPRa of the tumor suppressors Tsc1 and Tsc2 had minimal effects, Tsc1+Tsc2 showed a strong growth-suppressing genetic interaction (Fig. 4C). These results provide a clear demonstration of multiplexed in vivo CRISPRa.
Fig. 4.
In vivo multiplexed CRISPRa using flySAM. (A) Transcriptional activation of hh or ci in the wing (using MS1096-Gal4) results in patterning defects in the anterior and posterior compartments, respectively. The arrow indicates the hh phenotype; the arrowhead, the ci phenotype.) Multiplexed activation of ci and hh produces both individual phenotypes. (B) Multiplexed transcriptional activation of Hippo pathway components in the eye using eyeless-Gal4. (C) Simultaneous CRISPRa of Tsc1+Tsc2 in the eye. In B and C, two or three sgRNAs per target gene were used. (Scale bars: 250 μm.)
flySAM2.0, a Single Vector Containing UAS:flySAM and sgRNA for a Simplified Experimental Setup.
A major technical drawback of both the VPR system and the flySAM system described above is that they involve three separate transgenic elements: a Gal4 transgene, a UAS-CRISPRa transgene, and an sgRNA transgene. The requires the creation of a compound fly stock containing two of the components before this compound stock is crossed to a line expressing the third component. In our previous work with VPR, we attempted to overcome this technical bottleneck by generating a publicly available collection of more than 30 Gal4 + UAS:VPR compound stocks containing widely used Gal4 lines combined with UAS:VPR (2). However, using any additional Gal4 requires the generation of a compound Gal4 + UAS stock before performing CRISPRa experiments, which takes several weeks.
To simplify in vivo CRISPRa, we created a single vector containing both UAS:flySAM and an sgRNA, termed flySAM2.0 (Fig. 5A), reasoning that flySAM2.0 lines could simply be crossed to a Gal4 line to achieve tissue-specific CRISPRa in the offspring. We also included gypsy elements flanking the CRISPRa components, to insulate the CRISPRa components from surrounding regulatory features that could dampen their expression (14).
Fig. 5.
flySAM2.0 allows for inducible CRISPRa with a single genetic cross. (A) Diagram of flySAM2.0, which contains both the UAS:flySAM and the sgRNA in a single plasmid, between gypsy insulators. (B) Clonal CRISPRa using FLPout Gal4 > flySAM2.0 for ci and hh in L3 larval wing discs; GFP+ clones are indicated with dashed white lines. (Scale bar: 50 µm.) (C) Comparison of flySAM2.0 (one sgRNA per target gene) to an existing VPR collection (using two sgRNAs per target gene). See text for details.
To verify that flySAM2.0 functions as intended, we generated two flySAM2.0 lines targeting either hh or ci, crossed both lines to a FLPout-Gal4 stock (hsFlp; actin-FRT-STOP-FRT-Gal4; UAS-GFP), and examined CRISPRa clones in the larval wing disk using antibody staining for Ci or the hh target gene Ptc. In both cases, we detected strong, specific activation in GFP+ clones, indicating successful CRISRPa in a single genetic cross (Fig. 5B). These experiments also confirm the previously reported observation that CRISPRa is highly specific (2), as we did not observe CRISPRa outside of Gal4+ clones.
Having established that flySAM outperforms VPR for any given sgRNA, we next tested whether flySAM2.0 (with a single sgRNA) outperforms the existing collection of VPR sgRNA stocks (two sgRNAs per target gene). To do so, we compared CRISPRa wing phenotypes (nub-Gal4) using independent lines generated for each strategy that have differing protospacer sequences (Table S2). We first focused on a set of target genes for which existing VPR had previously failed to generate a phenotype (scw, aos, egr, WntD, and trk), or had been relatively weak (dpp). For all six of these genes, flySAM2.0 generated strong, specific phenotypes that were not observed using VPR (Fig. 5C).
We then tested four genes for which VPR did show phenotypes: upd2, upd3, pyr, and Wnt6. For upd3, pyr, and Wnt6, flySAM2.0 and VPR produced comparable phenotypes, whereas for upd2, VPR produced a stronger phenotype than flySAM2.0 (Fig. 5C). We attribute this result to the fact that because flySAM2.0 only uses a single sgRNA, this is likely a case in which that single sgRNA is suboptimal. Accordingly, a second independent flySAM2.0 line targeting upd2 generated a strong phenotype in the wing (Fig. 5C). Thus, taken together our results demonstrate that flySAM2.0 with a single sgRNA outperforms VPR with two sgRNAs in the large majority of cases. (Hereinafter, we refer to flySAM2.0 as simply “flySAM” for simplicity.)
We note that the flySAM construct is substantially larger than a typical sgRNA construct (15 kb vs. 7 kb), which is likely to reduce the efficiency of recovering transgenic lines. To address this concern, we directly compared the recovery of transformants from a pooled injection of 16 U6:2-sRNA2.0 constructs vs. 16 flySAM constructs. While the total number of transformants recovered was lower for flySAM (flySAM, 26 transformants from 117 surviving injected embryos; U6:2-sgRNA2.0, 83 transformants from 108 surviving injected embryos; Table S3), we were still able to recover 10 of the 16 constructs for flySAM, compared with 13 of the 16 constructs for U6:2-sgRNA2.0 (Table S3). Given the greatly improved downstream usefulness of flySAM constructs, and given that we can still recover transformants at a reasonable rate, we have begun large-scale production of flySAM constructs, toward a long-term goal of generating a genome-wide in vivo CRISPRa resource.
In summary, flySAM represents a major improvement over existing techniques for in vivo CRISPRa in terms of effectiveness, scalability, and ease of use. Regarding effectiveness, flySAM outperforms VPR in direct comparison holding sgRNA sequence constant (Fig. 2), and in fact flySAM using a single sgRNA outperforms VPR using two sgRNAs in the large majority of cases (Fig. 5). In terms of scalability, flySAM dramatically decreases the cost and time required to generate transgenic constructs. Specifically, while double-sgRNA constructs require PCR, gel purification, and Gibson cloning (15), single sgRNA constructs only require ligation of annealed oligos into a plasmid backbone. Finally, by combining the SAM components and sgRNA in a single vector, flySAM provides simplicity, allowing researchers to perform tissue-specific CRISPRa in a single genetic cross.
Given these improvements, we anticipate that flySAM will be widely useful to the Drosophila community for overexpressing specific genes in an inducible, tissue-specific manner, as well as in performing screens. We have begun transitioning our growing collection of transgenic TRiP-OE CRISPRa stocks from VPR to flySAM. Details of this collection are available at https://fgr.hms.harvard.edu/fly-in-vivo-crispr-cas.
Methods
Drosophila Stocks.
All Drosophila stocks were maintained at 25 °C with 60% humidity on standard cornmeal/sugar/agar media unless specified otherwise. CRISPRa lines (Table S1) and sgRNA lines (Table S2) were obtained from the Tsinhua Fly Center or the Bloomington Drosophila Stock Center. Additional fly stocks are listed in Table S4.
Transgene Constructs and Production of Transgenic Flies.
The flySAM1.0 vector was in the VALIUM20 backbone (16). Fragments were stitched together using a sequence- and ligation-independent cloning method (17). dCas9 was amplified from a nos-Cas9 vector (18), and two mutations in the RuvC domain (D10A) and HNH domain (H840A) were introduced separately. Activation domains were amplified from plasmid or genomic DNA using high-fidelity DNA polymerase and fused to the C terminus of dCas9. VP64, T2A, and MCP were synthesized by GENEWIZ. p65 was cloned from mouse cDNA for flySAM1.0 or Addgene 63798 (human) for flySAM2.0. HSF was amplified from Addgene 61426. Rta was amplified from Addgene 63798, and Dorsal and dHSF were cloned from Drosophila cDNA. The U6:2-sgRNA2.0 vector was cloned by replacing the sgRNA1.0 scaffold in U6b-sgRNA-short (18) with the sgRNA2.0 scaffold (9). The flySAM2.0 vector was made by inserting the U6B-sgRNA2.0 fragment digested with NheI and SpeI from the U6B-sgRNA2.0 vector into the flySAM1.0 vector.
For dpp-Gal4 > CRISPRa > wg experiments in larval wing disc, we used pCFD4SAM (2), which expresses two sgRNAs from U6:1 and U6:3, respectively. For all other multiplex experiments, we combined multiple U6:2-sgRNA fragments in a single plasmid, as follows. First, we cloned individual sgRNAs into the U6:2-sgRNA2.0 vector. One such U6:2-sgRNA2.0 vector was linearized by digesting with SpeI or NheI, while additional vectors were digested with both SpeI and NheI to isolate the fragment containing a U6b-sgRNA2.0 (∼1,000 bp). The resulting products were gel-purified (AxyPrep DNA Gel Extraction Kit) and ligated together.
To build the luciferase CRISPRa reporter, we cloned firefly luciferase downstream of the hsp70 basal promoter and upstream of an SV40 polyA tail. We inserted a synthesized sgRNA target site (sgRNA: CCTACAGCACGTCGCCGGCG) immediately upstream of the hsp70 promoter. This luciferase reporter fragment was inserted into U6:2-sgRNA2.0 vector using restriction digest cloning to generate the sgRNA2.0 luciferase reporter. UAS-hh and UAS-ci were created by first amplifying the coding sequences of these genes from Drosophila genomic DNA and then cloning them into a pVALIUM vector (16) with EcoRI and NheI restriction digests, using the Hieff Clone One-Step Cloning Kit (Yeasen).
Transgenic fly lines were generated by injecting the constructs into y sc v nanos-integrase; attP2 or y sc v nanos-integrase; attP40 embryos following standard procedures (16).
Viability and Phenotypic Analyses.
Initial viability assays were performed as described previously (2) by crossing flySAM1.0 to the ubiquitous driver y w; actin-Gal4/CyO and then counting the number of surviving offspring compared with siblings carrying the CyO balancer. Similar experiments were done using additional ubiquitous driver lines Ubi-Gal4 and tub-Gal4 and/or tissue-specific drivers. For all tissue-specific Gal4 tests, between 30 and 135 offspring per genotype were examined for tissue-specific morphological phenotypes and/or lethality.
Analyses of wings, nota, and eyes with flySAM1.0 were conducted using the Gal4 lines listed in Table S3. These drivers (combined with UAS:flySAM) were crossed to U6:2-sgRNA2.0 flies, and their offspring were analyzed for phenotypes.
For all phenotypic analyses, adult wings (10–20 per sex, per genotype) were mounted in a solution of either 1:1 ethanol/glycerol or 1:1 acetone to Permount (Thermo Fisher Scientific) and analyzed on either a Nikon Eclipse Ti microscope or a Zeiss Axioskop 2. Nota and eye phenotypes (5–10 per sex, per genotype) were imaged using a Leica MZ16FA stereomicroscope. Representative samples are shown for each genotype for the sex with the strongest phenotype (sex held constant for each comparison).
To directly compare flySAM2.0 collection (one sgRNA, one transgene) with dCas9-VPR (two sgRNAs, two transgenes), a published collection of double-sgRNA lines for VPR (2) was compared with a newly generated collection of single-sgRNA lines built in the flySAM2.0 backbone, with independently designed protospacer sequences. VPR lines were crossed to w; nubbin-Gal4; dCas9-VPR/SM5, TM6b, and flySAM2.0 lines were crossed to w; nubbin-Gal4, at 27 °C.
For dpp > CRISPRa > wg, we used previously described sgRNA-wg lines (2) in either pCFD4 or pCFD4SAM. These were crossed to w; dCas9-VPR; dpp-Gal4/TM6b or w; flySAM1.0; dpp-Gal4/TM6b, respectively, at 27 °C. Wing discs from approximately 20 larvae of each genotype were analyzed, and representative discs are shown. Note that this VPR experiment is the second replication of a published result (2, 7), and in no case was a duplicated wing pouch observed using VPR.
Cell culture experiments and quantitative PCR (qPCR) were performed as described previously (2, 7) with primers described previously (1).
FLP-out Induced Clonal Analysis and Immunostaining.
To generate FLP-out CRISPRa clones, the flySAM2.0-ci and flySAM2.0-hh lines were crossed with y, hs-FLP; act5C < STOP < Gal4, UAS-GFP/CyO flies and maintained at 25 °C. First instar larvae were heat-shocked at 37 °C for 1 h and maintained until L3, at which time wing discs were dissected, fixed, and stained using standard protocols.
The following primary antibodies were used: anti-Wg antibody [4D4, 1:100; Developmental Studies Hybridoma Bank (DSHB)], rat anti-ci antibody 2A1-c (1:10; DSHB), mouse monoclonal anti-ptc antibody ptc-c (1:50; DSHB), and rabbit polyclonal anti-GFP (290, 1:2,000; Abcam). Various secondary antibodies (Jackson ImmunoResearch Laboratories) conjugated with FITC or TRITC were used at 1:300.
Luciferase Assay.
Luciferase activity was measured using the Steady-Glo Luciferase Assay Kit (E2520; Promega) as described previously (19). In brief, five replicates of three 7-d-old adult female flies were collected separately in 100 μL or 50 μL Glo Lysis Buffer (E266A; Promega). Samples were homogenized and then centrifuged at 20,000 × g for 15 min at 4 °C. Then 35 μL or 55 μL of supernatant was transferred to a 96- well solid-white microplate and mixed with the same volume of Steady-Glo reagent. After incubation in the dark for 20 min, luminescence was measured on a spectral scanning multimode reader (Varioskan Flash; Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We thank Ryan Colbeth and Rich Binari for the expertise and assistance with fly work. This work was supported by the National Natural Science Foundation of China (Grant 31571320), the National Key Technology Research and Development Program of the Ministry of Science and Technology of the People’s Republic of China (Grants 2016YFE0113700 and 2015BAI09B03), the China Postdoctoral Science Foundation (Grant 2017M620747), and the National Institutes of Health (NIH; Grants R01 GM084947 and R24OD021997, to the N.P. laboratory). B.E.-C. was funded by Ruth L. Kirschstein National Research Service Award F32GM113395 from the NIH General Medical Sciences Division. N.P. is an investigator of the HHMI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800677115/-/DCSupplemental.
References
- 1.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewen-Campen B, et al. Optimized strategy for in vivo Cas9-activation in Drosophila. Proc Natl Acad Sci USA. 2017;114:9409–9414. doi: 10.1073/pnas.1707635114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Housden BE, et al. Loss-of-function genetic tools for animal models: Cross-species and cross-platform differences. Nat Rev Genet. 2017;18:24–40. doi: 10.1038/nrg.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowder LG, et al. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-act systems. Mol Plant. 2017;11:245–256. doi: 10.1016/j.molp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Liao H-K, et al. In vivo target gene activation via crispr/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171:1495–1507.e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wangensteen KJ, et al. Combinatorial genetics in liver repopulation and carcinogenesis with a novel in vivo CRISPR activation platform. Hepatology. 2017 doi: 10.1002/hep.29626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S, Ewen-Campen B, Ni X, Housden BE, Perrimon N. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics. 2015;201:433–442. doi: 10.1534/genetics.115.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep. 2017;7:2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng M, Diaz-Benjumea FJ, Vincent JP, Wu J, Cohen SM. Specification of the wing by localized expression of wingless protein. Nature. 1996;381:316–318. doi: 10.1038/381316a0. [DOI] [PubMed] [Google Scholar]
- 12.Bischof J, et al. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
- 13.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 14.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni J-Q, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li MZ, Elledge SJ. Gene Synthesis. Vol 852. Humana Press; Totowa, NJ: 2012. SLIC: A method for sequence- and ligation-independent cloning; pp. 51–59. [DOI] [PubMed] [Google Scholar]
- 18.Ren X, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni JQ, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]