Significance
In translation initiation, a preinitiation complex (PIC) of the 40S ribosomal subunit, eukaryotic initiation factors (eIFs), and initiator transfer RNA (tRNAi) scans the mRNA for an AUG codon in favorable context. AUG recognition evokes a closed PIC conformation and triggers dissociation of eIF1. Structural data predict that a clash between eIF1 β-hairpin Loop 2 and tRNAi impedes selection of non-AUG start codons in the open, scanning conformation of the PIC. Supporting this model, we show that alanine substitutions of Loop 2 residues increase initiation at suboptimal start codons (UUGs and AUGs in poor context) in vivo and destabilize tRNAi binding at UUG codons in reconstituted PICs in vitro. These results establish Loop 2–RNAi interaction as a key determinant of initiation accuracy.
Keywords: translation, initiation, eIF1, ribosome, yeast
Abstract
The eukaryotic 43S preinitiation complex (PIC), bearing initiator methionyl transfer RNA (Met-tRNAi) in a ternary complex (TC) with eukaryotic initiation factor 2 (eIF2)-GTP, scans the mRNA leader for an AUG codon in favorable context. AUG recognition evokes rearrangement from an open PIC conformation with TC in a “POUT” state to a closed conformation with TC more tightly bound in a “PIN” state. eIF1 binds to the 40S subunit and exerts a dual role of enhancing TC binding to the open PIC conformation while antagonizing the PIN state, necessitating eIF1 dissociation for start codon selection. Structures of reconstituted PICs reveal juxtaposition of eIF1 Loop 2 with the Met-tRNAi D loop in the PIN state and predict a distortion of Loop 2 from its conformation in the open complex to avoid a clash with Met-tRNAi. We show that Ala substitutions in Loop 2 increase initiation at both near-cognate UUG codons and AUG codons in poor context. Consistently, the D71A-M74A double substitution stabilizes TC binding to 48S PICs reconstituted with mRNA harboring a UUG start codon, without affecting eIF1 affinity for 40S subunits. Relatively stronger effects were conferred by arginine substitutions; and no Loop 2 substitutions perturbed the rate of TC loading on scanning 40S subunits in vivo. Thus, Loop 2–D loop interactions specifically impede Met-tRNAi accommodation in the PIN state without influencing the POUT mode of TC binding; and Arg substitutions convert the Loop 2–tRNAi clash to an electrostatic attraction that stabilizes PIN and enhances selection of poor start codons in vivo.
In the scanning mechanism of translation initiation, the small (40S) ribosomal subunit recruits initiator methionyl transfer RNA (Met-tRNAi) in a ternary complex (TC) with GTP-bound eukaryotic initiation factor 2 (eIF2) in a reaction stimulated by the factors eIF1, eIF1A, and eIF3, and the resulting 43S preinitiation complex (PIC) attaches to the 5′ end of mRNA and scans the mRNA leader for an AUG start codon. Nucleotides surrounding the AUG (the Kozak context) influence the efficiency of its selection. In the scanning PIC, eIF1 and eIF1A promote an open conformation of the 40S subunit, with TC bound in a relatively unstable conformation, “POUT”, suitable for inspecting successive triplets in the peptidyl (P) decoding site for complementarity with the anticodon of Met-tRNAi. The GTP in the TC can be hydrolyzed, stimulated by GTPase activating protein eIF5, but eIF1 blocks release of inorganic phosphate (Pi) at non-AUG codons. Start codon recognition triggers dissociation of eIF1 from the 40S subunit, enabling both Pi release from eIF2-GDP–Pi and more stable TC binding to the PIC, with Met-tRNAi accommodated in the “PIN” state (SI Appendix, Fig. S1A). Subsequent dissociation of eIF2-GDP and other eIFs from the 48S PIC enables eIF5B-catalyzed subunit joining and the formation of an 80S initiation complex ready to commence protein synthesis (1).
eIF1 plays a dual role in the scanning mechanism (SI Appendix, Fig. S1A), stabilizing the open conformation and promoting rapid TC loading in the POUT conformation while also destabilizing the closed/PIN state (2) and blocking Pi release at non-AUG codons (3), thus requiring eIF1 dissociation from the 40S subunit to permit selection of AUG codons (4–7). Scanning enhancer (SE) elements in the eIF1A C-terminal tail cooperate with eIF1 to promote TC binding in the open POUT conformation and impede rearrangement to the closed PIN state (8), whereas a scanning inhibitor element in the eIF1A N-terminal tail promotes rearrangement to the closed state, with dissociation of eIF1 and more-stable TC binding in the PIN conformation. Thus, in the current model of scanning (SI Appendix, Fig. S1A), both eIF1 and eIF1A SE elements must be displaced from the P site to accommodate Met-tRNAi binding in the PIN state for AUG recognition.
Basic residues in eIF1 located in helix α1 and the loop of β-hairpin 1 (Loop 1) directly contact 18S rRNA in various partial PICs whose structures have been resolved at atomic resolution (9–11), and we showed previously that substituting these residues with alanine or negatively charged amino acids weakens 40S binding by eIF1 in vitro and confers Gcd− and Sui− phenotypes in vivo, signifying a shift from the open/POUT conformation to the closed/PIN state (12). The Gcd− phenotype entails derepressed translation of GCN4 mRNA that results from a slower rate of TC binding to 40S subunits scanning the mRNA leader, allowing inhibitory upstream ORFs (uORFs) to be bypassed in favor of reinitiation downstream at the GCN4 AUG codon. The Sui− phenotype involves an increased frequency of aberrant initiation at UUG codons, which restores translation of a mutant HIS4 mRNA lacking the normal AUG start codon. The dual Gcd−/Sui− phenotypes of α1 and Loop 1 substitutions can be explained by aberrant dissociation of these eIF1 variants from the 40S subunit, reducing the occupancy of the open conformation to which TC binds rapidly (for the Gcd− phenotype) and also allowing inappropriate isomerization to the closed state/PIN conformation at non-AUG codons (for the Sui− phenotype) (12, 13) (SI Appendix, Fig. S1B). Supporting this interpretation, both phenotypes were suppressed by overexpressing the α1 and Loop 1 variants, which should compensate for their reduced 40S binding through mass action (12).
Each of the aforementioned eIF1 α1 and Loop 1 substitutions also conferred elevated expression of the mutant eIF1 variant itself, another phenotype indicating reduced accuracy of start codon selection (12, 13). The AUG start codon of the eIF1 gene (SUI1 in yeast) occurs in poor Kozak context, and its frequency of recognition is inversely related to eIF1 abundance, establishing a negative feedback loop that maintains eIF1 at proper levels (13, 14). Thus, overexpressing WT eIF1 suppresses initiation at its own, suboptimal AUG codon, and this autoregulation is eliminated by optimizing the Kozak context of the eIF1 start codon. In addition to increasing discrimination against the poor context of the eIF1 AUG, overexpressing WT eIF1 also suppresses aberrant initiation at UUG codons in strains harboring Sui− mutations in other initiation factors (8, 13–16). Both effects of eIF1 overexpression can be attributed to a reduced rate of eIF1 dissociation from the PIC (achieved by mass action) that prevents inappropriate isomerization to the closed conformation at UUG codons or at AUGs in poor context (13). The fact that eIF1 Loop 1 substitutions increase initiation at the eIF1 AUG codon in addition to elevating UUG initiation supports the conclusion that weakening eIF1 contact with the ribosome allows its inappropriate dissociation from the PIC at suboptimal start codons.
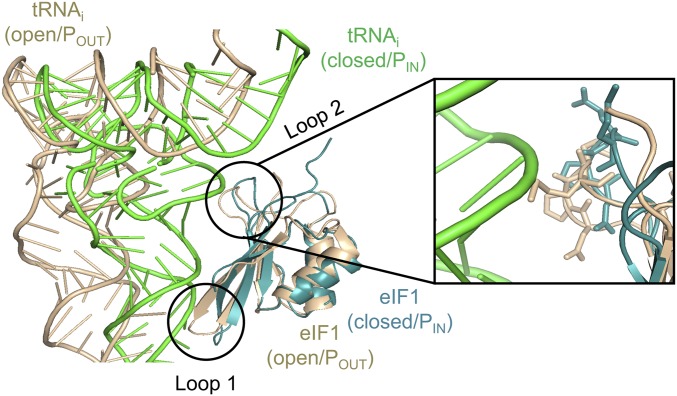
In addition to making direct contacts with the 40S subunit, eIF1 Loop 1 was predicted to clash with the anticodon loop of Met-tRNAi bound in the PIN state when eIF1 is bound in the open conformation of the PIC; and an additional clash with the Met-tRNAi D loop was predicted for eIF1 β-hairpin Loop 2 (9, 10). These predictions were supported by comparing two distinct py48S complexes (both containing eIF1, eIF1A, mRNA, and TC), which appear to represent different intermediates in scanning and start codon recognition, dubbed py48S-open and py48S-closed (17). The py48S-open complex exhibits an upward movement of the 40S head from the body that widens both the mRNA binding cleft and P site, eliminating a subset of 40S contacts with the mRNA and Met-tRNAi present in py48S-closed. Interestingly, Met-tRNAi in py48S-closed would clash with eIF1 in its py48S-open conformation, as Met-tRNAi moves relative to eIF1 during downward movement of the 40S head toward the body in the transition to py48S-closed. This predicted clash is avoided by a displacement of eIF1 on the 40S and altered conformations of eIF1 Loops 1 and 2 (Fig. 1). The remodeling of Loop 1 in py48S-closed disrupts certain eIF1 interactions with the 40S subunit, which should weaken its binding. These structural findings help to explain how eIF1 can coexist with tRNAi in the open, scanning conformation of the PIC while also impeding accommodation of tRNAi in the PIN state of the closed complex via steric or electrostatic clashes to thereby oppose recognition of non-AUG codons. They also suggest how the perfect codon:anticodon duplex formed at AUG and the attendant transition to the closed conformation can evoke the distortion and displacement of eIF1 that weakens its 40S contacts as a prelude to subsequent eIF1 dissociation, gating Pi release from eIF2-GDP–Pi.
Fig. 1.
Structural predictions of the eIF1 Loop 2 clash with Met-tRNAi. Overlay of Met-tRNAi–eIF1 in py48S-open (gold/tan) and py48S-closed (green/teal) from pdbs 3JAQ/3J81, respectively. Magnified portion shows predicted contacts between side chains of Loop 2 residues (shown as sticks) in the open complex and the Met-tRNAi D loop in the closed complex.
Based on the PIC structures just described, it can be predicted that substitutions in eIF1 Loop 1 or Loop 2 that diminish the predicted clashes with Met-tRNAi would favor isomerization to the closed state and increase its probability at either UUG or suboptimal AUG start codons. As noted above, these phenotypes are indeed conferred by Loop 1 substitutions, but these substitutions also diminish eIF1 binding to the 40S subunit; and a comparable binding defect conferred by substitutions in helix α1 was sufficient to elevate initiation at UUG or the poor-context eIF1 AUG codon (12). Thus, it remained unclear whether the predicted clash between eIF1 and Met-tRNAi contributes to initiation accuracy in vivo. To address this, we chose to examine substitutions in Loop 2, which does not contact the 40S subunit or interact with Met-tRNAi in the open complex (Fig. 1). Accordingly, these substitutions should differ from Loop 1 substitutions by having no impact on either 40S binding by eIF1 or the rate of TC loading to the open complex. As such, they should confer Sui−, but not Gcd−, phenotypes, and the Sui− phenotypes should not be overcome by overexpressing the Loop 2 variants. We also reasoned that combining Loop 2 substitutions with helix α1 mutations (mutations that weaken eIF1–40S binding without affecting eIF1–Met-tRNAi interaction) should further increase recognition of suboptimal initiation sites through the distinct mechanism of unscheduled eIF1 dissociation. These predictions have been realized, and we further provided biochemical evidence that Loop 2 substitutions facilitate isomerization to the closed conformation of the PIC at UUG codons. Interestingly, basic substitutions in Loop 2 were found to confer greater utilization of poor start codons compared with alanine substitutions at the same residues, suggesting that replacing a steric clash or repulsion with electrostatic attraction between Loop 2 and Met-tRNAi drives isomerization to the closed/PIN state without impeding subsequent eIF1 dissociation on start codon recognition.
Results
Substitutions of eIF1 Loop 2 Residues Decrease Discrimination Against Suboptimal Initiation Codons in Vivo.
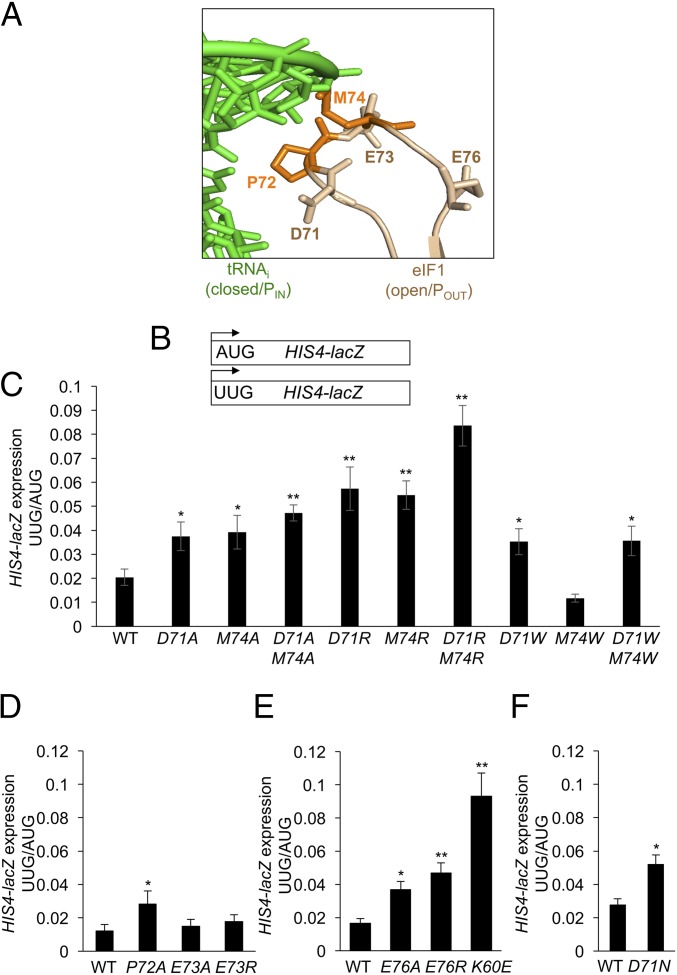
Comparison of the recent cryo-EM structures of yeast partial 48S PICs in open or closed conformations led to the prediction that Loop 2 residues of eIF1 would clash with the phosphodiester backbone of the D loop of tRNAi when eIF1 is bound to the 40S subunit in the conformation observed in the open complex, whereas Met-tRNAi is bound in the location seen in the closed complex (17) (Fig. 1). The Loop 2 sequence in Saccharomyces cerevisiae eIF1, 70KDPEMGE76, contains three acidic residues and a bulky hydrophobic residue (Met-74), which are substantially conserved in evolution (SI Appendix, Fig. S2). We reasoned that replacing the acidic or bulky side chains of D71 or M74 with the methyl group of alanine would diminish the clash between these Loop 2 residues and Met-tRNAi (Fig. 2A) and facilitate isomerization from an open to a closed conformation of the PIC at near-cognate UUG codons. However, it was also possible that mitigating the clash with tRNAi would impede subsequent eIF1 dissociation from the 40S subunit following isomerization to the closed state, and reduce rather than increase UUG initiation. We reasoned further that substituting these Loop 2 residues with basic residues might replace a steric/electrostatic clash with electrostatic attraction between eIF1 and tRNAi, which likewise could confer the counteracting effects of stabilizing the closed/PIN state versus impeding subsequent eIF1 release. In addition to D71 and M74, we also substituted additional Loop 2 residues (P72, E73, and E76), reasoning that their side chains might be instrumental in maintaining the conformation or position of Loop 2 required for the predicted clash with Met-tRNAi.
Fig. 2.
eIF1 Loop 2 substitutions decrease discrimination against UUG initiation codons. (A) eIF1 Loop 2 residues substituted in this study. (B) HIS4-lacZ reporters with AUG or UUG start codons assayed in remaining panels. (C–F) Derivatives of sui1Δ his4-301 strain JCY03 containing the indicated SUI1 alleles and harboring the HIS4-lacZ reporters in B were cultured in synthetic dextrose minimal medium supplemented with His and Trp at 30 °C to A600 of ∼1.0, and β-galactosidase activities (in units of nanomoles of o-nitrophenyl-β-d-galactopyranoside cleaved per minute per milligram) were measured in whole cell extracts. The ratio of expression of the UUG to AUG reporter was calculated from six to nine different transformants, and the mean and SEMs were plotted. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
Mutations introducing amino acid substitutions into Loop 2 residues were generated in a SUI1 allele under its native promoter on a single-copy (sc) LEU2 plasmid, and the resulting mutant alleles were used to replace WT SUI1 on a URA3 plasmid (plasmid-shuffling) by counterselection on medium containing 5-fluoroorotic acid (FOA). To examine the effects of the substitutions on the fidelity of start codon selection, the resulting mutant strains were first assayed for expression of otherwise identical HIS4-lacZ reporters containing an AUG or UUG start codon (Fig. 2B). Significant, approximately twofold increases in expression of the UUG relative to AUG reporter (UUG:AUG ratio) were observed for Ala substitutions in D71, P72, M74, and E76, and a somewhat greater increase was evident for the D71A-M74A and D71A-E76A double-Ala substitutions (Fig. 2 C–E and SI Appendix, Fig. S3). Interestingly, Arg substitutions in D71, E76, and M74 conferred larger increases in the UUG:AUG initiation ratio versus the corresponding Ala replacements (Fig. 2C and SI Appendix, Fig. S3), and the effect of D71R-M74R and D71R-E76R double-Arg substitutions also exceeded those of double- and triple-Ala replacements of these residues, increasing the UUG:AUG ratio by greater than fourfold (Fig. 2C and SI Appendix, Fig. S3). Notably, the triple-Arg substitution D71R-M74R-E76R conferred the largest increase in UUG:AUG initiation ratio, greater than sixfold (SI Appendix, Fig. S3). These findings suggested that replacing acidic or bulky side chains with the small, nonpolar methyl group of alanine increases selection of a near-cognate start codon, and that introducing the large, basic side chain of Arg produces an even greater reduction in fidelity.
Because an Ala for Asp substitution eliminates a negative charge in addition to reducing the size of the side chain, we examined the effect of an Asn replacement of D71, which should eliminate the charge without decreasing the dimensions of the side chain. The D71N substitution mimicked the Ala replacement, increasing the UUG:AUG ratio by approximately twofold (Fig. 2F), thus suggesting that eliminating the negative charge at this Loop 2 residue is sufficient to decrease initiation accuracy.
Realizing that the Arg side chain is considerably larger than that of Asp or Met, we asked whether the increased UUG:AUG ratio conferred by Arg substitutions resulted from the positively charged side chain or, rather, whether the unusually bulky side chain of Arg at these positions provoked aberrant eIF1 dissociation from the PIC following transition to the PIN state by a steric clash with Met-tRNAi. If increasing the dimension of the side chain contributed to the strong increase in UUG initiation conferred by Arg substitutions, then tryptophan replacements at D71/M74 should likewise exceed the effects of Ala substitutions, as the Trp side chain is larger than that of Asp or Met. However, the M74W substitution actually conferred a small decrease, not increase, in the UUG:AUG ratio compared with WT eIF1 (Fig. 2C), as might be expected from an increased steric clash resulting from substituting one bulky hydrophobic side chain (Met) with an even larger one (Trp). In addition, the D71W and D71W/M74W substitutions led to UUG:AUG ratios indistinguishable from those conferred by D71A (Fig. 2C), as expected if elimination of the negative charge of D71 is primarily responsible for the phenotype of the D71A substitution. These findings suggest that the relatively stronger effects of replacing D71/M74 with Arg versus Ala or Trp result from introducing positive charges at the eIF1–tRNAi interface, conferring an electrostatic attraction with tRNAi that stabilizes the closed/PIN conformation at UUG codons without appreciably interfering with subsequent eIF1 dissociation from the PIC.
The yeast strain employed for these studies contains the his4-301 allele, lacking an AUG start codon. The histidine auxotropy conferred by his4-301 can be suppressed by Sui− mutations that enhance initiation at the third, in-frame UUG codon, with attendant increased synthesis of the histidine biosynthetic enzyme His4 (18). However, none of the single or double Loop 2 substitutions affecting residues D71 or M74 conferred detectable growth on medium containing only 1% of the usual histidine supplement (SI Appendix, Fig. S4 A–C, odd-numbered rows). Based on the phenotypes of Sui− substitutions in eIF1 helix α1, the absence of a His+ phenotype for the Loop 2 substitutions can likely be attributed to an insufficient increase in the UUG:AUG ratio, which appears to be at or below the threshold (approximately fourfold increase in UUG:AUG ratio) required for a detectable His+ phenotype (12). Supporting this, the D71R-M74R-E76R triple substitution, conferring the largest increase in UUG:AUG among Loop 2 mutants, and comparable to that given by helix α1 substitution K60E (Fig. 2E and SI Appendix, Fig. S3), displayed a weak His+ phenotype (SI Appendix, Fig. S4D). It is noteworthy that the His+ phenotype is dependent on Gcn4-mediated derepression of HIS4 transcription (19), and most Sui− mutations, including eIF1 K60E (12), confer constitutive derepression of HIS4 mRNA owing to their Gcd− phenotypes. However, as shown in eIF1 Loop 2 Substitutions Do Not Impair eIF1–40S Interaction in Vitro or Reduce TC Recruitment in Vivo, the Loop 2 mutations do not confer a Gcd− defect, which might also contribute to their general lack of His+ phenotypes.
We showed previously that overexpressing eIF5 from a high-copy (hc) TIF5 plasmid accentuates the His+ phenotype of eIF1 Sui− mutations (6). Consistent with this, introducing the hc TIF5 plasmid into the subset of Loop 2 mutants with D71 or M74 substitutions conferred weak to moderate His+ phenotypes specifically for the mutants exhibiting the largest increases in UUG:AUG ratio, namely D71A-M74A and the single- and double-Arg substitutions (SI Appendix, Fig. S4A, row 8 and SI Appendix, Fig. S4B, rows 4, 6, and 8). These findings support the conclusion that the Arg substitutions confer stronger Sui− phenotypes compared with the corresponding Ala substitutions, and confirm the additive effects of D71 and M74 substitutions in relaxing discrimination against UUG codons. Together, our results support the notion that Loop 2 residues D71 and M74 impose electrostatic or steric impediments to accommodation of Met-tRNAi in the closed/PIN conformation, and further suggest that introducing basic side chains at these positions stabilizes rather than impedes the PIN state to enhance selection of near-cognate UUG codons. Presumably, the side chain of E76 maintains Loop 2 in the conformation or position necessary for the clash between D71/M74 and Met-tRNAi.
eIF1 Loop 2 Sui− Substitutions Reduce Discrimination Against the eIF1 AUG Codon in Suboptimal Context.
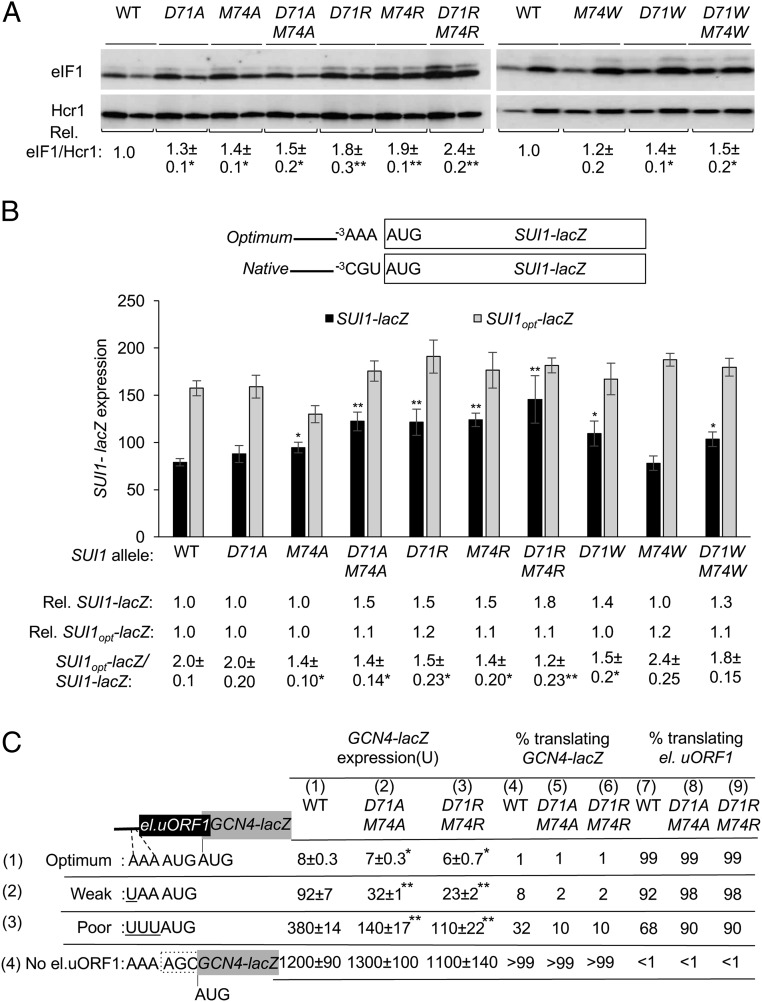
In addition to discriminating against near-cognate start codons, eIF1 also discriminates against the poor Kozak context present at its own start codon at the SUI1 gene, which underlies negative autoregulation of eIF1 expression (13, 14). Thus, in addition to increasing initiation at near-cognate UUG codons, many Sui− mutations also increase eIF1 abundance by enhancing initiation at its poor-context AUG codon (13). Consistent with these previous findings, the Loop 2 substitutions of D71 or M74 that increase the UUG:AUG ratio also increase eIF1 abundance, with the double-Ala and double-Arg substitutions having greater effects than the corresponding single mutants, and D71W and D71W/M74W exceeding the effect of M74W, which is closest to WT in eIF1 expression (Fig. 3A). Furthermore, all of these substitutions, except D71A and M74W, increase expression of the SUI1-lacZ fusion containing the native, poor context of the SUI1 AUG codon, −3CGU−1, but not that of a modified SUI1-lacZ fusion containing optimum context, −3AAA−1 (Fig. 3B). In WT cells, the latter SUI1opt-lacZ fusion is expressed at approximately twofold-higher levels than the native SUI1-lacZ fusion. Importantly, the SUI1opt-lacZ:SUI1-lacZ expression ratio is significantly diminished by all six Loop 2 eIF1 substitutions found to increase expression of the native SUI1-lacZ reporter (Fig. 3B). Similar reductions in discrimination against poor context were obtained for Loop 2 substitutions E76A and E76R, but not for E73A and E73R (SI Appendix, Fig. S5A), in accordance with the ability of E76, but not E73, replacements to increase UUG initiation (Fig. 2 D and E). It is noteworthy that the strongest reductions in the SUI1opt-lacZ:SUI-lacZ expression ratio were given by the double-Arg replacement D71R-M74R and the E76R substitution (Fig. 3B and SI Appendix, Fig. S5A), which also conferred the greatest increases in UUG initiation among this group of Loop 2 mutants (Fig. 2 C–E). We conclude that the Loop 2 substitutions reduce discrimination against the poor context of the eIF1 start codon to an extent that largely parallels their ability to increase UUG initiation.
Fig. 3.
eIF1 Loop 2 substitutions decrease discrimination against AUG codons in suboptimal context. (A) Derivatives of sui1Δ his4-301 strain JCY03 containing the indicated SUI1 alleles were cultured in synthetic dextrose minimal medium supplemented with His, Trp, and Ura at 30 °C to A600 of ∼1.0, and whole cell extracts were subjected to Western blot analysis using antibodies against eIF1 and Hcr1 (loading control). Two amounts of each extract, differing by a factor of two, were loaded in successive lanes. eIF1 Western signals were normalized to those for Hcr1, and mean values (±SEM) were calculated from three biological replicates. (B) Same strains as in A but harboring sc plasmids (pPMB24 or pPMB25) with SUI1-lacZ fusions with native suboptimal (−3CGU−1) or optimum (−3AAA−1) AUG contexts were cultured and assayed for β-galactosidase activities as in Fig. 2C. Mean expression levels and SEMs were calculated from six transformants, and relative (Rel.) mean expression levels normalized to that of the WT strain are listed Below, along with expression ratios for the SUI1-lacZ versus SUI1-opt-lacZ reporters. (C) Transformants of JCY03 harboring WT SUI1, sui1-D71A-M74A or sui1-D71R-M74R, and el.uORF1 GCN4-lacZ reporters (pC3503, pC3502, or pC4466) containing the depicted optimum (row 1), weak (row 2), or poor (row 3) context of uAUG-1, or an uORF-less GCN4-lacZ reporter with a mutated uAUG-1 (pC3505, row 4), were assayed for β-galactosidase activities as in Fig. 2C. Mean expression values with SEMs were determined from six transformants (columns 1, 2 and 3). The percentages of scanning ribosomes that translate el.uORF1 (columns 7, 8, and 9) or leaky-scan uAUG-1 and translate GCN4-lacZ instead (columns 4, 5, and 6) were calculated from results in columns 1, 2, and 3 by comparing the amount of expression observed for each uORF-containing reporter to the uORF-less construct, as described in SI Appendix, Fig. S4 C and D. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
To confirm that eIF1 Loop 2 substitutions reduce discrimination against AUGs in poor context, we asked whether they increase recognition of the suboptimal AUG codon of a uORF, and thereby decrease expression of the downstream ORF encoded on the same mRNA. To this end, we assayed expression of GCN4-lacZ reporters containing a modified uORF1 elongated to overlap the GCN4 ORF (el.uORF1). With the WT (optimal) context −3AAA−1 at el.uORF1, virtually all scanning ribosomes recognize its AUG codon (uAUG-1) and, because reinitiation at the downstream GCN4 ORF following el.uORF translation is nearly nonexistent, GCN4-lacZ expression is extremely low (20). In WT cells, replacing only the optimal A with U at the -3 position of el.uORF1 increases leaky scanning of uAUG-1 to produce an approximately eightfold increase in GCN4-lacZ translation. Introducing the poor context of −3UUU−1 further increases leaky scanning, for an approximate 32-fold increase in GCN4-lacZ expression; and eliminating uAUG-1 altogether increases GCN4-lacZ expression by >100-fold (Fig. 3C, column 1, rows 1, 2, 3, and 4 and SI Appendix, Fig. S5 B and D). From these results, the percentages of scanning ribosomes that either translate el.uORF1 or bypass (leaky-scan) uAUG-1 and translate GCN4-lacZ instead can be calculated, revealing that >99%, ∼92%, and ∼68% of scanning ribosomes recognize uAUG-1 in optimum, weak, and poor context, respectively, in WT cells (Fig. 3C, columns 4 and 7 and SI Appendix, Fig. S5 C and D).
The potent eIF1 Loop 2 substitution D71R-M74R reduces leaky scanning of uAUG-1, as indicated by significantly reduced GCN4-lacZ expression, for all three reporters containing el.uORF1, but not for the uORF-less reporter (Fig. 3C, cf. columns 1 and 3, rows 1, 2, 3, and 4 and SI Appendix, Fig. S5 B and D). However, calculating the percentages of ribosomes that recognize uAUG-1 revealed that D71R-M74R confers a substantial increase in recognition of uAUG-1 only for the poor-context (UUU) reporter, from ∼68 to ∼90%, while producing a moderate increase for the weak-context (UAA) reporter, from ∼92 to ∼98%, but <1% increase for the optimal-context (AAA) reporter, (Fig. 3C, cf. columns 7 and 9, rows 1, 2, and 3 and SI Appendix, Fig. S5C). Very similar results were obtained for the D71A-M74A substitutions (Fig. 3C, cf. columns 7 and 8, rows 1, 2, and 3 and SI Appendix, Fig. S5C); moreover, D71R, M74R, and E76R increased recognition of uAUG-1 in both poor context and weak context to an extent comparable to that observed for the two D71-M74 double substitutions (cf. Fig. 3C and SI Appendix, Fig. S6 A–D). Together, the results show that the Loop 2 substitutions reduce discrimination against poor context, both at the SUI1 AUG codon and uAUG-1 of GCN4 mRNA, to an extent that generally parallels their effects in relaxing discrimination against UUG start codons at HIS4.
eIF1 Loop 2 Substitutions Do Not Impair eIF1–40S Interaction in Vitro or Reduce TC Recruitment in Vivo.
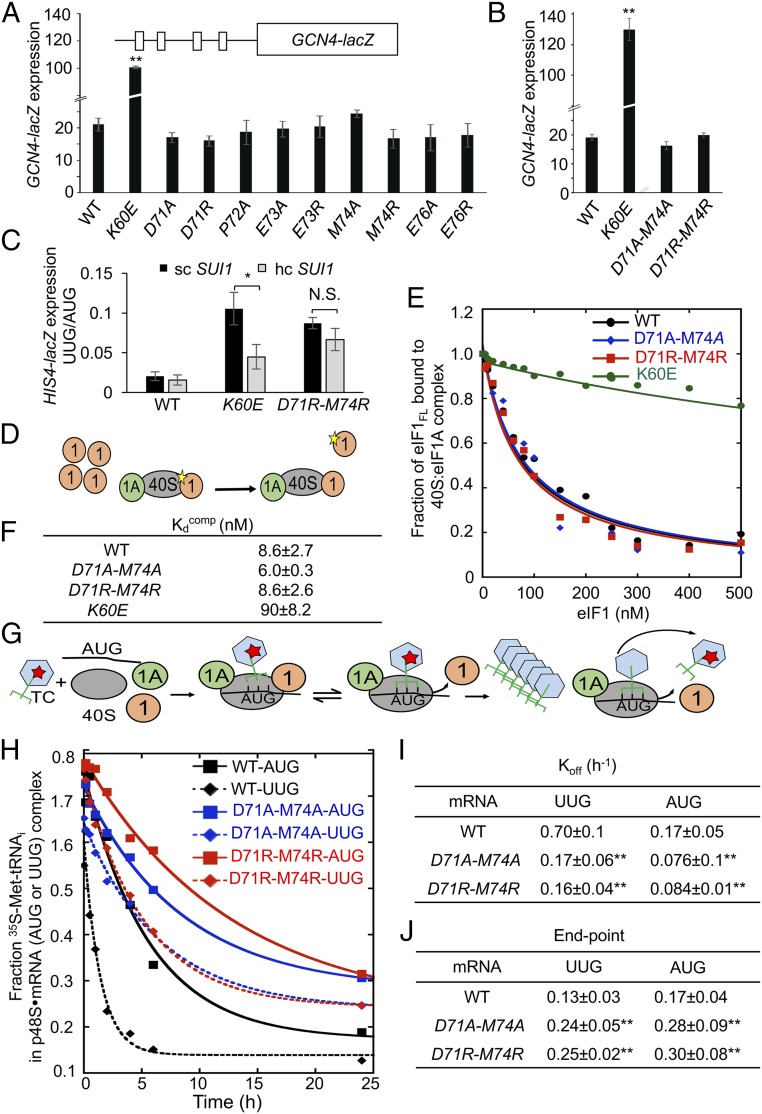
We showed previously that substituting basic residues in helix α1 or Loop 1 that impair eIF1 binding to 40S subunits in vitro confer the dual Gcd− and Sui− phenotypes in vivo that signify a shift from the open to closed conformation of the PIC. The Gcd− phenotype—derepressed expression of GCN4—results from a reduced rate of TC binding to 40S subunits scanning the GCN4 mRNA leader after translating uORF1, which allows them to bypass the remaining inhibitory uORFs and reinitiate at the GCN4 start codon at elevated levels, even in the absence of eIF2α phosphorylation and attendant reduced TC formation triggered by amino acid starvation (21). Because eIF1 enhances TC binding to the open conformation of the PIC (2), the Gcd− phenotype of eIF1 α1/Loop 1 variants has been attributed to diminished eIF1 occupancy on 40S subunits and the resulting slower rate of TC binding to 40S subunits scanning the GCN4 mRNA leader. The weaker eIF1 interaction with 40S subunits also allows more frequent eIF1 dissociation during scanning and inappropriate transition to the closed/PIN conformation at UUG codons for the Sui− phenotype. Supporting these interpretations, overexpressing eIF1 α1 and Loop 1 variants suppresses both their Gcd− and Sui− phenotypes (12).
We reasoned that if the eIF1 Loop 2 substitutions increase UUG initiation by removing a barrier to the closed/PIN state, rather than weakening eIF1 binding to the 40S subunit, then they should not reduce the rate of TC binding to scanning 40S subunits in the open conformation and confer a Gcd− phenotype. Moreover, overexpressing Loop 2 variants should not substantially diminish the elevated UUG initiation observed in these Sui− mutants. Indeed, none of the Loop 2 substitutions conferred a detectable increase in GCN4-lacZ expression (Fig. 4 A and B), including D71R-M74R that produced the largest increase in UUG initiation for this group of mutants (Fig. 2C). By contrast, as shown previously (12), the α1 substitution K60E strongly derepresses GCN4-lacZ expression (Fig. 4B) while conferring an increase in UUG:AUG initiation comparable to that of D71R-M74R (Fig. 2 C and E). Furthermore, overexpressing the D71R-M74R mutant had relatively little effect on the increased UUG:AUG ratio conferred by this Loop 2 variant, in contrast to the more extensive suppression of the Sui− phenotype of the K60E variant on its overexpression (Fig. 4C). These genetic findings support the idea that Loop 2 substitutions increase UUG initiation primarily by decreasing the barrier to the PIN conformation imposed by the eIF1–tRNAi clash rather than by weakening eIF1 binding to the 40S subunit.
Fig. 4.
eIF1 Loop 2 substitutions do not impair eIF1–40S interaction or reduce TC recruitment in vivo but stabilize the closed/PIN conformation of the 48S PIC at UUG codons in vitro. (A and B) Transformants of JCY03 containing the indicated SUI1 alleles and a GCN4-lacZ reporter (p180) were assayed for β-galactosidase activities as in Fig. 2C. Mean expression levels and SEMs calculated from six to eight transformants of each strain are plotted. (C) Transformants of JCY03 containing WT SUI1 on an sc (pJCB01) or hc (pCFB04), K60E on an sc (pPMB02) or hc (pPMB39), and D71R-M74R on an sc (ATP98) or hc (ATP169) plasmid, and HIS4-lacZ reporters with AUG or UUG start codons, were assayed for β-galactosidase activities as in Fig. 2C. Asterisks in A–C indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01). (D–F) Measurement of eIF1 binding constants. Fluorescein-labeled WT eIF1 (5 nM) was prebound to 40S subunits (15 nM) in the presence of eIF1A (1 µM), mixed with increasing concentrations of unlabeled WT eIF1, eIF1-D71A-M74A, or eIF1-D71R-M74R, and the change in fluorescence anisotropy measured (D). One of two replicate experiments (E) from which mean Kdcomp values and average deviations were calculated (F). (G–J) Measurement of TC dissociation kinetics. Partial 48S complexes were assembled with radiolabeled WT TC, eIF1A, and model mRNA containing an AUG or UUG start codon, and with WT or mutant eIF1 proteins; they were chased with excess unlabeled TC for increasing periods of time; and the fraction of labeled Met-tRNAi bound to the PIC was determined (G). One of four replicate experiments (H) from which mean rate constants (I) and end points (J) (with SEMs) were calculated. Asterisks in I and J indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
To provide biochemical evidence that Loop 2 substitutions do not perturb eIF1–40S interaction, we compared the ability of purified recombinant forms of these eIF1 variants to compete with WT eIF1 for 40S binding in vitro. Partial PICs containing fluorescently labeled WT eIF1 (eIF1FL) and eIF1A bound to 40S subunits were challenged with increasing concentrations of unlabeled mutant or WT eIF1 proteins, and the fraction of eIF1FL bound to 40S–eIF1A complexes at each competitor concentration was determined by monitoring changes in fluorescence anisotropy (Fig. 4D). In accordance with previous results (12), the helix α1 variant K60E competed poorly with WT eIF1FL for binding to 40S–eIF1A complexes, increasing the eIF1 dissociation constant (Kdcomp) by ∼10-fold. By contrast, the Loop 2 mutants D71A-M74A and D71R-M74R competed with WT eIF1FL indistinguishably from that seen for WT unlabeled eIF1, indicating no significant change in their affinity for 40S–eIF1A complexes (Fig. 4 E and F). These findings support the conclusion that the relaxed discrimination against poor initiation sites conferred by Loop 2 substitutions does not involve weaker eIF1 interaction with the 40S subunit.
eIF1 Loop 2 Substitutions D71A-M74A and D71R-M74R Promote Met-tRNAi Accommodation in the PIN Conformation of the 48S PIC at UUG Codons in Vitro.
To provide biochemical evidence that the Loop 2 substitutions facilitate isomerization of Met-tRNAi to the PIN state, we measured the rate of TC dissociation from reconstituted PICs in vitro. Partial 43S–mRNA complexes (lacking eIF3 and eIF5; henceforth dubbed p48S PICs) were formed by incubating WT TC (assembled with [35S]-Met-tRNAi and the nonhydrolyzable GTP analog GDPNP) with saturating amounts of eIF1A; WT or mutant eIF1; an uncapped, unstructured model mRNA containing either AUG or UUG start codon [mRNA(AUG) or mRNA(UUG)]; and 40S subunits. p48S PICs containing [35S]-Met-tRNAi were incubated for increasing time periods in the presence of excess unlabeled TC (chase) and resolved via native gel electrophoresis to separate 40S-bound and unbound fractions of TC (Fig. 4G). Previous work indicated that TC bound in the POUT conformation is too unstable to remain associated with the PIC during native gel electrophoresis, such that the measured rate of TC dissociation in these assays largely reflects the proportion of complexes in the PIN state and the stability of that state (22, 23). In agreement with previous findings (22), we observed that TC dissociates more rapidly from the mRNA(UUG) versus mRNA(AUG) PICs in reactions containing WT eIF1 (Fig. 4 H and I), reflecting the reduced formation and relative instability of the PIN state at near-cognate UUG versus AUG codons. Strikingly, replacing WT eIF1 with Loop 2 variants D71A-M74A or D71R-M74R decreased the rate of TC dissociation (Koff) by greater than fourfold from PICs assembled on mRNA(UUG) (Fig. 4 H and I). Additionally, the extent of TC dissociation from the UUG complexes, reflected in end points of the reactions, was increased by the Loop 2 substitutions, indicating increased formation of a hyperstable complex from which no TC dissociation occurs on the time scale of these experiments (23) (Fig. 4 H and J). Using mRNA(AUG), the Loop 2 substitutions also increased formation of the hyperstable complex but produced a relatively smaller reduction in Koff (approximately twofold) compared with that observed for the corresponding mRNA(UUG) PICs (Fig. 4 H–J). Together, these findings support the notion that the Loop 2 substitutions reduce a barrier to isomerization from POUT to PIN, particularly at near-cognate UUG codons, in accordance with the increased initiation at UUG codons they produce in vivo.
eIF1 Loop 2 and α1 Substitutions Confer Additive Increases in Selection of Suboptimal Start Codons in Vivo.
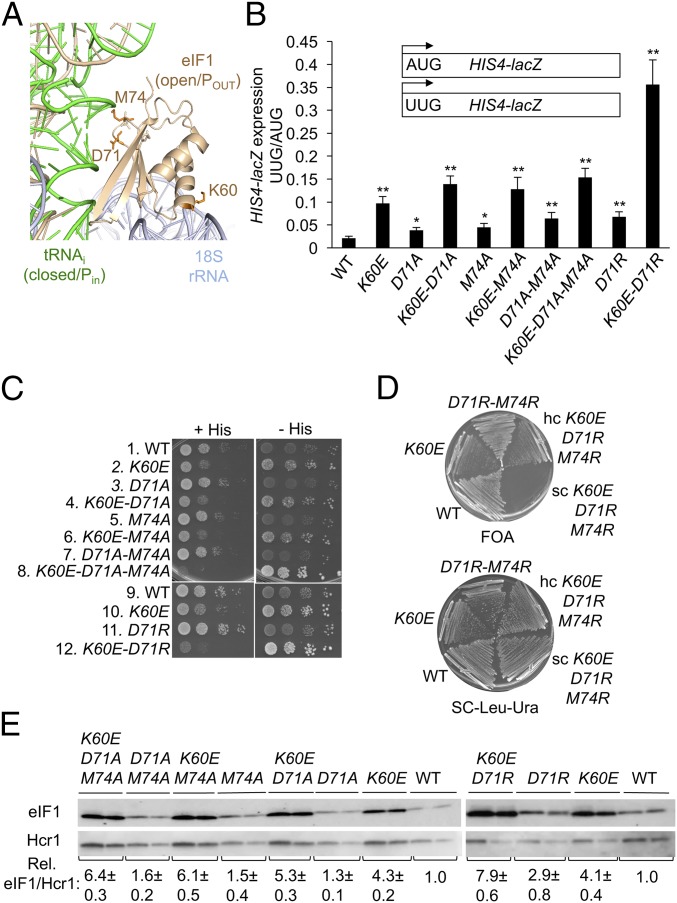
The results presented above indicate that the eIF1 Loop 2 substitutions reduce discrimination against suboptimal initiation codons by altering eIF1–Met-tRNAi interactions to facilitate transition of the scanning PIC to the closed/PIN conformation required for start codon recognition. This mechanism differs from that proposed for other eIF1 substitutions analyzed previously, such as K60E, which eliminate eIF1 contacts with the 40S subunit and thereby increase inappropriate eIF1 dissociation from the scanning PIC at suboptimal start sites (12). We reasoned, therefore, that combining Loop 2 substitutions with K60E would increase selection of both UUG codons and AUGs in poor context by compounding perturbations of eIF1–Met-tRNAi interactions with defects in eIF1–40S interaction mediated by distinct eIF1 interfaces in the PIC (Fig. 5A).
Fig. 5.
eIF1 Loop 2 and α1 substitutions confer additive increases in suboptimal start codon utilization in vivo. (A) Side chains of eIF1 residues in Loop 2 (D71, M74) or helix α1 (K60) substituted in this study shown as sticks. (B) Transformants of JCY03 containing the indicated SUI1 alleles and HIS4-lacZ reporters were assayed for β-galactosidase activities as in Fig. 2C. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01). (C) Tenfold serial dilutions of JCY03 derivatives with the indicated SUI1 alleles were spotted on synthetic complete medium lacking leucine (SC-Leu) supplemented with 0.3 mM His and incubated at 30 °C for 2 d (+His), or on SC-Leu plus 0.003 mM His (−His) for 5 to 6 d. (D) JCY03 derivatives containing the indicated SUI1 alleles sc or hc plasmids were streaked on SC-Leu supplemented with 5.75 mM FOA (Upper), or SC-Leu-Ura (Lower), and incubated at 30 °C for 5 d or 2 d, respectively. (E) eIF1 expression in derivatives of JCY03 with the indicated SUI1 alleles measured by Western blot analysis as in Fig. 3A.
These expectations were borne out by our findings that combining K60E with single- or double-Ala substitutions of D71 and M74 increased the HIS4-lacZ UUG:AUG initiation ratio above that conferred by K60E, D71A, M74A, or D71A-M74A alone (Fig. 5B). This effect was also observed on combining K60E and D71R, which dramatically increased the UUG:AUG ratio to ∼0.40 compared with ratios of 0.05 to 0.1 for the two single mutants (Fig. 5B). Combining K60E with D71A-M74A or D71R also reduced growth on medium containing histidine while enhancing growth on –His medium, signifying a marked increase in the Sui− phenotype of the resulting triple and double mutants compared with strains harboring the K60E or Loop 2 substitutions alone (Fig. 5C, rows 2, 7, 8, 10, 11, and 12). Furthermore, combining the K60E and D71R-M74R mutations was lethal, as the triple-mutant allele did not permit eviction of the WT SUI1, URA3 plasmid on FOA medium (Fig. 5D). The lethality was overcome by introducing the triple-mutant allele on an hc plasmid; although the resulting strain still grew poorly following eviction of WT SUI1 on FOA medium (Fig. 5D). The rescue of lethality by overexpression can be explained as the result of mitigating the 40S binding defect produced by K60E through mass action, as observed for other lethal substitutions at the eIF1–40S interface (12). Lastly, combining K60E with M74A-D71A and D71R led to higher levels of eIF1 expression than observed for any of these substitutions on their own (Fig. 5E).
Discussion
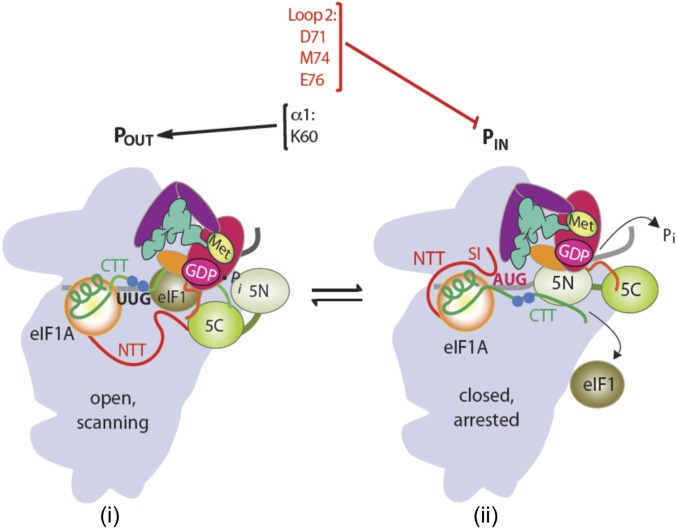
The eIF1 plays a central role in promoting ribosomal scanning and reducing selection of both non-AUG start codons and AUGs in poor context, both in reconstituted systems and in vivo. At the biochemical level, eIF1 promotes TC recruitment in the POUT state of the scanning PIC while destabilizing Met-tRNAi binding in the PIN conformation to help restrict Pi release from eIF2-GDP–Pi (an irreversible step) to AUG codons in optimum context (Fig. 6). These impediments to start codon recognition require the dissociation of eIF1 from the PIC (1), an assertion supported by the fact that eIF1 substitutions at its 40S interface that weaken 40S binding invariably increase utilization of poor initiation sites in cells, whereas eIF1 substitutions that strengthen 40S binding confer the opposite hyperaccuracy phenotype (7, 12). Structural evidence suggests that eIF1 function in restricting the PIN state involves a physical clash between eIF1 and Met-tRNAi (9, 10), and overlays of eIF1 in partial yeast 48S PICs deemed to represent open or closed PIC conformations suggest that PIN binding requires lateral displacement of eIF1 on the 40S platform and deformation of its β-hairpin Loops 1 and 2 to avoid clashes with the anticodon and D loops of Met-tRNAi, respectively (11, 17) Until now, genetic and biochemical support for this appealing model has been lacking.
Fig. 6.
Distinct eIF1 domains mediate its dual roles of promoting the open conformation and impeding the closed conformation within the scanning PIC. Transition from the open, scanning conformation with Met-tRNAi bound in the POUT state to the closed/PIN conformation is depicted as in Fig. S1A. Residues including K60 in helix α1 promote the open conformation by anchoring eIF1 to the 40S subunit, whereas Loop 2 residues D71, M74, and E76 oppose transition to the closed/PIN state by promoting the clash between eIF1 Loop 2 bound in the open conformation and Met-tRNAi bound in the closed/PIN state.
Our previous work demonstrating that replacing conserved basic residues in eIF1 Loop 1 increases initiation at poor start sites in vivo (12) is consistent with the foregoing model; however, these substitutions also disrupt eIF1 contacts with the 40S subunit, which is sufficient to decrease initiation accuracy (12). Because Loop 2 does not contact the 40S subunit, we could test the functional importance of the predicted eIF1 clash with Met-tRNAi without the complication of impaired eIF1–40S interaction. Importantly, Ala substitutions of conserved acidic Loop 2 residues D71 and E76, and conserved bulky hydrophopic residue M74, all conferred the hypoaccuracy phenotypes found previously for Loop 1 or α1 substitutions, increasing initiation at both a UUG codon at HIS4 and the AUG codons at SUI1 or GCN4 uORF1 when they reside in poor context. These are the results expected from diminishing a steric clash or electrostatic repulsion of Loop 2 with the Met-tRNAi D loop. The fact that D71A and D71N have similar phenotypes suggests that electrostatic repulsion is the critical contribution of the D71 acidic side chain to initiation accuracy. Our finding that even stronger hypoaccuracy phenotypes were conferred by Arg replacements of D71 and M74 suggests that replacing a clash/repulsion with electrostatic attraction between Loop 2 and Met-tRNAi promotes, rather than antagonizes, rearrangement to the PIN state. We demonstrated in the reconstituted system that exemplar Loop 2 substitutions do not reduce eIF1 affinity for 40S–eIF1A complexes, eliminating this complication to the interpretation of hypoaccuracy phenotypes. They additionally increased the stability of Met-tRNAi binding to partial 48S complexes, especially at UUG codons, as expected for increased accommodation of Met-tRNAi in the closed/PIN state in the absence of a perfect codon:anticodon match, helping to explain their increased UUG initiation in vivo. Consistent with their robust 40S interaction, the Loop 2 variants do not confer the Gcd− phenotype, which signifies a slower rate of TC recruitment to the open conformation of the PIC, and is conferred by the reduced 40S occupancy of Loop 1 and α1 eIF1 variants (12). Thus, whereas Loop 1/α1 substitutions, like K60E, specifically disfavor TC binding in the open/POUT state and indirectly shift the system toward the closed/PIN state, the Loop 2 substitutions remove a barrier to the closed/PIN conformation without affecting the rate or stability of TC binding in the open/POUT state (Fig. 6). These findings place eIF1 Loop 2 in the same functional category as the conserved G31:C39 base pair in the anticodon stem-loop of tRNAi whose substitution with different Watson–Crick base pairs confers Sui−, but not Gcd−, phenotypes (23).
It is possible that the Loop 2 clash with Met-tRNAi is instrumental in not only restricting accommodation of Met-tRNAi in the PIN state, but in subsequent dissociation of eIF1 from the 40S subunit to gate Pi release. If so, our Ala substitutions in Loop 2 would have counteracting effects on start codon selection. The fact that we observe increased initiation at poor start codons in these mutants implies that removing the impediment to the PIN state has a greater effect than the possible reduction in eIF1 dissociation conferred by diminishing the clash/repulsion of Loop 2 with Met-tRNAi. However, it remains possible that the hypoaccuracy phenotypes of the Loop 2 mutations are being dampened by their opposing effects on eIF1 release. This possibility is consistent with our finding that the hypoaccuracy phenotypes of Loop 2 substitutions are exacerbated by the α1 K60E substitution, which eliminates a key 40S contact made by eIF1 (12) and, hence, should neutralize the putative effect of Loop 2 substitutions in reducing eIF1 dissociation from the 40S subunit. Alternatively, the stronger hypoaccuracy phenotypes on combining K60E and Loop 2 substitutions could arise simply from the compound effects of mutations that enable poor start codon selection by independent mechanisms. If so, the fact that the Loop 2 substitutions appear to have a greater effect on the transition to PIN versus subsequent eIF1 release might indicate that other perturbations in the eIF1–40S interaction that accompany transition to PIN are sufficient to trigger eIF1 dissociation from the closed/PIN state. These include the distortion of Loop 1, expected to eliminate a subset of its interactions with the 40S subunit (11), and conformational changes in h44 near the eIF1 binding site (10).
Recent high-resolution structures of eIF2D bound to 40S ribosomes containing Met-tRNAi base-paired to the AUG codon of the hepatitis C virus (HCV) internal ribosome entry site (IRES) reveal that its SUI domain, related to eIF1 in sequence and tertiary structure, occupies the same position as eIF1 on the 40S subunit (24). The same was found for the SUI domain in the C-terminal half of the DENR protein (25) that forms a heterodimer with MCT-1 to constitute a functional homolog of eIF2D. eIF2D and DENR/MCT-1 can substitute for eIF2 in recruitment of Met-tRNAi to specialized mRNAs in vitro, such as the HCV IRES, which place the AUG codon directly in the P site without scanning (26, 27), and they might function primarily in cells under stress conditions when eIF2 is inactivated by phosphorylation. Interestingly, the SUI domains of both eIF2D and DENR contain a basic Loop 1 counterpart positioned near the codon:anticodon duplex (24), which provides one of several contacts with different portions of Met-tRNAi that appear to stabilize an orientation of tRNAi distinct from that seen in the yeast partial 48S PIC (11). The SUI domain of eIF2D also contains a counterpart of Loop 2 that is larger and less acidic than eIF1 Loop 2, which likewise seems to stabilize the distinctive orientation of Met-tRNAi in this complex (24). By contrast, Loop 2 is essentially missing in DENR (25). These findings, plus the fact that these proteins are dependent on having the AUG codon positioned in the P site by the mRNA, suggest that Loops 1 and/or 2 of eIF2D and Loop 1 of DENR act primarily to stabilize Met-tRNAi binding to the P site when they are substituting for canonical eIFs during initiation and may have no role in discriminating against non-AUG start codons in the manner we established for their counterparts in eIF1.
In summary, our results provide genetic and biochemical evidence that the physical juxtaposition of eIF1 Loop 2 and D loop of tRNAi visualized in structures of partial yeast 48S PICs has no role in anchoring eIF1 or in recruiting TC to the open, scanning conformation of the PIC—functions attributed to eIF1 Loop 1 and helix α1. Rather, a clash between Loop 2 and the D loop predicted from comparing PICs in different conformational states acts to oppose accommodation of Met-tRNAi in the PIN state and restrict its occurrence to optimal initiation sites. This function exceeds any possible contribution of the Loop 2–D loop clash in driving eIF1 dissociation from the PIC, which promotes rather than impedes start codon selection. The Loop 2–D loop clash can be added to the network of molecular interactions occurring among different initiation factors, ribosomal proteins, rRNA residues, mRNA, and tRNAi that collaborate to ensure the optimum level of initiation accuracy in living cells (28).
Materials and Methods
Plasmid and Yeast Strain Constructions.
Yeast strains used in this study are listed in SI Appendix, Table S1. Derivatives of strain JCY03 [MATa ura3-52 leu2-3 leu2-112 trp1Δ-63 his4-301(ACG) sui1Δ::hisG p1200 (sc URA3 SUI1)], strains ATY100 to ATY106, ATY112 to ATY119, ATY130 to ATY137, and ATY160 to ATY164, were constructed by transforming JCY03 to Leu+ with sc or hc LEU2 plasmids harboring the appropriate SUI1 alleles (indicated in SI Appendix, Table S2) on synthetic complete medium (SC) lacking leucine (SC-Leu), and the resident SUI1+ URA3 plasmid (p1200) was evicted by selecting for growth on FOA medium. Derivatives of strain ATY138 through ATY157 containing plasmid-borne TIF5 or empty vector were generated by transformation and selection on SC lacking leucine and tryptophan (SC-Leu-Trp). The QuikChange site-directed mutagenesis system (Stratagene) was employed with primers indicated in SI Appendix, Table S3 to generate all of the corresponding plasmids shown in SI Appendix, Table S1 using as templates plasmids pJCB101, pCFB04, and pTYB2-eIF1.
Biochemical Assays Using Yeast Cell Extracts.
Assays of β-galactosidase activity in whole cell extracts (WCEs) were performed as described previously (29). For Western blot analysis, WCEs were prepared by trichloroacetic acid extraction as previously described (30), and immunoblot analysis was conducted as previously described (6) using antibodies against eIF1/Sui1 (31) and Hcr1 (31). Enhanced chemiluminiscence (Amersham) was used to visualize immune complexes, and signal intensities were quantified by densitometry using NIH ImageJ software.
Biochemical Analysis in the Reconstituted Yeast System.
WT eIF1 and eIF1 variants D71A-M74A, D71R-M74R, and K60E were expressed in bacterial strain BL21(DE3) Codon Plus cells (Agilent Technologies) and purified using the IMPACT system (New England Biolabs) as described previously (32). His6-tagged WT eIF2 was overexpressed in yeast and purified as described in ref. 32. 40S subunits were purified as described previously from strain YAS2488 (32). Model mRNAs with sequences 5′-GGAA[UC]7UAUG[CU]10C-3′ and 5′-GGAA[UC]7UUUG[CU]10C-3′ were purchased from Thermo Scientific. Yeast tRNAiMet was synthesized from a hammerhead fusion template using T7 RNA polymerase, charged with [35S]-methionine, and used to prepare radiolabeled eIF2–GDPNP–[35S]-Met-tRNAi ternary complexes ([35S]-TC), all as previously described (32). Yeast Met-tRNAiMet was purchased from tRNA Probes; LLC. For eIF1 binding competition experiments, WT eIF1 protein was labeled at its C terminus with Cys-Lys-ε-fluorescein dipeptide, using the expressed protein ligation system, as previously described (33).
TC dissociation rate constants (Koff) were measured by monitoring the amount of [35S]-TC that remains bound to 40S–eIF1–eIF1A–mRNA (43S–mRNA) complexes over time, in the presence of excess unlabeled TC (chase), using a native gel shift assay to separate 40S-bound from unbound [35S]-TC. 43S–mRNA complexes were preassembled for 2 h at 26 °C in reactions containing 40S subunits (20 nM), eIF1 (1 µM), eIF1A (WT or mutant variants, 1 µM), mRNA (10 µM), and [35S]-TC (0.25 µM eIF2, 0.1 mM GDPNP, and 1 nM [35S]-Met-tRNAi) in 60 µL of reaction buffer [30 mM Hepes–KOH (pH 7.4), 100 mM potassium acetate (pH 7.4), 3 mM magnesium acetate, and 2 mM DTT]. To initiate each dissociation reaction, a 6 µL-aliquot of the preassembled 43S–mRNA complexes was mixed with 3 µL of threefold-concentrated unlabeled TC chase (comprising 2 µM eIF2, 0.3 mM GDPNP, and 0.9 µM Met-tRNAi), representing a 300-fold excess over labeled TC in the final dissociation reaction, and incubated for the prescribed period of time. A converging time course was employed so that all dissociation reactions were terminated simultaneously by the addition of native-gel dye and loaded directly on a running native gel. The fraction of [35S]-Met-tRNAi remaining in 43S complexes at each time point was determined by quantifying the 40S-bound and unbound signals by Phosphor Imaging, normalized to the ratio observed at the earliest time point, and the data were fit with a single exponential equation (22).
Fluorescence anisotropy measurements of equilibrium binding constants (Kd) for eIF1 binding to 40S–eIF1A complexes were performed using a T-format Spex Fluorolog-3 (Horiba/Jobin Yvon) as described previously (33). The excitation and emission wavelengths were 497 and 520 nm, respectively. The data were fit with a quadratic equation describing the competitive binding of two ligands to a receptor, as previously described (12).
Supplementary Material
Acknowledgments
We thank Jagpreet Nanda, Jinsheng Dong, Laura Marler, Fan Zhang, and Jon Lorsch for advice and assistance in using the yeast reconstituted system; all members of our laboratory; and members of the Jon Lorsch and Tom Dever laboratories for invaluable suggestions. This work was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800938115/-/DCSupplemental.
References
- 1.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 5.Cheung YN, et al. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanda JS, et al. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Marcos P, et al. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA. 2014;20:150–167. doi: 10.1261/rna.042069.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 10.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain T, et al. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Marcos P, et al. β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J Biol Chem. 2013;288:27546–27562. doi: 10.1074/jbc.M113.498642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Marcos P, Cheung YN, Hinnebusch AG. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol. 2011;31:4814–4831. doi: 10.1128/MCB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci USA. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alone PV, Cao C, Dever TE. Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding. Mol Cell Biol. 2008;28:6877–6888. doi: 10.1128/MCB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llácer JL, et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donahue T. Genetic approaches to translation initiation. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Saccharomyces cerevisiae. Translational Control of Gene Expression. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2000. pp. 487–502. [Google Scholar]
- 19.Yoon HJ, Donahue TF. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant CM, Miller PF, Hinnebusch AG. Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA varies with the downstream cistron. Mol Cell Biol. 1994;14:2616–2628. doi: 10.1128/mcb.14.4.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 22.Kolitz SE, Takacs JE, Lorsch JR. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA. 2009;15:138–152. doi: 10.1261/rna.1318509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong J, et al. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev. 2014;28:502–520. doi: 10.1101/gad.236547.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisser M, et al. Structural and functional insights into human re-initiation complexes. Mol Cell. 2017;67:447–456.e7. doi: 10.1016/j.molcel.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 25.Vaidya AT, Lomakin IB, Joseph NN, Dmitriev SE, Steitz TA. Crystal structure of the C-terminal domain of human eIF2D and its implications on eukaryotic translation initiation. J Mol Biol. 2017;429:2765–2771. doi: 10.1016/j.jmb.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dmitriev SE, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skabkin MA, et al. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch AG. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci. 2017;42:589–611. doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid GA, Schatz G. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem. 1982;257:13056–13061. [PubMed] [Google Scholar]
- 31.Olsen DS, et al. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 33.Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]