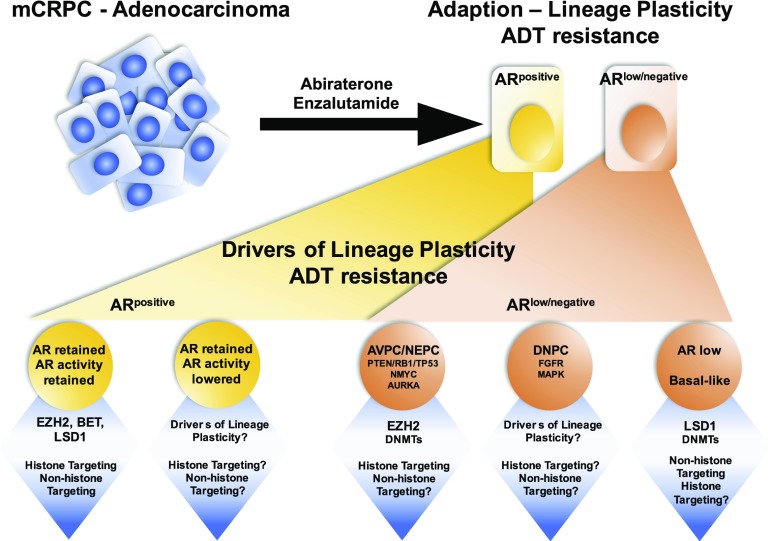
Prostate cancer initiation and progression to a lethal metastatic castration-resistant (mCRPC) phenotype remains largely dependent on the function of the androgen receptor (AR) (1). Recently, however, more potent androgen-deprivation therapy (ADT) regimens that target the AR and the androgen biosynthesis pathways have led to the emergence, in a subset of cases, of resistance mechanisms independent of AR activity (CRPC-AI). These lethal mCRPC appear to adapt to ADT via lineage plasticity rather than as a result of the emergence of resistant mutations, and adopt a phenotype no longer reliant on AR expression and signaling. These tumors may display neuroendocrine features, a stem or basal cell-like phenotype, altered kinase signaling, and characteristic epigenetic alterations (2–5) (Fig. 1).
Fig. 1.
As therapeutic targeting of the AR becomes more precise, a growing number of patients are emerging with resistance mechanisms to AR blockade independent of AR activity. Clinical and preclinical evidence highlights that these patients exhibit low AR activity, independent of AR expression, with increased activity of epigenetic modifiers that drive cellular reprogramming, resulting in lineage plasticity, independent of their canonical catalytic function. This insight provides new understanding and rationale targeting epigenetic modifiers, irrespective of their canonical enzymatic function, providing potential to reverse resistance, or prolong durability of ADT. DNPC, double-negative prostate cancer.
The lysine-specific demethylase 1 (LSD1), a histone demethylase also known as KDM1A, is a regulator of gene expression in stem cells. Prior literature demonstrated that LSD1 acts by demethylation of both H3K4me1/2 and H3K9me1/2, providing dual functionality of gene repression and activation, respectively (6). The latter includes AR target genes, suggesting a functional role for LSD1 in androgen-sensitive disease (7). In PNAS, Sehrawat et al. (8) show that LSD1 promotes survival of castration-resistant prostate cancer cells, independently of its demethylase function and of the AR. In fact, inactivation of LSD1 increased the expression of a subset of AR target genes. Furthermore, the authors show that LSD1’s effects on cell survival are explained in part by activation of a lethal prostate cancer gene network in collaboration with the binding protein ZNF217. Importantly, LSD1 suppression reduces survival of prostate cancer cells grown in the absence of androgens, resistant to the AR antagonist enzalutamide, or that do not express AR. Enrichment of embryonic stem cell gene sets was also linked to LSD1 function, again corroborating previously published data highlighting reversal of programming to a more primitive state and consequent lineage plasticity as an increasingly important mechanism of therapeutic resistance to ADT that is worthy of targeting.
Recent preclinical and clinical studies have provided mechanistic insight on what governs the emergence of lineage plasticity and ADT resistance, involving combinatorial loss-of-function/expression of key tumor-suppressor genes, namely PTEN, RB1, and TP53 (3, 4). In addition, progression to CRPC-AI involves a significant rewiring of the epigenome, providing novel therapeutic targets to treat this lethal phenotype (3, 9, 10). Distinct DNA methylation patterns, together with enhancer of zeste-homolog 2 (EZH2) overexpression, are seen in patients who have progressed to CRPC-AI compared with the adenocarcinoma from which they originated (3). This is an interesting observation, as EZH2 recruits DNA methlytransferases (DMNT) to repress gene expression (11), highlighting the possibility of EZH2 as a master regulator of epigenome rewiring. Furthermore, this raises important consideration that a combination of DNMT and EZH2 inhibition should be considered for CRPC patients exhibiting ADT resistance. In support of EZH2 as a master regulator of epigenome rewiring, Dardenne et al. (12) and Ku et al. (10) recently highlighted that in a genetically engineered mouse model of prostate adenocarcinoma driven by the loss that of Pten, concurrent overexpression of NMyc or loss of Rb1 resulted in lower AR expression, and increased expression of neuroendocrine, stem, and epigenetic gene sets, along with increased expression of EZH2. These tumors were sensitive to EZH2 inhibition, and EZH2 inhibition reactivated AR expression, sensitizing tumors to ADT (10). In prostate cancer patients with the AR− phenotype but in the absence of neuroendocrine differentiation, dependence on FGF and MAPK pathway activation was identified (2). Both FGF and MAPK signaling are known to be critical for stem cell self-renewal and differentiation, and attenuation of either resulted in significant reduction of cell survival (2). Collectively, these data on CRPC-AI suggest that the loss of AR expression/function is a key molecular event that occurs as prostate cancer cells acquire a stem-like phenotype.
Very recent data from Small et al. (13) described an intermediate adenocarcinoma pathologic subtype identified in mCRPC patients that retains high AR expression. Transcriptional signatures from intermediate adenocarcinoma patients overlapped with datasets from neuroendocrine prostate cancer patients, suggesting that in the context of retained AR function, tumors can also activate lineage plasticity networks to evade ADT without shutting down AR expression. Evidence of this exists where enzalutamide-resistant preclinical models with AR expression have shown benefit from disruption of bromo- and extraterminal domain (BET) bromodomain proteins by BET inhibitors (14, 15). Overall, BET inhibitors enhanced efficacy and reversed resistance to AR antagonists. Similarly, inhibition of LSD1 expression in the paper by Sehrawat et al. (8) resulted in greater antitumor effect when combined with ADT in vitro, suggesting the potential for LSD1 targeting to enhance ADT, or reverse resistance to ADT. Importantly, targeting of LSD1 may be effective in both AR+ and AR− prostate cancer, where cellular reprogramming drives resistance to ADT.
An important finding from Sehrawat et al. (8) is that LSD1’s activation of a lethal prostate cancer gene network comprised of specific master regulator transcription factors was independent of its demethylase function. This was demonstrated by the absence of change in H3K4me2 and H3K9me2 marks following LSD1 RNAi and by rescue of LSD1-deficient cells with a catalytically deficient mutant form of LSD1. Instead, LSD1 activates this lethal prostate cancer gene network through binding ZNF217. These collective data have important implications when targeting epigenetic marks. In EZH2-expressing tumors, it has been demonstrated that both histone and nonhistone targeting is important for progression to ADT resistance. Xu et al. (16) had previously demonstrated that EZH2 could be phosphorylated at serine 21, resulting in nonhistone targeting. Overall, global loss of H3K27me3 was accompanied by EZH2 complexed with the AR. Importantly, this AR–EZH2 complex driving AR target genes was still dependent on EZH2 catalytic activity, implying that current catalytic inhibitors of EZH2 will successfully target EZH2 action in either scenario. Because it was demonstrated that LSD1 action was independent of its catalytic function, it is no surprise that catalytic inhibitors targeting the FAD domain of LSD1 did not impact survival of prostate cancer cell lines that were sensitive to LSD1 knockdown. However, treatment with a compound SP-2509, which targeted a H3 pocket within LSD1, antagonized its binding to ZNF217 and replicated the antitumor effects noted with LSD1 RNAi (8). These data suggest that protein–protein interactions, many of which are cell-type–specific, may be key mediators of noncanonical functions of chromatin-modifying proteins.
Overall, Sehrawat et al. (8) provide a novel mechanism underlying the progression of lethal prostate cancer and may provide a road forward to target a subset of these lethal cases. LSD1 appears to promote prostate cancer cell survival in part through the activation of gene networks, including those that regulate a stem cell phenotype. Thus, targeting these pluripotential prostate cancer cells with LSD1 inhibitors may be an attractive approach to resensitize tumors to androgen deprivation. While epigenetic modifiers such as EZH2 already represent therapeutic targets in castration-resistant disease, Sehrawat et al. show that noncanonical demethylase-independent functions of LSD1 are important. Similar studies may clarify critical functions of other chromatin-modifying enzymes, providing the basis for approaches like those outlined by Sehrawat et al. to block noncanonical functions through allosteric inhibition or protein degradation.
Footnotes
The authors declare no conflict of interest.
See companion article on page E4179.
References
- 1.Yuan X, et al. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bluemn EG, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio AM, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res. 2016;22:1520–1530. doi: 10.1158/1078-0432.CCR-15-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith BA, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA. 2015;112:E6544–E6552. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hino S, Kohrogi K, Nakao M. Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci. 2016;107:1187–1192. doi: 10.1111/cas.13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai C, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sehrawat A, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci USA. 2018;115:E4179–E4188. doi: 10.1073/pnas.1719168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clermont PL, et al. Polycomb-mediated silencing in neuroendocrine prostate cancer. Clin Epigenetics. 2015;7:40. doi: 10.1186/s13148-015-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ku SY, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viré E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 12.Dardenne E, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small EJ, et al. Clinical and genomic characterization of metastatic small cell/neuroendocrine prostate cancer (SCNC) and intermediate atypical prostate cancer (IAC): Results from the SU2C/PCF/AACRWest Coast Prostate Cancer Dream Team (WCDT) J Clin Oncol. 2016;34:5019. [Google Scholar]
- 14.Asangani IA, et al. BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol Cancer Res. 2016;14:324–331. doi: 10.1158/1541-7786.MCR-15-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welti J, et al. Targeting bromodomain and extra-terminal (BET) family proteins in castration resistant prostate cancer (CRPC) Clin Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-17-3571. [DOI] [PubMed] [Google Scholar]
- 16.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]