Significance
Drug interactions, including drug–drug interactions (DDIs) and drug–food constituent interactions, can trigger unexpected pharmacological effects such as adverse drug events (ADEs). Several existing methods predict drug interactions, but require detailed, but often unavailable drug information as inputs, such as drug targets. To this end, we present a computational framework DeepDDI that accurately predicts DDI types for given drug pairs and drug–food constituent pairs using only name and structural information as inputs. We show four applications of DeepDDI to better understand drug interactions, including prediction of DDI mechanisms causing ADEs, suggestion of alternative drug members for the intended pharmacological effects without negative health effects, prediction of the effects of food constituents on interacting drugs, and prediction of bioactivities of food constituents.
Keywords: deep learning, structural similarity profile, DeepDDI, drug–drug interactions, drug–food interactions
Abstract
Drug interactions, including drug–drug interactions (DDIs) and drug–food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. Several computational methods have been developed to better understand drug interactions, especially for DDIs. However, these methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction. Here, we report development of a computational framework DeepDDI that uses names of drug–drug or drug–food constituent pairs and their structural information as inputs to accurately generate 86 important DDI types as outputs of human-readable sentences. DeepDDI uses deep neural network with its optimized prediction performance and predicts 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. DeepDDI is used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects. Furthermore, DeepDDI is applied to 3,288,157 drug–food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents are predicted. These results suggest that DeepDDI can provide important information on drug prescription and even dietary suggestions while taking certain drugs and also guidelines during drug development.
Intended efficacy of a drug can be substantially altered when coadministered with another drug or consumed together with specific food constituents. Therefore, understanding drug interactions, including drug–drug interaction (DDI) and drug–food constituent interaction (DFI), is critical to minimize unexpected adverse drug events (ADEs) (1) and to maximize synergistic benefits when treating a disease. This motivation has grown larger as the number of prescriptions of multiple drugs (e.g., at least two drugs) for disease treatment continues to increase (2, 3). According to a recent study in 2010–2011, 67% of elderly Americans were taking five or more medications, including prescription drugs, over-the-counter drugs, and dietary supplements (3). A problem is that DDIs have been estimated to be associated with 30% of all of the reported ADEs (4). In addition, ADEs due to DDIs are one of the major reasons for drug withdrawal from the market (5). Despite the importance, clinical examination of the effects of all possible drug interactions, including DDIs and DFIs, is highly constrained due to the difficulties of performing the actual study with the patients who are taking different food with the drugs, which also requires much time and high costs.
To solve this problem, various computational methods have been developed using methods based on structural and other similarities (6–11) or drug-target associations (12–14). These computational methods have greatly contributed to better understanding of DDIs, but they have two major limitations. The computational methods developed so far predict the chance of interactions between the given drugs in pair, but do not provide specific descriptions on DDI in terms of pharmacological effects. Also, many of the computational methods often require a large volume of detailed drug information such as drug targets, interacting drugs, and side effects, which are often unavailable, as input to predict DDIs (7, 9–14). Thus, the DDI prediction methods previously developed are largely applicable to drugs with known mechanisms of action (e.g., approved or investigational drugs). Finally, the DDI prediction methods developed so far have not been examined as to whether they could be used to analyze DFIs, another category of drug interactions equally important as DDIs.
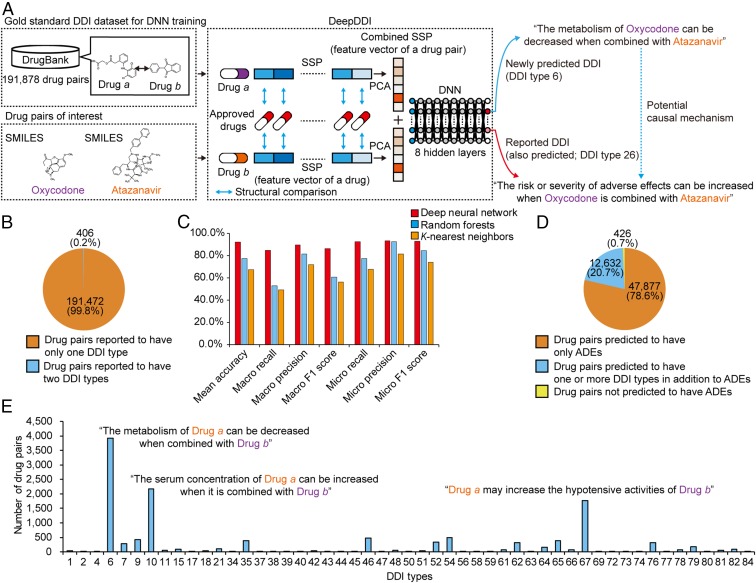
Here, we present a computational framework DeepDDI that takes structural information and names of two drugs in pair as inputs, and accurately predicts relevant DDI types for the input drug pair (Fig. 1A). Input structural information is provided in the simplified molecular-input line-entry system (SMILES) that describes the structure of a chemical compound. DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair; to the best of our knowledge, DDI prediction methods developed so far do not generate information at this level of detail and at a sufficiently high mean accuracy. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “the risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir” (Fig. 1A). The DeepDDI output sentences were generated from the predefined 86 important general sentence structures, representing 86 DDI types that are described at DrugBank (15) (SI Appendix, Materials and Methods and Dataset S1). These general output sentence structures were prepared using the gold standard DDI dataset from DrugBank, which covers 192,284 DDIs contributed by 191,878 drug pairs also presented in the form of sentences (Dataset S2). Importantly, DeepDDI can also be applied to drug–food constituent pairs as long as their structural information is available. This extended application is possible because the gold standard DDI dataset used to develop DeepDDI covers a wide range of compound types including natural products that could be found as food constituents (SI Appendix, Materials and Methods).
Fig. 1.
Overall scheme, performance evaluation, and application of DeepDDI. (A) DeepDDI consists of the structural similarity profile (SSP) generation pipeline and deep neural network (DNN). DeepDDI accepts chemical structures (in SMILES describing the structure of a chemical compound) and names of drugs in pairs as inputs, and predicts their potential drug–drug interaction (DDI) types as outputs in human-readable sentences having the input drug names. DNN of DeepDDI is a multilabel classification model that can predict multiple DDI types at the same time for a given drug pair. To develop DeepDDI, a gold standard DDI dataset covering 191,878 drug pairs was obtained from DrugBank, and used to train the DNN of DeepDDI. A single, combined SSP (feature vector of a drug pair) is generated for each input drug pair (SI Appendix, Materials and Methods). DeepDDI has many implications such as prediction of potential causal mechanism for the adverse drug evens (ADEs) of a drug pair of interest (blue dotted arrow) using the output sentences. It should be noted that the use of input data on the same drug pairs, but with different drug orders, results in different DeepDDI output sentences. For example, the use of input data in the order of atazanavir and oxycodone generated a DeepDDI output sentence corresponding to the DDI type 26 with atazanavir appearing before oxycodone; and the output sentence for DDI type 6 was not generated in this case. (B) Number (percentage) of drug pairs in the gold standard DDI dataset having a single DDI type or two. The dataset does not have drug pairs having more than two DDI types. (C) Prediction performance of DeepDDI for classifying DDI types for drug pairs in the gold standard DDI dataset using three different machine learning algorithms (SI Appendix, Materials and Methods). (D) DeepDDI prediction results for the drug pairs reported to have ADEs in the gold standard DDI dataset. (E) Number of drug pairs having additionally predicted DDI types using DeepDDI in addition to the reported ADEs.
Results
For the development of DeepDDI to effectively classify the DDI types for a given drug pair, deep neural network (DNN) was employed among other machine learning approaches due to its proven outstanding performance in classification without the need of feature extraction (16, 17). To implement DNN, structural information (SMILES) of each drug in the input drug pair was first used to generate a feature vector called structural similarity profile (SSP); SSP was devised to effectively capture a unique structural feature of a given drug and to associate this feature with a set of the reported DDI types (Fig. 1A and SI Appendix, Fig. S1 and Materials and Methods). To predict the DDI types for a given drug pair, two SSPs were generated for each drug pair, subjected to the dimension reduction and combined as a single vector (Fig. 1A and SI Appendix, Fig. S2 and Materials and Methods); the combined SSP is a feature vector of a drug pair. The combined SSPs of all of the DDIs in the gold standard DDI dataset were created, and the entire set was used to develop the DNN for the accurate prediction of DDI types (Fig. 1A and SI Appendix, Fig. S3 and Materials and Methods). The DNN of DeepDDI was designed to be a multilabel classification model that can predict multiple DDI types for a given drug pair (i.e., simultaneous activation of multiple output neurons, each representing DDI type) because drug pairs can have multiple DDI types as suggested by such 406 drug pairs in the gold standard DDI dataset (Fig. 1B). The DNN of DeepDDI has 86 output neurons, which represent 86 DDI types considered in this study; these output neurons’ activity values range between 0 (no interaction between the pair) and 1 (interaction between the pair with the highest confidence), which can be considered as a probability (18). A given drug pair was considered to have a specific DDI type if the corresponding output neuron became activated by having its activity greater than a predetermined threshold of 0.47 (see below). DNN was trained by minimizing prediction errors in comparison with the gold standard DDI dataset using cross entropy as loss function and the Adam method for optimization (SI Appendix, Materials and Methods). DNN was trained up to 100 epochs. For architecture optimization, all of the combinations of one to nine hidden layers and 128, 256, 512, 1,024, and 2,048 nodes in each hidden layer were tested (SI Appendix, Fig. S3). Using the optimal DNN architecture after training and validation (SI Appendix, Figs. S4 and S5), the complete DeepDDI showed reasonably accurate performance, attaining 84.8–93.2% for the seven different performance metrics using the DrugBank gold standard DDI dataset (Fig. 1C and SI Appendix, Materials and Methods); here, the threshold of 0.47 was used to determine the activation of an output neuron (see SI Appendix, Fig. S6 for this value and SI Appendix, Materials and Methods). DeepDDI was further evaluated with respect to the use of SSP versus 685 molecular descriptors as a feature vector of the drug pair (SI Appendix, Fig. S7). DeepDDI using SSP achieved greater accuracies for all of the 86 DDI types. Also, SSP showed a slightly greater mean accuracy than feature vectors generated by two state-of-the-art vectorization methods, namely Molecular Autoencoder (19) and Mol2vec (20), when used with the DeepDDI framework (SI Appendix, Fig. S7). For the comparison of DNN performance, two other machine learning approaches, random forests and K-nearest neighbors, were employed for the DDI type classification. DeepDDI with DNN was better than the others by achieving greater accuracies for 82 of 86 DDI types (SI Appendix, Fig. S8), suggesting that DNN was indeed better than the other machine learning methods. Although outstanding performance of DNN in processing a large volume of complex data are well known (16, 17), understanding precise reasons for the observed performance differences needs further in-depth studies. Finally, DeepDDI was compared with another DDI prediction method called heterogeneous network-assisted inference (HNAI) (7), which uses information on chemical structures, drug targets, drug side effects, and the Anatomical Therapeutic Chemical (ATC) classification system as inputs; DeepDDI again showed a greater mean accuracy (SI Appendix, Fig. S9).
DeepDDI was then applied to various drug–drug pairs and drug–food constituent pairs to generate sentences describing relevant interactions. In particular, the DeepDDI output sentences can be used to provide in-depth inferences associated with the drug interactions. In actual prediction of DDI and DFI, 57 among the 86 DDI types were considered to ensure reliability of prediction. These 57 DDI types are those having at least 10 drug pairs in the gold standard DDI dataset, and those showing accuracies of 70% or greater by DeepDDI (SI Appendix, Fig. S7). It should be noted that five drug pairs were considered for a DDI type during the development (training and validation) of DeepDDI (SI Appendix, Materials and Methods).
The gold standard DDI dataset in DrugBank contains DDI information for 191,878 drug pairs. However, only 406 drug pairs (0.2%) are associated with two DDI types, while the rest (99.8%) of them are associated with only single DDI type (Fig. 1B). Since DeepDDI systematically suggests all possible DDI types, it was reasoned that the outputs of DeepDDI can be used to suggest unknown (1) causal mechanisms of ADEs reported for the drug pairs in the gold standard DDI dataset. For the 60,935 drug pairs reported to show ADEs in the gold standard DDI dataset, 60,509 drug pairs (99.3%) were correctly predicted to have ADEs, which had the expression of “the increased risk or severity of adverse effects” in the DeepDDI output sentences (DDI type 26) (Fig. 1D). Among the 60,509 drug pairs, 12,632 drug pairs were assigned with multiple DeepDDI output sentences suggesting specific pharmacological effects (e.g., “the decreased metabolism” and “the increased serum concentration”) in addition to “the increased risk or severity of adverse effects” (Fig. 1E).
For further validation, the DeepDDI outputs were compared with the consistent descriptions on the DDIs of 182 drug pairs present in the Drugs.com database (https://www.drugs.com/), which provides additional information regarding DDIs beyond those described in DrugBank (Dataset S3). The most frequently observed DDIs of the 182 drug pairs were involved in decreasing metabolism of interacting drugs (69 drug pairs for DDI type 6), followed by DDIs increasing serum concentration (37 drug pairs for DDI type 10), increasing corrected QT interval (QTc)-prolonging activity (23 drug pairs for DDI type 76), and increasing anticoagulant activity of interacting drugs (13 drug pairs for DDI type 46). These results suggest that abnormally increased bioavailability (DDI types 6 and 10) could be one of the major causes for the observed ADEs of various drug pairs (21) (Fig. 1E).
Other than 182 drug pairs examined above by comparing with the information presented in the Drugs.com database, the possible causal mechanisms of ADEs for the remaining 12,450 drug pairs are not available elsewhere to the best of our knowledge. Therefore, DeepDDI output sentences describing additional DDI types for the drug pairs with the reported ADEs can serve as the likely causal mechanisms of DDIs for further validation.
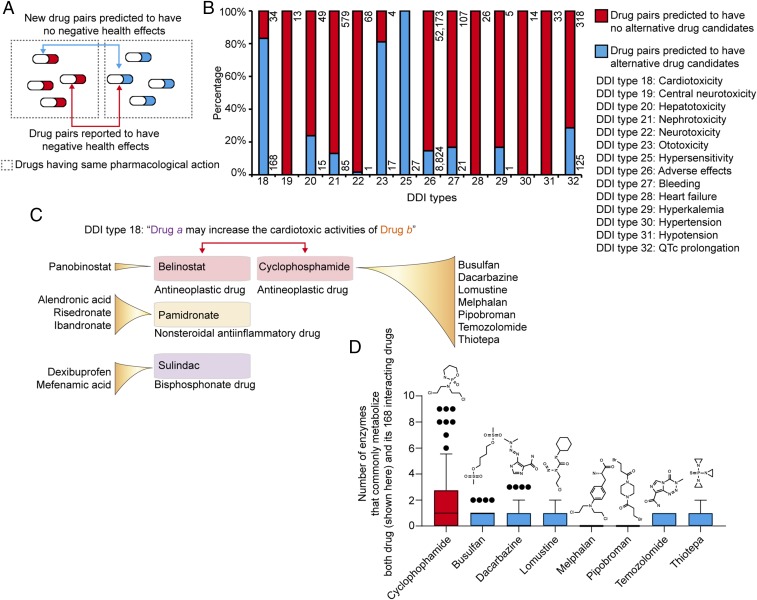
Since DeepDDI could predict potential causal mechanisms of a given drug pair, we extended our work of employing DeepDDI to suggest alternative drug members for drug pairs reported to have negative health effects so as to achieve only the intended beneficial pharmacological effects of each drug member (Fig. 2A); the drug pairs having “negative health effects” are those in the gold standard DDI dataset described with “the increased risk or severity of adverse effects” and/or explicit expression such as “cardiotoxic activity,” “nephrotoxic activity,” and “the increased risk or severity of bleeding.” In the gold standard DDI dataset, 62,707 drug pairs were found to have clear “negative health effects” and corresponded to one of the 14 DDI types. Other than these 14 DDI types, the remaining DDI types were not considered in this analysis because health effects of their pharmacological effects, such as “the decreased absorption” (DDI type 1) and “the decreased serum concentration” (DDI type 9), are rather ambiguous and depend on physiological condition of individuals. To find an alternative drug that does not exert “negative health effects,” the 62,707 drug pairs were redesigned by changing an original drug member to another approved drug from DrugBank, having the same pharmacological effects, one at a time (bidirectional blue line in Fig. 2A). The DDI types of candidate drug pairs were subsequently predicted using DeepDDI. If none of the 14 DDI types was predicted for the candidate drug pair (i.e., corresponding output neuron’s activity in the DNN of DeepDDI less than 0.47), the new drug pair was considered to have a low chance of the reported negative health effects; the new drug pair with the output neuron’s activity value closer to zero was considered more beneficial in this case (Fig. 2A). A total of 9,284 drug pairs were predicted to have alternative drug members that would help avoid corresponding negative health effects for the intended pharmacological effects of each original drug member (Fig. 2B and see Dataset S4 for details). For example, an anticancer drug cyclophosphamide was reported to interact with 168 drugs in the gold standard DDI dataset, and predicted to be replaceable with seven different anticancer drugs (i.e., busulfan, dacarbazine, lomustine, melphalan, pipobroman, temozolomide, and thiotepa) with lower chance of cardiotoxicity (Fig. 2C). Cyclophosphamide has been known to exert cardiotoxicity, which might be increased when coadministered with one of 168 drugs. An interesting observation here is that cyclophosphamide and its 168 drugs are commonly metabolized by a greater number of enzymes (e.g., cytochrome P450) compared with seven alternative anticancer drugs and the same set of interacting drugs (Fig. 2D). This means that the serum concentration of cyclophosphamide would be higher than desired (22) when administered with one of 168 drugs. If cyclophosphamide has to be used to treat a cancer despite its cardiotoxicity, its interacting drugs (i.e., belinostat, pamidronate, and sulindac) can be replaced with alternative drugs having the same pharmacological effects to minimize the chance of cardiotoxicity (Fig. 2C). Despite the importance, there were few systematic methods or resources to design prescription or suggest treatment strategies for diseases involving more than one drug compound without “negative health effects.” DeepDDI can meet this demand by suggesting alternative drug members for a given drug pair of interest, or even new drug pairs to examine. When more information on DDIs of drug pairs becomes available, beyond the 14 DDI types explicitly known to exert “negative health effects,” the use of DeepDDI will serve as a more powerful tool suggesting alternative drug compounds and designing drug pairs to be used. Finally, although alternative drug members may be prioritized using output neuron activity values for a given drug pair as described above, additional important factors, such as an individual’s genetic makeup and lifestyle (e.g., diet), should be carefully considered because they can siginficantly affect DDIs. Personal multiomics data profiling (23), for example, can be very useful in this respect.
Fig. 2.
Prediction of new drug members for a drug pair to avoid the reported negative health effects. (A) For a drug pair with DDI reported to have negative health effects (bidirectional red arrow, see Fig. 2B and Dataset S1 for the 14 relevant DDI types), new drug pairs (bidirectional blue arrow) having alternative drug members were predicted using DeepDDI. (B) Percentage (number) of the reported drug pairs that were predicted to have new drug members that would lower the chance of each DDI type having negative health effects. Key toxicity terms are listed for each DDI type next to the graph. (C) Alternative drug members predicted for cyclophosphamide and its three interacting drugs (among the 168 interacting drugs), which could lower the chance of cardiotoxic activity (DDI type 18). If cyclophosphamide has to be used to treat a cancer despite its cardiotoxicity, its interacting drugs (i.e., belinostat, pamidronate, and sulindac) can be replaced with alternative drugs having the same pharmacological effects to minimize the chance of cardiotoxicity. (D) Number of enzymes that commonly metabolize cyclophosphamide and each of its 168 interacting drugs (red box), and seven new drug members predicted in place of cyclophosphamide and each of its 168 interacting drug (blue box). Boxes represent the 25th–75th percentiles, while whiskers represent the 5th–95th percentiles. Drug pairs with new drug members were predicted to have lower chance of cardiotoxic activity, while achieving the intended anticancer efficacy.
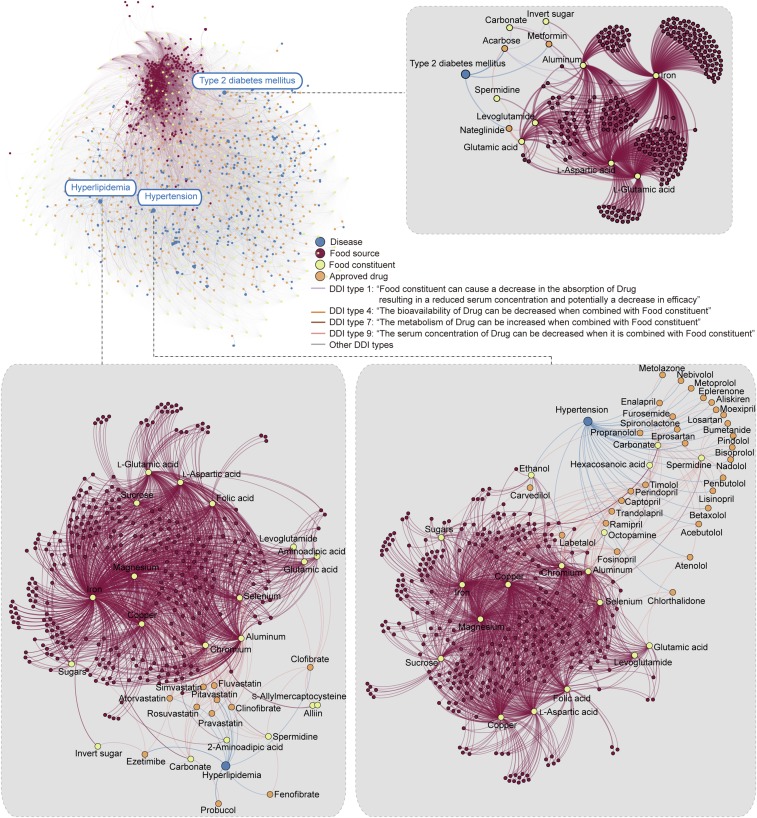
As an extended application, DeepDDI was applied to drug–food constituent pairs to understand the effects of food constituents on pharmacological effects of interacting drugs. This information is important in designing diet that helps avoid ADEs from unwanted DFIs during medication. For this, information on 1,523 well-characterized food constituents from 377 food sources was obtained from FooDB (foodb.ca/). Subsequently, the DDI types of 3,288,157 drug–food constituent pairs (2,159 approved drugs individually paired with 1,523 food constituents) were predicted using DeepDDI (SI Appendix, Materials and Methods). Although we are examining the DFI type, the output is given as the DDI type by DeepDDI; thus, the DDI type reported below should be considered as the DFI type. The same DNN generated above was used because the gold standard DDI dataset used to develop it also covers some, but not all, known natural products found in food sources. Since not all of the known natural products (food constituents) to examine here were covered in the DNN trained above, another validation step was added. The predicted DDI type of a drug–food constituent pair was considered valid if there exist drug pairs in the gold standard DDI dataset showing the same DDI type as the observed drug–food constituent pair, and at the same time at least one of the drugs in the pair is structurally similar (Tanimoto coefficient >0.75) (8, 11) to the food constituent under examination. This inference was based on a well-known assumption that a drug pair A is likely to have the same DDI as another drug pair B if both drug pairs have structurally similar drug members (8). As a result, 319,993 drug–food constituent pairs were predicted to have a total of 690,956 DDI types, while the remaining drug–food constituent pairs were not assigned with any DDI types from DeepDDI. The names of 274 food constituents appeared as the “subject” in 358,995 out of the 690,956 DeepDDI output sentences (e.g., alliospiroside A in the sentence of “Alliospiroside A may decrease the cardiotoxic activities of Mianserin”) (SI Appendix, Fig. S10A). From a grammatical perspective, such 274 food constituents could be considered to affect pharmacological effects of their interacting drugs (see Dataset S5 for full list). In particular, attention was paid to those food constituents that might decrease in vivo concentration of interacting drugs, which could potentially reduce the therapeutic efficacies. Among the 358,995 output sentences with food constituents as “subject,” 73 food constituents were predicted to decrease in vivo concentrations of 430 drugs used to treat a total of 357 diseases by decreasing absorption, bioavailability, or serum concentration of drugs, or by increasing metabolism of drugs (Fig. 3 and Dataset S6 for details). For example, in vivo concentrations of 30 drugs for treating hypertension could be potentially decreased by at least one of 18 food constituents such as l-glutamic acid, levoglutamide, and spermidine (abundant in oat according to FooDB; names of the food source shown in parentheses hereafter), and levoglutamide (red beetroot). Similarly, in vivo concentrations of 11 drugs for treating hyperlipidemia could be reduced by at least one of 20 food constituents such as aminoadipic acid and levoglutamide (broccoli), and alliin and S-allylmercaptocysteine (garlic), while three drugs for type 2 diabetes mellitus were affected by at least one of nine food constituents such as l-glutamic acid and l-aspartic acid (abundant in many food sources including soybean, wheat, and gelatin; Fig. 3 and see Dataset S6 for details). Furthermore, polyvalent cations such as aluminum, calcium, chromium, iron, and magnesium were found to reduce the in vivo concentrations of the drugs for the above-mentioned three chronic diseases; polyvalent cations were reported to cause treatment failures by binding to drugs (24, 25). These results altogether suggest that DeepDDI can be useful in suggesting which food or its food constituents might need to be avoided during specific medication. In particular, information on the relationship among diseases, drugs to treat the disease, food sources, and food constituents generated in this study (Fig. 3 and Dataset S6) can serve as an essential consideration for medication guideline.
Fig. 3.
Prediction of food constituents that reduce the in vivo concentration of approved drugs. A network showing relationships among 357 diseases, 430 approved drugs, 274 food constituents, and 356 food sources was created using the DeepDDI output sentences obtained from 358,995 drug-food constituent pairs (Datasets S5 and S6). As representative examples, local networks for hypertension, hyperlipidemia, and type 2 diabetes mellitus are presented in gray boxes. In vivo concentration of drugs was predicted to be reduced by the decreased absorption (DDI type 1), decreased bioavailability (DDI type 4), increased metabolism (DDI type 7), and decreased serum concentration (DDI type 9) through drug–food constituent interactions (DFIs). Networks were drawn using Gephi (33).
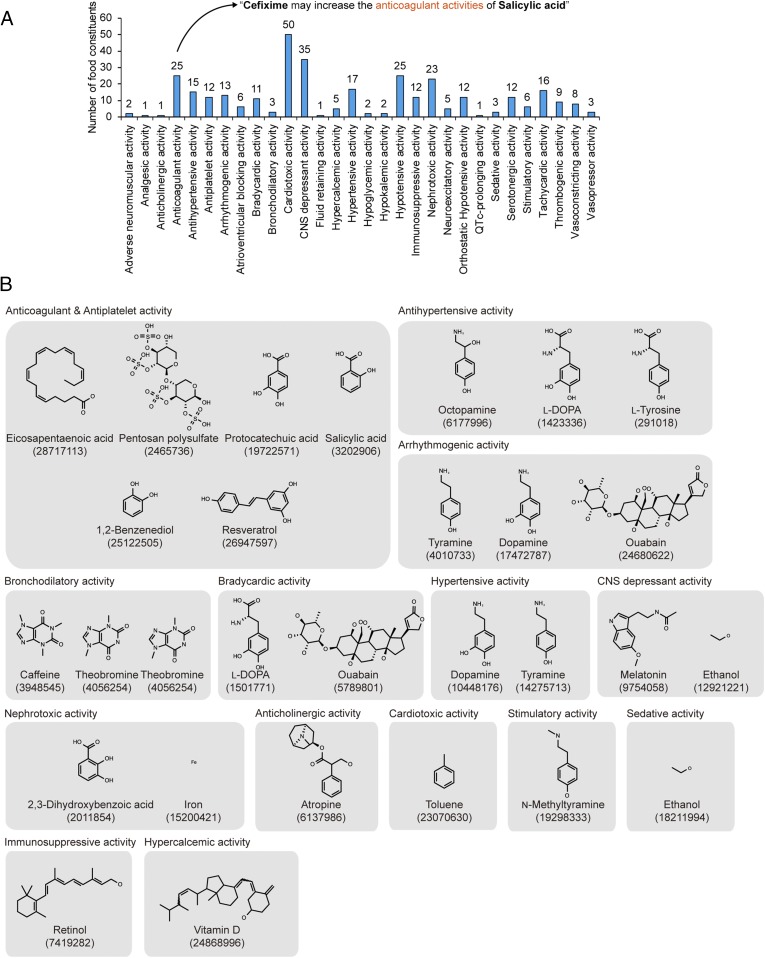
Finally, bioactivities of food constituents were inferred from the above-mentioned 690,956 DeepDDI output sentences for the whole set of 3,288,157 drug–food constituent pairs analyzed. Food constituents were assigned with bioactivities if they were described with the expression form of “[pharmacological effects] activities of [food constituent]” in the 690,956 DeepDDI output sentences (Fig. 4A and SI Appendix, Fig. S10B); for example, salicylic acid could be assigned to have “anticoagulant activity” based on the DeepDDI output sentence of “Cefixime may increase the anticoagulant activities of Salicylic acid”). Consequently, 149 food constituents could be assigned with at least one of the 30 types of bioactivities using 29,423 DeepDDI output sentences having such expression form (Fig. 4A and Dataset S7). Among the 149 food constituents, 23 food constituents were found to have relevant experimental evidences previously reported for the predicted bioactivities; for example, antihypertensive activities of octopamine (mandarin orange), l-DOPA (broad bean), and l-tyrosine (red bell pepper) could be predicted (Fig. 4B). This analysis demonstrates another strategy of deciphering useful information from the DeepDDI output sentences; DeepDDI could be used to predict bioactivities of relatively unknown food constituents involved in drug interactions, provided that their structures are available.
Fig. 4.
Bioactivity prediction of food constituents. (A) Number of food constituents predicted to have each bioactivity based on the DeepDDI output sentences (SI Appendix, Fig. S10B). A unique set of 149 unique food constituents were predicted to have at least one of the 30 bioactivities. (B) Twenty-three food constituents grouped in gray boxes based on their predicted bioactivities among the 149 food constituents. These food constituents have reported evidences. Number below each chemical name refers to PubMed identifier (PMID) of relevant literature (see Dataset S7 for details).
Discussion
In this paper, we report development of DeepDDI that accurately predicts DDI types for given drug pairs and drug–food constituent pairs simply by using names and chemical structures as inputs. DeepDDI employs an optimized DNN along with SSP as a feature vector of drugs, which showed high accuracies (84.8–93.2%) according to the seven standard performance metrics (Fig. 1C). We then showcased four applications of DeepDDI to better understand drug interactions in the context of DDIs and DFIs, including prediction of DDI mechanisms causing ADEs, suggestion of alternative drug members for the intended pharmacological effects without negative health effects, prediction of the effects of food constituents on interacting drugs, and prediction of bioactivities of food constituents. These applications suggest that DeepDDI provides more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date (7). When the DNN used in this study is upgraded based on more training with more data on drug pair interactions, the accuracy of prediction will be further increased. An interesting future study will be performing the above predictions by DeepDDI on a group of multiple (more than two) drugs and/or food constituents. In the real world, we obviously intake many more than two food constituents. Also, such a study will be essential to examine DDIs and DFIs when traditional oriental medicine is used (26). Another important future study will be considering the effects of the concentrations of drugs and metabolites and also in vivo affinities of two given molecules in pair to their targets on DDIs and DFIs, which will require development of new algorithms.
So far, the best DDI database offered by DrugBank only provides DDI types for two drugs only. Once DDI data for multiple drugs and/or food constituents become available, the DNN can be upgraded by training for DeepDDI analysis of DDIs and DFIs of multiple drugs and food constituents. Also, to complement DeepDDI in understanding the mechanisms of action of multiple drugs, increasingly available complementary data and methods can be used, including transcriptome data under drug treatment conditions (27), and the use of drug target association-based methods (12–14), text mining-based methods (28, 29), molecular docking (30), and/or human genome-scale metabolic models (31, 32). For example, the method by Huang et al. (14) provides additional information on the potential of two drugs in pair to interact with the same protein target, which helps better understand mechanisms of action associated with the predicted DDI type (SI Appendix, Fig. S11). In this context, DeepDDI will serve as an essential tool to analyze the pairs of drugs and/or food constituents, and can be further extended to the “real world” DDI and DFI studies on multiple compounds in the future.
Materials and Methods
All of the materials and methods conducted in this study are detailed in SI Appendix, Materials and Methods: preparation of the gold standard DDI dataset for DeepDDI, calculation of structural similarity profile used as an input for DNN, optimization of DNN architecture, DNN training, DNN evaluation, and prediction of drug–food constituent interaction. Source code for DeepDDI is available at https://bitbucket.org/kaistsystemsbiology/deepddi.
Supplementary Material
Acknowledgments
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea. This work was also supported by the Fourth Industrial Revolution AI Flagship initiative from the Korean Advanced Institute of Science and Technology.
Footnotes
Conflict of interest statement: The authors and sponsor declare the technology described here is patent filed (KR-10-2017-0164115) for potential commercialization.
Data deposition: The data and source code reported in this paper have been deposited in the Zenodo and Bitbucket repositories, https://zenodo.org/record/1205795 and https://bitbucket.org/kaistsystemsbiology/deepddi, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803294115/-/DCSupplemental.
References
- 1.Edwards IR, Aronson JK. Adverse drug reactions: Definitions, diagnosis, and management. Lancet. 2000;356:1255–1259. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 2.Kantor ED, Rehm CD, Haas JS, Chan AT, Giovannucci EL. Trends in prescription drug use among adults in the United States from 1999-2012. JAMA. 2015;314:1818–1831. doi: 10.1001/jama.2015.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qato DM, Wilder J, Schumm LP, Gillet V, Alexander GC. Changes in prescription and over-the-counter medication and dietary supplement use among older adults in the United States, 2005 vs 2011. JAMA Intern Med. 2016;176:473–482. doi: 10.1001/jamainternmed.2015.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pirmohamed M, Orme M. Drug Interactions of Clinical Importance. Chapman & Hall; London: 1998. pp. 888–912. [Google Scholar]
- 5.Onakpoya IJ, Heneghan CJ, Aronson JK. Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: A systematic review of the world literature. BMC Med. 2016;14:10. doi: 10.1186/s12916-016-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilar S, Uriarte E, Santana L, Tatonetti NP, Friedman C. Detection of drug-drug interactions by modeling interaction profile fingerprints. PLoS One. 2013;8:e58321. doi: 10.1371/journal.pone.0058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng F, Zhao Z. Machine learning-based prediction of drug-drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J Am Med Inform Assoc. 2014;21:e278–e286. doi: 10.1136/amiajnl-2013-002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilar S, et al. Drug-drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc. 2012;19:1066–1074. doi: 10.1136/amiajnl-2012-000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb A, Stein GY, Oron Y, Ruppin E, Sharan R. INDI: A computational framework for inferring drug interactions and their associated recommendations. Mol Syst Biol. 2012;8:592. doi: 10.1038/msb.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P, Wang F, Hu J, Sorrentino R. Label propagation prediction of drug-drug interactions based on clinical side effects. Sci Rep. 2015;5:12339. doi: 10.1038/srep12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilar S, et al. Similarity-based modeling in large-scale prediction of drug-drug interactions. Nat Protoc. 2014;9:2147–2163. doi: 10.1038/nprot.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 13.Park K, Kim D, Ha S, Lee D. Predicting pharmacodynamic drug-drug interactions through signaling propagation interference on protein-protein interaction networks. PLoS One. 2015;10:e0140816. doi: 10.1371/journal.pone.0140816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, et al. Systematic prediction of pharmacodynamic drug-drug interactions through protein-protein-interaction network. PLoS Comput Biol. 2013;9:e1002998. doi: 10.1371/journal.pcbi.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart DS, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2017;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 17.Angermueller C, Pärnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol Syst Biol. 2016;12:878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan EA. Neural network classification: A Bayesian interpretation. IEEE Trans Neural Netw. 1990;1:303–305. doi: 10.1109/72.80269. [DOI] [PubMed] [Google Scholar]
- 19.Gómez-Bombarelli R, et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger S, Fulle S, Turk S. Mol2vec: Unsupervised machine learning approach with chemical intuition. J Chem Inf Model. 2018;58:27–35. doi: 10.1021/acs.jcim.7b00616. [DOI] [PubMed] [Google Scholar]
- 21.Ferslew KE, Hagardorn AN, Harlan GC, McCormick WF. A fatal drug interaction between clozapine and fluoxetine. J Forensic Sci. 1998;43:1082–1085. [PubMed] [Google Scholar]
- 22.Huitema AD, Mathôt RA, Tibben MM, Rodenhuis S, Beijnen JH. A mechanism-based pharmacokinetic model for the cytochrome P450 drug-drug interaction between cyclophosphamide and thioTEPA and the autoinduction of cyclophosphamide. J Pharmacokinet Pharmacodyn. 2001;28:211–230. doi: 10.1023/a:1011543508731. [DOI] [PubMed] [Google Scholar]
- 23.Price ND, et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35:747–756. doi: 10.1038/nbt.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suda KJ, Garey KW, Danziger LH. Treatment failures secondary to drug interactions with divalent cations and fluoroquinolone. Pharm World Sci. 2005;27:81–82. doi: 10.1007/s11096-004-7040-0. [DOI] [PubMed] [Google Scholar]
- 25.Palleria C, et al. Pharmacokinetic drug-drug interaction and their implication in clinical management. J Res Med Sci. 2013;18:601–610. [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HU, Ryu JY, Lee JO, Lee SY. A systems approach to traditional oriental medicine. Nat Biotechnol. 2015;33:264–268. doi: 10.1038/nbt.3167. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. A next generation connectivity map: L1000 Platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raja K, Patrick M, Elder JT, Tsoi LC. Machine learning workflow to enhance predictions of adverse drug reactions (ADRs) through drug-gene interactions: Application to drugs for cutaneous diseases. Sci Rep. 2017;7:3690. doi: 10.1038/s41598-017-03914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tari L, Anwar S, Liang S, Cai J, Baral C. Discovering drug-drug interactions: A text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010;26:i547–i553. doi: 10.1093/bioinformatics/btq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavecchia A, Cerchia C. In silico methods to address polypharmacology: Current status, applications and future perspectives. Drug Discov Today. 2016;21:288–298. doi: 10.1016/j.drudis.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu JY, Kim HU, Lee SY. Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism. Proc Natl Acad Sci USA. 2017;114:E9740–E9749. doi: 10.1073/pnas.1713050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bastian M, Heymann S, Jacomy M. International AAAI Conference on Weblogs and Social Media. Association for the Advancement of Artificial Intelligence; Palo Alto, CA: 2009. Gephi: An open source software for exploring and manipulating networks; pp. 361–362. [Google Scholar]