Significance
The main constituent of the ribosomal stalk is L12, a multicopy protein of the ribosomal 50S subunit that interacts with the translational GTPases in different stages of translation. Using mutagenesis, fast kinetics, and molecular dynamics simulations, we show that charge complementarity between a conserved region on the C-terminal domain of L12 and a conserved region on the G domain of initiation factor 2 (IF2) forms the basis of L12–IF2 interaction, which is crucial for fast association of the ribosomal subunits. Our analysis also suggests that L12 possibly interacts with other G factors by a similar mechanism. This study provides significant mechanistic insights into the interaction of L12 and the translational GTPases at the molecular level.
Keywords: protein synthesis, ribosomal protein L7/12, protein–protein interaction, ribosome, translation initiation
Abstract
The interaction between the ribosomal-stalk protein L7/12 (L12) and initiation factor 2 (IF2) is essential for rapid subunit association, but the underlying mechanism is unknown. Here, we have characterized the L12–IF2 interaction on Escherichia coli ribosomes using site-directed mutagenesis, fast kinetics, and molecular dynamics (MD) simulations. Fifteen individual point mutations were introduced into the C-terminal domain of L12 (L12-CTD) at helices 4 and 5, which constitute the common interaction site for translational GTPases. In parallel, 15 point mutations were also introduced into IF2 between the G4 and G5 motifs, which we hypothesized as the potential L12 interaction sites. The L12 and IF2 mutants were tested in ribosomal subunit association assay in a stopped-flow instrument. Those amino acids that caused defective subunit association upon substitution were identified as the molecular determinants of L12–IF2 interaction. Further, MD simulations of IF2 docked onto the L12-CTD pinpointed the exact interacting partners—all of which were positively charged on L12 and negatively charged on IF2, connected by salt bridges. Lastly, we tested two pairs of charge-reversed mutants of L12 and IF2, which significantly restored the yield and the rate of formation of the 70S initiation complex. We conclude that complementary charge-based interaction between L12-CTD and IF2 is the key for fast subunit association. Considering the homology of the G domain, similar mechanisms may apply for L12 interactions with other translational GTPases.
Ribosomal-stalk protein L7/L12 (referred to hereafter as L12) is a two-domain protein connected by a flexible hinge that allows it to acquire multiple conformations on the ribosome (1, 2). While the helical N-terminal domain (NTD) of L12 is involved in the formation of tight L12 dimers (1, 3), the globular C-terminal domain (CTD) of L12 (4) interacts with the translational GTPase factors (5). Because of the elastic nature of the hinge region, the L12-CTD can stretch out from the ribosome (1, 3) and interact with the translational GTPases (referred to as tr-GTPases), thereby facilitating their recruitment to the ribosome (6). Earlier NMR mapping suggests that L12 interacts with four major tr-GTPases, namely initiation factor 2 (IF2), elongation factor Tu (EF-Tu), elongation factor G (EF-G), and release factor 3 (RF3) through a highly conserved region of its CTD (5). The L12-CTD comprises two α-helices—helix 4 and helix 5 (referred to as helices 4/5)—which contain multiple conserved residues. Among these, V66, K70, L80, and E82 (Escherichia coli) were directly mapped by NMR for interactions with the translation factors (5). Moreover, mutation of some of the residues on helices 4/5 decreased the rate of A-site binding of EF-Tu ternary complex (7) and inhibited EF-G turnover on the ribosome (8). Collectively, this evidence suggests that helices 4/5 of the L12-CTD are crucial for tr-GTPase factor interaction.
On the basis of multiple-turnover GTP hydrolysis experiments with EF-Tu and EF-G on the ribosome, it has been proposed that L12 may be the GTPase-activating protein for these tr-GTPases (9). In an earlier study, even isolated L12 protein was claimed to stimulate intrinsic GTP hydrolysis by EF-G (10). It is now generally accepted that L12 protein does not per se have any stimulatory action on the GTPase activity of the tr-GTPases. We have reported that the removal of L12 proteins from the large subunit of the ribosome causes a dramatic decrease in the rate of IF2-mediated subunit association, although the actual rate of GTP hydrolysis is only marginally affected (11). Furthermore, reduction in the number of the L12 dimers on the ribosome causes slower subunit association in the presence of IF2 (12). These results suggest that a specific interaction between L12 and IF2 is essential for efficient subunit association.
The role of IF2 in initiation of protein synthesis has been extensively studied (13, 14). Its main role is to promote association of the ribosomal subunits in the GTP-bound form, especially when an initiator tRNA is present in the P-site of the mRNA programmed 30S subunit, also referred to as 30S preinitiation complex (30S preIC) (15–17). GTP hydrolysis by IF2 is needed for dissociation of IF2 from the fully formed 70S initiation complex (70S IC) (18–20). However, it is not understood how IF2•GTP promotes association of 30S preIC with 50S. Reports from our group (11, 12), as well as the cryo-EM structure of the 70S IC from E. coli (21), identified flexible protein L12 on the periphery of the 50S subunit as the likely interaction partner of IF2. The interaction could initiate subunit joining by bringing the 50S subunit closer to the IF2-bound 30S subunit. However, the molecular mechanism of L12–IF2 interaction is not known.
In this study, we introduced 15 point mutations on the helices 4/5 of L12 and reconstituted 50S subunits by adding those L12 variants to the L12-depleted 50S core. Since the helices 4/5 contain mostly positively charged amino acids, we hypothesized that the L12 interaction region on IF2 probably possesses negative charges and is homologous to helix D of EF-Tu previously identified as interacting with L12-CTD (7). With extensive bioinformatics analysis, we identified a region between the G4 and G5 motifs on the G domain of IF2 as the possible candidate region for L12 interaction. Notably, an earlier cryo-EM structure of the 70S IC (21) and a cross-linking study (22) also suggested that L12 interacts with IF2’s G domain. Following our hypothesis, we introduced 15 individual mutations on IF2 in the G4–G5 peptide.
All L12 and IF2 variants were tested in subunit association assay using Rayleigh light scattering in a stopped-flow instrument, which allowed detection of the amino acids that are crucial for L12–IF2 interaction. Further, molecular dynamics (MD) simulations of IF2 docked onto the L12-CTD pinpointed the exact interacting residues. These experiments were further supported by testing ionic-strength dependence of subunit association reaction, and by restoration of the rate and the yield of IF2-mediated subunit association with charge-reversed mutants of L12 and IF2. Our results unravel the molecular basis of interaction between L12 and IF2 and demonstrate that it is the key for fast subunit association during the initiation of protein synthesis in bacteria.
Results
Mutations in the Potential GTPase Factor Interaction Sites on Helices 4/5 of L12-CTD.
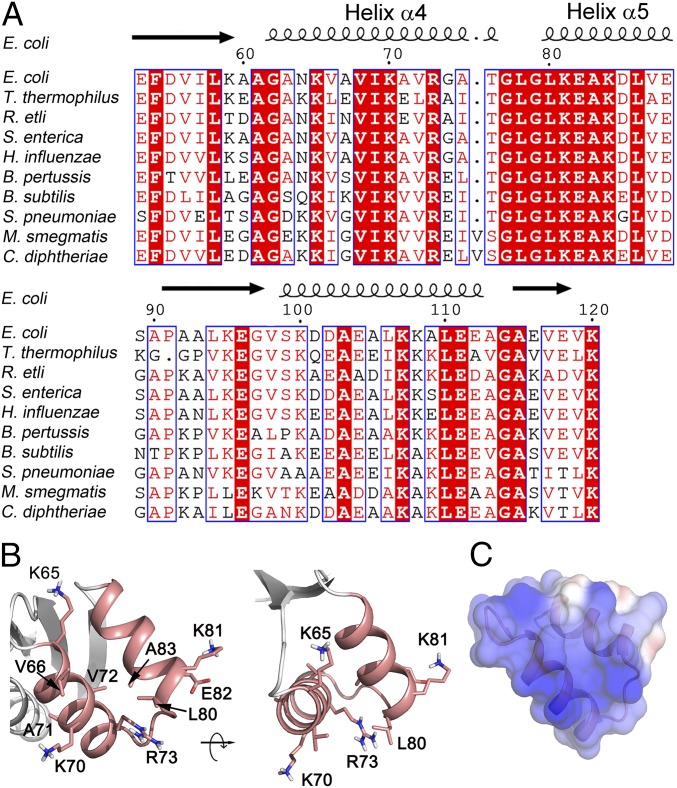
Alignment of the amino acid sequences of the L12-CTDs (amino acids 53 to 120 in E. coli) shows more than 60% identity and 70% similarity across a wide range of bacteria (Fig. 1A), including a number of strictly conserved residues in helix 4 (K65, V66, I69, K70, A71, and R73) and helix 5 (L80, K81, and E82) (Fig. 1A). The arrangement of these residues in the L12-CTD is shown in Fig. 1B. Among these, V66, K70, L80, and E82 were identified by NMR mapping as the tr-GTPase interaction sites (5). Moreover, the surface electrostatic potential of L12-CTD (Fig. 1C) reveals a highly positively charged surface on helices 4/5, which might be crucial for GTPase factor interaction.
Fig. 1.
Sequence and structure analysis of E. coli L12-CTD with focus on helices 4/5. (A) Sequence alignment of L12-CTD from the bacteria E. coli, Thermus thermophilus, Rhizobium etli, Salmonella enterica, Haemophilus influenzae, Bordetella pertussis, Bacillus subtilis, Streptococcus pneumoniae, Mycobacterium smegmatis, and Corynebacterium diphtheriae. Strictly conserved amino acids are highlighted in red. Moderately conserved residues are in red letters. The secondary structure of L12-CTD is indicated above the alignment. (B) Ribbon illustration of the helices 4/5 (salmon) from the crystal structure of L12-CTD (4) (PDB 1CTF) showing the side chains of the conserved residues suggested to be involved in tr-GTPase interactions (5). (C) Electrostatic surface potential representation of the helices 4/5 on the L12-CTD. Blue and red in C correspond to positive and negative potentials, respectively.
To identify the amino acids on L12-CTD involved in IF2 interaction, the following mutations were created in the helices 4/5: K65A, V66A, V66C, V66D, I69A, K70A, A71C, A71D, V72A, R73A, G79A, L80A, K81A, E82A, and A83C. The mutants were purified to homogeneity and then reconstituted onto the L12-depleted 50S core as described in Huang et al. (11). The depletion and reconstitution of L12 was confirmed by Western blotting using anti-L12 antibody (SI Appendix, Fig. S1).
Prediction of the L12 Interaction Site on IF2 by Comparison with EF-Tu.
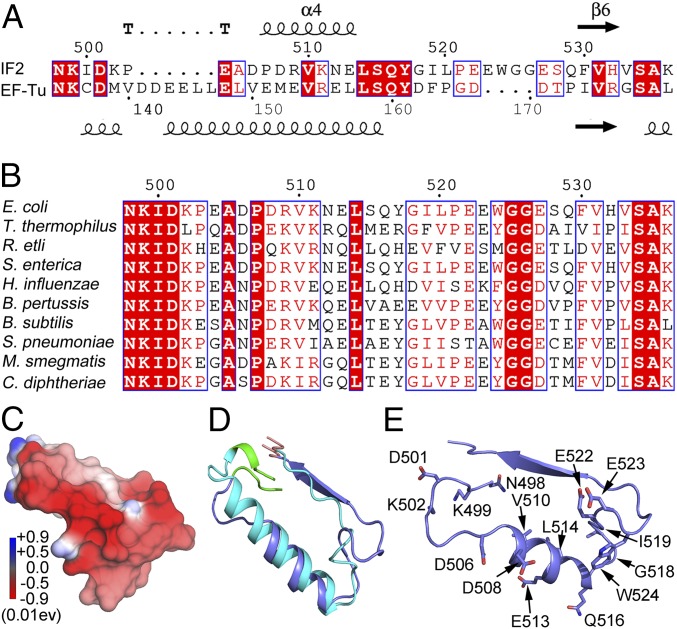
Earlier mutagenesis experiments have identified helix D of EF-Tu as the potential interaction site for L12-CTD (7). A homologous region on IF2 between G4 (NKID) and G5 (SAK) motifs (Fig. 2A) was identified by a homology search. This region is highly conserved (Fig. 2B) and contains multiple acidic amino acids with negatively charged surface potentials (Fig. 2C). Also, similar to helix D of EF-Tu [Protein Data Bank ID code (PDB) 1EFU], this region of E. coli IF2 is folded into a tight helix flanked by loose loop regions (23) (PDB 3JCN) (Fig. 2D). Therefore, we hypothesized that the helix between the G4 and G5 motifs (G4–G5 helix) is the potential interaction site for the positively charged helices 4/5 of L12-CTD (Fig. 1C). This view is further supported by close inspection of the cryo-EM maps of IF2 in the 30S preIC (PDB 3JCN and 5ME0) (23, 24), which display the G4–G5 helix of IF2 exposed toward the subunit interface (SI Appendix, Fig. S2).
Fig. 2.
Sequence and structural analysis of the G4–G5 peptide of E. coli IF2. (A) Comparison of the amino acid sequences of the peptides flanked by the G4 (NKID) and G5 (SAK) motifs of IF2 and EF-Tu from E. coli. The secondary structures based on IF2 (PDB 3JCN) and EF-Tu (PDB 1EFU) are shown on above and below the alignment, respectively. (B) Multiple sequence alignment of G4–G5 peptide of IF2 from the bacteria E. coli, T. thermophilus, R. etli, S. enterica, H. influenzae, B. pertussis, B. subtilis, S. pneumoniae, M. smegmatis, and C. diphtheria. Strictly conserved amino acids are highlighted in red. (C) The electrostatic surface potential plot (blue indicates positive and red indicates negative) for the G4–G5 helix of IF2. (D) Structural comparison of the G4 (bright green) to G5 (red) motifs of E. coli IF2 (PDB 3JCN indicated in blue) and EF-Tu (PDB 1EFU indicated in cyan). (E) The residues in the G4 and G5 motifs of IF2 selected for mutagenesis.
Characterizing the IF2 Mutants for GTP Binding and Formation of 30S preIC.
Fifteen point mutations were introduced in the G4–G5 helix of IF2 for most of the conserved and acidic residues. These are NK498/499A, D501A, K502A, D506A, D508A, V510A, K511A, E513A, L514A, Q516A, G518A, I519A, E522A, E523A, and W524A (Fig. 2E). The IF2 mutants were purified to homogeneity and were checked for GTP binding. Further, their ability to form 30S preIC was tested by nitrocellulose filter binding assay using [3H]GTP and [35S]fMet-tRNAfMet (SI Appendix, Materials and Methods). Also, IF2 retention on the 30S preIC was checked by cosedimentation of the complexes through a sucrose cushion by ultracentrifugation at 280,000 × g for 2 h at 4 °C.
None of the IF2 mutants showed reduced occupancy [35S]fMet-tRNAfMet on the 30S preIC (SI Appendix, Fig. S3A). Also, comparison with the S1 band on SDS/PAGE clearly showed that all IF2 variants bind to the 30S preIC to a similar extent (SI Appendix, Fig. S3B). However, when tested for [3H]GTP binding, NK498/499A, D501A, and K502A IF2s showed about fourfold lesser counts compared with the wild-type IF2 (WT), suggesting that these mutants are defective in GTP binding (SI Appendix, Fig. S3C). This was further confirmed by estimating the equilibrium dissociation constant (Kd) for GTP binding to the IF2 variants using a fluorescent GTP-analog (mant-GTP) (25). As summarized in SI Appendix, Table S2, these three IF2 mutants showed about twofold higher Kd values for mant-GTP than all other IF2 variants, confirming that they bind GTP poorly. Given that the residues NK498/499 and D501 constitute the G4 motif important for GTP binding (26) and that K502 is just next to G4, this result is not too surprising. All other IF2 mutants had affinity to GTP similar to that of the WT (SI Appendix, Table S2).
Mutations on Helices 4/5 of L12 Decrease the Rate of IF2-Mediated Ribosomal Subunit Association.
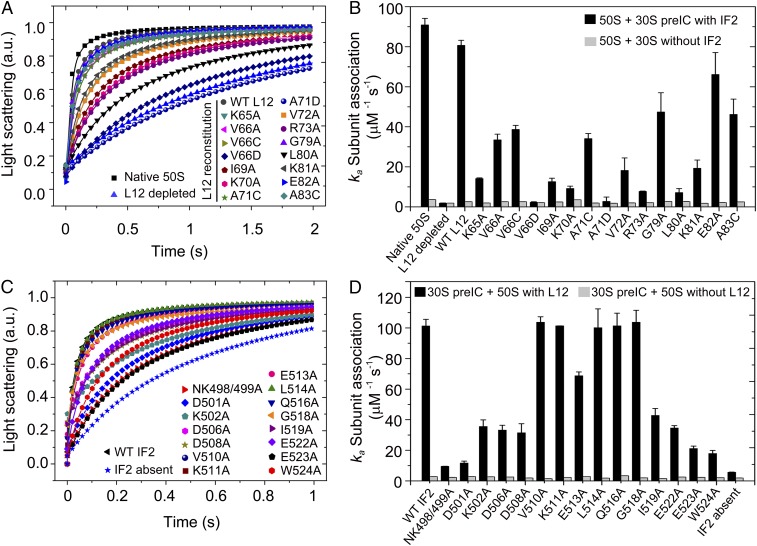
The reconstituted 50S subunits containing either WT or L12 mutants were subjected to subunit association assay with either naked 30S or mRNA programmed 30S preIC containing fMet-tRNAfMet, IF1, and IF2. The formation of 70S or 70S IC was followed by monitoring the increase in light scattering at 430 nm in a stopped-flow instrument (Fig. 3A). The association rate constants (ka) (Fig. 3B and SI Appendix, Table S1) were determined by fitting the data with the equation for the 30S and 50S subunit joining reaction as in Antoun et al. (17).
Fig. 3.
The effect of L12 and IF2 mutations on subunit association. The kinetics of subunit association monitored by the change in Rayleigh light scattering (430 nm) in a stopped-flow instrument. The averaged data from three to five individual experiments were fitted according to Antoun et al. (17) (solid lines in A and C) using Origin 8.0, and the association rate constants (ka) (presented in B and D) with SDs (error bars) were estimated. (A and C) The time course of 70S IC formation by association of 50S reconstituted with WT or mutant L12s (A) and 30S preIC containing WT or mutant IF2s (C) as indicated. Native and L12-depleted 50S were used as positive and negative controls, respectively, in A. 30S preIC without IF2 was also tested (C). (B) The ka values for association of 50S (native or reconstituted with WT or mutant L12) with 30S preIC with WT IF2 (black bars) or with 30S alone without IF2 (gray bars). (D) The ka values for association of 30S preIC containing IF2 variants with native 50S (black bars) or L12-depleted 50S (gray bars).
Consistent with a previous report (11), native 50S and the 50S reconstituted with WT L12 associated with 30S preIC with similar rate constants of 90.89 ± 3.26 μM−1s−1 and 80.7 ± 2.54 μM−1s−1, respectively. In comparison, the L12-depleted 50S was about 50 times slower (ka = 1.91 ± 0.02 μM−1s−1) in 70S IC formation (Fig. 3B and SI Appendix, Table S1). As expected, the 50S subunits reconstituted with the L12 mutants showed different degrees of defect in 70S IC formation. The strongest defect was for an Ala substitution at K70 (ka = 9.19 ± 1.2 μM−1s−1), R73 (ka = 7.73 ± 0.17 μM−1s−1), and L80 (ka = 7.16 ± 1.97 μM−1s−1) in which the ka decreased more than 10-fold compared with the native 50S. Significant defects were also seen with the mutations K65A, I69A, V72A, and K81A (see SI Appendix, Table S1 for ka values); other mutations were less deleterious. Interestingly, substitution of the nonpolar V66 and A71 with negatively charged Asp caused major defects, yielding ka values of 2.27 ± 0.29 μM−1s−1 and 2.73 ± 2.18 μM−1s−1, respectively, which are comparable to the ka value for the L12-depleted 50S. It should be noted that Ala or Cys mutations of V66 and A71 affected the rate of 70S IC formation only moderately. Importantly, none of the L12 mutations, or even the depletion of L12, affected the rate of IF2-free subunit association. This reconfirms the earlier conclusion that L12 is not needed for naked subunit association, nor is L12 itself involved in interaction with the 30S (11). Our result demonstrates that the substitution of positively charged residues to nonpolar ones or the introduction of negatively charged residues on helices 4/5 of L12 is clearly unfavorable for IF2-mediated association of 70S IC. It also suggests that K65, K70, R73, L80, and K81 on L12 are possibly engaged in interaction with IF2.
Defective Subunit Association with IF2 Bearing Mutations in the G4–G5 Region.
The 30S preIC containing IF2 variants were tested in subunit association assay (Fig. 3C). All IF2s were at saturating concentration as determined by IF2 titration (SI Appendix, Fig. S4). Other than the NK498/499A, D501A, and K502A mutants, which are defective in GTP binding, the IF2 mutants showing moderate (60%) to significant (80 to 90%) defect in subunit association were D506A, D508A, I519A, E522A, E523A, and W524A (Fig. 3D and SI Appendix, Table S2). Since none of these mutant IF2s showed defect in GTP binding or 30S preIC binding, the defects in subunit association denoted by ka (SI Appendix, Table S2) must arise from the defect in IF2’s interaction with 50S. Significantly, no difference in the ka was seen compared with the WT when these mutants were tested with L12-depleted 50S (Fig. 3D and SI Appendix, Table S2). This result suggests that the defects in subunit association with IF2 mutants at D506, D508, I519, E522, E523, and W524 are most likely due to their impaired interaction with L12 on 50S.
Ionic Strength Dependence of Subunit Association with IF2.
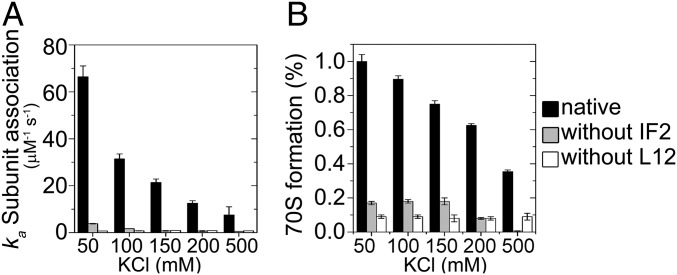
Our mutagenesis-based scanning indicated that several charged residues on the helices 4/5 of L12 and the G4–G5 helix of IF2 are involved in the intermolecular interaction. This would mean that the L12 and IF2 interaction is largely based on ionizable amino acid side chains. To test this, we followed subunit association at different ionic strengths and estimated ka values. The IF2-mediated subunit association with native 50S showed strong salt concentration dependence (Fig. 4). The ka decreased gradually from 66.4 ± 4.6 μM−1s−1 to 7.5 ± 3.5 μM−1s−1 as the KCl concentration increased from 50 to 500 mM. Simultaneously, the formation of 70S IC was reduced, forming only 40% 70S with 500 mM KCl. Interestingly, the ionic strength dependence of the subunit association reaction disappeared or reduced significantly upon removal of either L12 from the 50S subunit or IF2 from 30S preIC (Fig. 4). This confirms that L12 and IF2 interact with each other in a charge-dependent fashion, and furthermore, that the L12–IF2 interaction is the key to fast subunit association.
Fig. 4.
Ionic strength dependence of subunit association. The association of 30S preIC (with or without IF2) and 50S (native or L12 depleted) was monitored in stopped-flow in 10 mM Tris-HCl (pH 7.5) containing 5 mM MgCl2 and 50–500 mM KCl. (A) The rate constants of subunit association. (B) The relative yield of 70S IC. The data are the average of at least three independent experiments. The error bars indicate SD.
Characterization of L12–IF2 Interaction by MD Simulations.
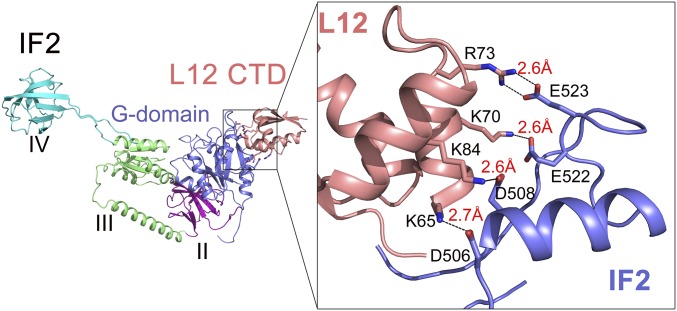
To characterize the molecular basis of the L12–IF2 interaction, a model was built by docking the G4–G5 helix of IF2 (PDB 3JCN) (23) onto the helices 4/5 of L12-CTD (PDB 1CTF) (4) with ClusPro (27, 28). The model was refined by energy minimization programs and MD simulations were performed with the Q program (29) using the refined structure as the initial conformation. The resulting model revealed four well-defined electrostatic interactions (with typical distances of <3 Å) between helices 4/5 of L12-CTD and the G4–G5 helix of IF2. These interactions involve the highly conserved and oppositely charged K65 (L12) and D506 (IF2), K70 (L12) and E522 (IF2), R73 (L12) and E523 (IF2), and K84 (L12) and D508 (IF2)—all forming tight salt bridges between them (Fig. 5). These results, in complete agreement with the mutagenesis and salt-dependence experiments, strongly suggest that IF2 interacts with L12-CTD by electrostatic complementarity.
Fig. 5.
MD simulations-based identification of the interacting amino acids on L12-CTD and IF2. Illustration of the molecular interaction between helices 4/5 of L12 (salmon) and the G4–G5 helix of IF2 (blue) based on MD simulations. Enlarged view (Right) shows that the interacting partners forming salt bridges are K65 on L12 and D506 on IF2, K84 on L12 and D508 on IF2, K70 on L12 and E522 on IF2, and R73 on L12 and E523 on IF2.
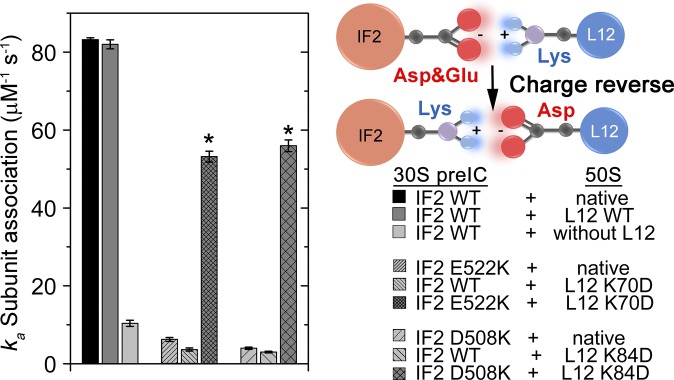
Charge-Reversed Mutations of L12 and IF2 Rescue Defects in Subunit Association.
To confirm our model of complementary charge-based interaction between L12-CTD and IF2, we performed the classical charge-reversal experiment involving the interacting amino acid pairs identified by MD simulations (Fig. 5). For that, we created charge-reversed mutants of L12 (K84D and K70D) and IF2 (D508K and E522K) (Fig. 5) and tested those individually and pairwise in subunit association assay. As expected, all four L12 and IF2 mutations showed a large defect in subunit association when tested singly; the rate of subunit association was only 5 to 10% of the rate of the native ones (ka = 83.2 ± 0.5 μM−1s−1, 100%) (Fig. 6 and SI Appendix, Tables S1 and S2). In contrast, the rate of subunit association was recovered to about 70% when the charge-swapped mutants of L12 and IF2 were used pairwise: 50S (L12 K70D)–30S preIC (IF2 E522K) (ka = 53.2 ± 1.4 μM−1s−1) and 50S (L12 K84D)–30S preIC (IF2 D508K) (ka = 56.5 ± 1.5 μM−1s−1). This result confirms that L12–IF2 interaction depends on charge complementarity of the interacting interfaces.
Fig. 6.
The rate constants (ka) of subunit association with the native and charge-reversed mutants of L12 and IF2 (Left) as indicated by the experimental design (Right). The bars with an asterisk represent ka with charge-reversed mutant pairs of L12 and IF2 as indicated (Right). Data represent mean ± SD of at least three independent experiments.
Discussion
The aim of this work was primarily to identify the molecular markers for L12–IF2 interaction on the ribosome, and further, to understand the underlying principle guiding the interaction. Although we had some idea from NMR mapping work (5) about which amino acids on L12-CTD are possibly involved in IF2 interaction, there was no prior knowledge regarding which residues on IF2 interact with L12-CTD. The search began with the facts that (i) the helices 4/5 constituting the common tr-GTPase interaction site on L12-CTD contain mostly positively charged and highly conserved residues (5), (ii) L12-CTD could be cross-linked to the G domain of IF2 on the ribosome (22), (iii) the cryo-EM reconstruction of the E. coli 70S IC suggested a direct interaction between the L12-CTD and the G domain of IF2 (21), (iv) the L12-CTD interacts with helix D of EF-Tu (7), and (v) the L12-CTD is a structural homolog of EF-Ts (7), which strongly interacts with EF-Tu for guanine nucleotide exchange. We therefore hypothesized that the L12 interaction site on IF2 should be a highly conserved region (and thus most likely located in the G domain) and should contain mainly negative charges. Using helix D of EF-Tu as a search sequence, we identified the G4–G5 helix of IF2 as the potential L12 interaction site. Structure and sequence analysis confirmed that this helical region contains several negatively charged and strictly conserved amino acids (Fig. 2).
By testing the L12 and IF2 mutants in subunit association assay and further analyzing the L12-CTD and IF2 interaction with MD simulations, we have identified the exact residues involved in the L12–IF2 interaction. These include the highly conserved K65, K70, R73, and K81 residues on helices 4/5 of L12-CTD, which match perfectly with the earlier NMR mapping result (5). It is noteworthy that the helices 4/5 of L12-CTD are also suggested to interact with other tr-GTPases such as EF-G (30), EF-Tu (7), RF3 (31), and BipA (32) on the ribosome. Our current results, together with recent structures (30–32), confirm that helices 4/5 indeed constitute the general tr-GTPase interaction site on L12-CTD.
We have successfully predicted and identified the crucial residues on the G4–G5 helix on IF2 involved in L12 interaction. These are D506, D508, E522, and E523, substitutions of which cause significant defect in subunit association. Interestingly, the mutations are not effective if L12 is removed from 50S, suggesting that these residues act specifically toward L12 on the 50S. Inspection of the cryo-EM structure of E. coli IF2 on 30S preIC shows these residues as exposed on the 30S surface (SI Appendix, Fig. S2) (23, 24, 33). This region of IF2 was also seen in close proximity to L12-CTD on 70S IC (21).
On the basis of a cryo-EM structure of Thermus thermophilus 70S IC with IF2 (PDB 3J4J), it was claimed that L12 probably interacts with the NTD of IF2 (34), which contradicts all earlier cryo-EM and chemical footprinting-based reports (21, 35). However, the authors stated that the electron density in the potential interaction area is not defined well enough in the 11.5-Å resolution structure and that deletion of 69 residues from the IF2 NTD caused only twofold reduction in subunit association (34). Moreover, the NTD of IF2 is the least conserved region of IF2. This region varies in length and amino acid composition (36), which makes it unlikely to be the interaction partner for the highly conserved, general tr-GTPase interaction site on L12-CTD.
Our results demonstrate the key interacting residues between the helices 4/5 of L12-CTD and the G4–G5 helix of IF2. These amino acids, being basic and acidic in nature, respectively, on L12-CTD and IF2, form salt bridges with each other, thereby suggesting a strong and specific charge-based interaction between them. Furthermore, high ionic strength inhibits this interaction, and charge swapping between the interacting partners largely rescues the defect in subunit association. Thus, we conclude that charge complementarity forms the basis of interaction between L12 and IF2, which in turn, is the key to fast subunit interaction.
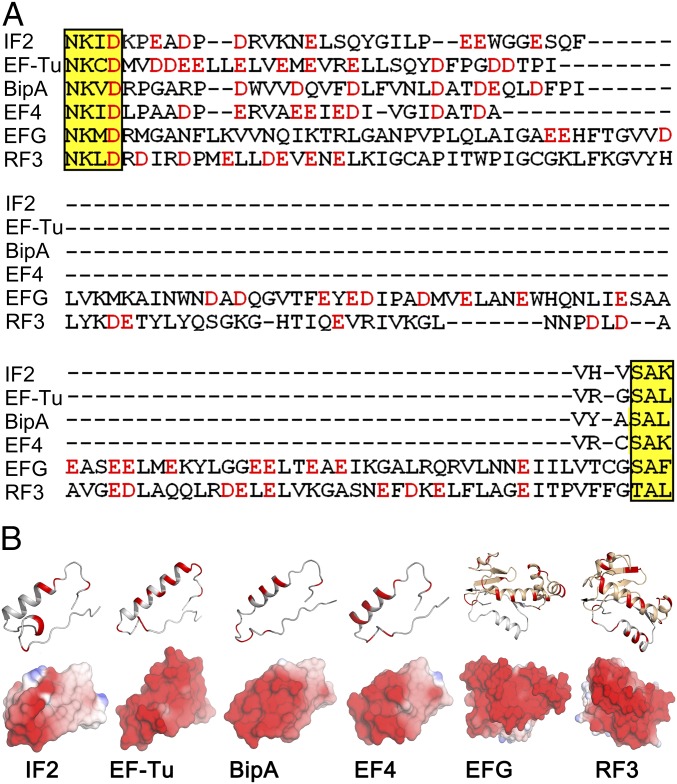
On the basis of our current results and available sequence, biochemical, and structural data, we speculate that L12 interaction sites are probably located between the G4 and G5 motifs of the G domain in the tr-GTPases. Recent high-resolution structure of a tr-GTPase BipA supports the hypothesis (Fig. 7) (32). This region is dominated by acidic amino acids, resulting in a negatively charged surface (Fig. 7). Moreover, with exceptions of EF-G and RF3, which contain an additional G′ domain between the G4 and G5 motifs, the amino acid sequence of this region is highly conserved among tr-GTPases. Interestingly, based on cryo-EM structures and biochemical analysis, it has been proposed that L12-CTD interacts with the G′ domains of EF-G (37–39) and RF3 (31), which are also electronegative (Fig. 7). From this analysis, we propose that L12-CTD interacts with all major tr-GTPases in a charge-dependent manner, probably involving the G4–G5 region of the highly conserved G domain of the tr-GTPases. This means that the interactions are not only significant for ribosome function but are also significant from an evolutionary perspective. Further experiments will clarify the functional significance of L12 interaction for individual tr-GTPases.
Fig. 7.
Comparison of the sequences and electrostatic surface potentials of tr-GTPases. (A) Multiple amino acid sequence alignments of E. coli IF2, EF-Tu, BipA, EF4, EF-G, and RF3 between G4 and G5 motifs (yellow highlight). Electronegative amino acids are in red. (B) The ribbon diagram (Upper) and the surface potentials (Lower) of the peptide between G4 and G5 motifs in IF2 (PDB 3JCN), EF-Tu (PDB 4PC7), BipA (PDB 5A9V), EF4 (PDB 3JCD), EF-G (PDB 4V9O), and RF3 (PDB 4V89). The electronegative amino acids and surfaces are in red. Blue indicates electropositive surface potential.
Materials and Methods
Subunit Association Assay.
Two reaction mixes were prepared in Hepes-polymix buffer (pH 7.5) (40). Mix A [containing 0.5 μM 30S subunit, 1 μM XR7 MLF mRNA with sequence UAAGGAGGUAUUAAAUGCUGUUUUAA (Shine-Dalgarno sequence underlined; coding sequence in bold), 2 μM IF1, 2 μM IF2 (WT or mutants), and 2 μM fMet-tRNAfMet] and Mix B (containing 0.5 μM 50S subunit carrying WT or mutant L12) were rapidly mixed in the stopped-flow instrument (Applied Photophysics) at 37 °C. The kinetics of subunit association was followed by an increase in Rayleigh light scattering at 430 nm. The data (average of three to five individual experiments) were fitted using subunit association equation (17) using Origin 8.0 (OriginLab Corp).
Ribosome Reconstitution with WT and L12 Variants.
The 50S and 30S subunits were purified from E. coli MRE600 using a standard protocol (11, 12). For L12 depletion, 50S was treated with 1 M NH4Cl for 1 h on ice and then precipitated with 50% ethanol. Further, L12 variants were reconstituted by adding a 10× excess of WT or mutant L12 proteins to the L12-depleted 50S core at 37 °C for 30 min (11). The L12-depleted and -reconstituted 50S subunits were purified by ultracentrifugation at 250,000 × g through a 37% sucrose cushion and confirmed by Western blotting using anti-L12 antibody (SI Appendix, Fig. S1).
Detailed descriptions of materials and methods (mutagenesis, protein expression and purification, and MD simulations) are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work is funded by the Swedish Research Council (Grant 2014-4423 and Research Environment Grant 2016-06264), a Knut and Alice Wallenberg Foundation grant to RiboCORE (2011.0081), and a joint Department of Biotechnology (Ministry of Science and Technology, India)–VINNOVA (Swedish Governmental Agency for Innovation Systems)/Swedish Research Council Grant (2013-8778) (to S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802001115/-/DCSupplemental.
References
- 1.Wahl MC, Bourenkov GP, Bartunik HD, Huber R. Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12. EMBO J. 2000;19:174–186. doi: 10.1093/emboj/19.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandra Sanyal S, Liljas A. The end of the beginning: Structural studies of ribosomal proteins. Curr Opin Struct Biol. 2000;10:633–636. doi: 10.1016/s0959-440x(00)00143-3. [DOI] [PubMed] [Google Scholar]
- 3.Mulder FA, et al. Conformation and dynamics of ribosomal stalk protein L12 in solution and on the ribosome. Biochemistry. 2004;43:5930–5936. doi: 10.1021/bi0495331. [DOI] [PubMed] [Google Scholar]
- 4.Leijonmarck M, Liljas A. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 A. J Mol Biol. 1987;195:555–579. doi: 10.1016/0022-2836(87)90183-5. [DOI] [PubMed] [Google Scholar]
- 5.Helgstrand M, et al. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Diaconu M, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Kothe U, Wieden HJ, Mohr D, Rodnina MV. Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J Mol Biol. 2004;336:1011–1021. doi: 10.1016/j.jmb.2003.12.080. [DOI] [PubMed] [Google Scholar]
- 8.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 10.Savelsbergh A, Mohr D, Wilden B, Wintermeyer W, Rodnina MV. Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J Biol Chem. 2000;275:890–894. doi: 10.1074/jbc.275.2.890. [DOI] [PubMed] [Google Scholar]
- 11.Huang C, Mandava CS, Sanyal S. The ribosomal stalk plays a key role in IF2-mediated association of the ribosomal subunits. J Mol Biol. 2010;399:145–153. doi: 10.1016/j.jmb.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Mandava CS, et al. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 2012;40:2054–2064. doi: 10.1093/nar/gkr1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marintchev A, Wagner G. Translation initiation: Structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 14.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunberg-Manago M, et al. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J Mol Biol. 1975;94:461–478. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- 16.Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antoun A, Pavlov MY, Tenson T, Ehrenberg M MN. Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol Proced Online. 2004;6:35–54. doi: 10.1251/bpo71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai A, et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487:390–393. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goyal A, Belardinelli R, Maracci C, Milón P, Rodnina MV. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res. 2015;43:10700–10712. doi: 10.1093/nar/gkv869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 22.Heimark RL, Hershey JW, Traut RR. Cross-linking of initiation factor IF2 to proteins L7/L12 in 70 S ribosomes of Escherichia coli. J Biol Chem. 1976;251:7779–7784. [PubMed] [Google Scholar]
- 23.Sprink T, et al. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci Adv. 2016;2:e1501502. doi: 10.1126/sciadv.1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Alonso JP, et al. Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways. Nucleic Acids Res. 2017;45:2179–2187. doi: 10.1093/nar/gkw1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koripella RK, et al. Mechanism of elongation factor-G-mediated fusidic acid resistance and fitness compensation in Staphylococcus aureus. J Biol Chem. 2012;287:30257–30267. doi: 10.1074/jbc.M112.378521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dever TE, Glynias MJ, Merrick WC. GTP-binding domain: Three consensus sequence elements with distinct spacing. Proc Natl Acad Sci USA. 1987;84:1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozakov D, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marelius J, Kolmodin K, Feierberg I, Aqvist J. Q: A molecular dynamics program for free energy calculations and empirical valence bond simulations in biomolecular systems. J Mol Graph Model. 1998;16:213–225, 261. doi: 10.1016/s1093-3263(98)80006-5. [DOI] [PubMed] [Google Scholar]
- 30.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallesen J, et al. Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1. eLife. 2013;2:e00411. doi: 10.7554/eLife.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar V, et al. Structure of BipA in GTP form bound to the ratcheted ribosome. Proc Natl Acad Sci USA. 2015;112:10944–10949. doi: 10.1073/pnas.1513216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julián P, et al. The cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011;9:e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonetti A, et al. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc Natl Acad Sci USA. 2013;110:15656–15661. doi: 10.1073/pnas.1309578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzi S, et al. Ribosomal localization of translation initiation factor IF2. RNA. 2003;9:958–969. doi: 10.1261/rna.2116303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caserta E, et al. Ribosomal interaction of Bacillus stearothermophilus translation initiation factor IF2: Characterization of the active sites. J Mol Biol. 2010;396:118–129. doi: 10.1016/j.jmb.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 37.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G′ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 38.Nechifor R, Murataliev M, Wilson KS. Functional interactions between the G′ subdomain of bacterial translation factor EF-G and ribosomal protein L7/L12. J Biol Chem. 2007;282:36998–37005. doi: 10.1074/jbc.M707179200. [DOI] [PubMed] [Google Scholar]
- 39.Carlson MA, et al. Ribosomal protein L7/L12 is required for GTPase translation factors EF-G, RF3, and IF2 to bind in their GTP state to 70S ribosomes. FEBS J. 2017;284:1631–1643. doi: 10.1111/febs.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koripella RK, et al. A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate. Sci Rep. 2015;5:12970. doi: 10.1038/srep12970. [DOI] [PMC free article] [PubMed] [Google Scholar]