Abstract
Planar cell polarity (PCP) signaling is well known to play a critical role during prenatal brain development; whether it plays specific roles at postnatal stages remains rather unknown. Here, we investigated the role of a key PCP-associated gene scrib in CA1 hippocampal structure and function at postnatal stages. We found that Scrib is required for learning and memory consolidation in the Morris water maze as well as synaptic maturation and NMDAR-dependent bidirectional plasticity. Furthermore, we unveiled a direct molecular interaction between Scrib and PP1/PP2A phosphatases whose levels were decreased in postsynaptic density of conditional knock-out mice. Remarkably, exposure to enriched environment (EE) preserved memory formation in CaMK-Scrib−/− mice by recovering synaptic plasticity and maturation. Thus, Scrib is required for synaptic function involved in memory formation and EE has beneficiary therapeutic effects. Our results demonstrate a distinct new role for a PCP-associated protein, beyond embryonic development, in cognitive functions during adulthood.
Keywords: planar cell polarity, synapse, LTD, consolidation memory, phosphatases
Introduction
Planar cell polarity (PCP) signaling controls an array of early developmental processes including asymmetric cell division, collective cell movement and tissue organization, and embryonic disruption of PCP signaling leads to severe developmental defects in mammals (Montcouquiol et al. 2006a; Wang and Nathans 2007; Bayly and Axelrod 2011; Wallingford 2012; Ezan and Montcouquiol 2013; Tissir and Goffinet 2013). In the mammalian brain, existing evidence has highlighted a key role of PCP proteins in early brain development, during which they modulate axonal guidance, dendritic extension and branching and synaptogenesis (Shima et al. 2007; Tissir and Goffinet 2010; Feng et al. 2012a; Liu et al. 2014; Sowers et al. 2013; Hagiwara et al. 2014; Nagaoka et al. 2014; for review see Sans et al. 2016). Previous studies involving embryonic mutations of PCP proteins have suggested that the PCP signaling pathway might affect brain function in adulthood (Moreau et al. 2010; Feng et al. 2012a; Sowers et al. 2013). However, although these mutations affect prenatal brain development, the potential direct role of PCP proteins during postnatal stages still remains unknown. For example, a recent report from our group showed that a spontaneous mutation with a dominant negative effect affecting one of the key PCP-associated genes, Scribble (Scrib) in mice (circletail mice; Scribcrc), during embryonic stages leads to alterations in cognitive processes in a hippocampus-dependent memory task (Moreau et al. 2010). While this suggests that the contribution of the PCP signaling pathway to the nervous system might not be restricted to early prenatal development, whether Scrib is involved in memory processes in adulthood remains to be addressed.
Scrib was originally characterized as a tumor suppressor belonging to the LAP (LRR and PDZ) protein family combining in their structure both LRR (Leucine Rich Repeats) at their N-terminus and one to four PDZ (PSD-95/Dlg/ZO-1) domains (Bilder et al. 2000). Subsequently, Scrib has been implicated in the establishment of apical/basal and planar cell polarities in different tissues and systems (Montcouquiol et al. 2003, 2006b; Dow et al. 2007; Courbard et al. 2010; Verghese et al. 2012). Recent observations in the mammalian brain have revealed that Scrib accumulates at dendritic spines and postsynaptic densities in the hippocampus (Audebert et al. 2004; Moreau et al. 2010; Richier et al. 2010; Piguel et al. 2014). However, whether Scrib contributes to hippocampal structure and function following the major developmental processes of postnatal stages is not known.
Here, to address this question, we developed conditional knock-out mice for Scrib (CaMK-Scrib−/−) that target postnatal excitatory neurons of the hippocampus using Cre-recombinase under the CaMKIIα promoter to avoid deletion of Scrib during prenatal brain development. We found that the absence of postnatal Scrib in hippocampal excitatory neurons led to impaired NMDAR-dependent synaptic plasticity underlying spatial learning and memory consolidation. Remarkably, exposure to an enriched environment (EE) preserved memory formation in the CaMK-Scrib−/− mice and recovered synaptic plasticity and maturation in the adult hippocampus. We show that the PCP protein Scrib allows for synaptic maturation and the bidirectional plasticity required for memory formation in the hippocampus. In conclusion, this study unveils a fundamental role of Scrib-dependent-signaling not only in prenatal brain development but also during postnatal stages in regulating higher-order brain function such as learning and memory.
Experimental Procedures
Animals
To generate the Scrib flox line a mouse bacterial artificial chromosome (BAC) containing 11 994 bp of Scrib genomic DNA was identified by screening a 129-based BAC library (CJ7 ES cell DNA, CITB, Research Genetics). After generating an intermediate targeting allele (TA) with loxP sites upstream of Scrib exon 2 and downstream of exon 8, and frt sites that flank a positive selection neomycin (NEO) marker, mice containing the TA were mated to mice expressing Flpase, which removed the neo marker and generated the floxed (f/f) allele. In Fig. S1, arrows indicate locations of PCR primers F (forward), R1 (reverse 1), and R2 (reverse 2). Scrib mutant mice were genotyped by PCR using the following primers: F-gcacactgggtatcatggcta; R1-gcaatctccagagccttacaga; R2-cccttggaaacctacatcccaa. The amplified products were 437 bp for WT band (F+R1), 541 bp for flox band (F+Wt); 193 bp for cKO band (F+R1) used to identify whether exons 2–8 had been deleted (Supplementary Fig. S1A). Cre genotyping was done using the following primers: F-CGGCATGGTGCAAGTTGAATA; R-GCGATCGCTATTTTCCATGAG. The resultant band was 300 bp. Scrib conditional knock-out (cKOs) mice were generated by crossing Scribf/f mice that carry a floxed Scribble gene and CaMKIIαCre/+ mice that express Cre-recombinase in principal neurons of the brain (Casanova et al. 2001). The Scrib flox line was previously described (Yamben et al. 2013).
To profile CaMKIIa-Cre-directed gene expression throughout the mouse brain, we crossed homozygote B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J (thereafter called Ai6 mice) with CaMKIIα-Cre/+ mice, in order to obtain a compound Ai6f/+/CaMKIIα-Cre/+.
Adult Scribf/f and CaMK-Scrib−/− male mice were housed collectively in groups of five to eight in polypropylene cages for biochemical experiments and EE behavioral experiment and in individual cages for others behavioral experiments. Room temperature (RT) was set at 23 ± 1°C, lights were on from 7:00 a.m. to 7:00 p.m. and food and water were available ad libitum. Male mice littermates were used in all experiments.
This study was performed in full accordance with recommendations of the European Communities Council Directives (86/609/EEC), the French national Committee (87/848), and the requirements of the United Kingdom Animals (Scientific Procedures) Act 1986, AWERB Newcastle University (ID: 374).
Histology and Immunohistochemistry
Mice were anesthetized using a Phenobarbital i.p injection (lethal dose). Physiological saline followed by 4% paraformaldehyde (PFA) in 0.1 M PBS at pH 7.4 were next perfused transcardially, and fixed brains were removed and stored in 4% PFA in PBS at 4°C for 1 week. Serial brain sections of 40 µm were obtained using a vibratome and stained with cresyl violet. Cre-recombinase expression in the hippocampus was detected using Ai6 transgenic mice. About 40 µm fixed adult-brain sections were processed accordingly to the manufacturer's instructions of the BrainStain Imaging kit (Life Technologies). Neuronal staining was obtained using NeuroTrace 530/615 red fluorescent Nissl stain (1/300; Life Technologies) and was coupled with DAPI (4′,6-diamidino-2-phenylindole), a nucleic acid stain (1/300).
For Scrib immunofluorescence staining, brains were frozen in isofluorane, and cryosectioned at 20 µm. After a short fixation in 4% PFA in PBS, non-specific sites were blocked with 3% normal goat serum (NGS)/0.1% Triton X-100 in PBS for 1 h at RT. Anti-Scrib rabbit polyclonal antibody (Montcouquiol et al. 2006b) (1/300) was incubated with brain sections overnight at 4°C. Following washing, secondary Cy3-coupled anti-rabbit (1/500; Jackson ImmunoResearch) and DAPI (1/20 000) in 0.1% Triton X-100 in PBS were applied for 1 h at RT. Labeled brain sections were mounted on slides using Prolong Gold Antifade (Life Technologies) and conserved at 4°C for analysis with fluorescence microscope (Zeiss). No fluorescence was found in control experiments in which primary antibody was omitted.
Electron Microscopy Analysis of CA1 Hippocampal Synapses
Adult male mice were terminally anesthetized by a brief inhalation of isoflurane (0.05% in air) and intramuscular injection of ketamine (100 mg/kg) and xylazine (10 mg/kg), and then transcardially perfused with 4% PFA and 2% glutaraldehyde in 0.1 M PBS, pH 7.2. The brains were dissected and stored in 4% PFA in PBS, at 4°C, overnight. Vibratome sections (100 µm) were cut, collected in PBS, postfixed in 1% osmium tetroxide, dehydrated in ascending scale of ethanol, infiltrated, and flat embedded in Durcupan epoxy resin as previously described (Moreau et al. 2010; Piguel et al. 2014). CA1 hippocampi were trimmed and embedded in resin blocks for further semithin and ultrathin sectioning with a Leica UC6 ultramicrotome. Ultrathin sections (70–90 nm) were counter-colored with uranyl acetate and lead citrate, and visualized with a Philips CM100 transmission electron microscope (FEI) at 100 kV. The images were captured with an AMT XR40 4 megapixel side mounted CDD camera at a magnification between 7900 and 92 000×. Only identified synapses on dendritic spines of apical dendrites of pyramidal cells in CA1 “stratum radiatum” were included in the analysis. No tangentially cut synapses were analyzed. To determine the spine density (number of spine/µm2), we utilized 14–17 images per animal (7 900× magnification, single image area = 176.95 µm2) in which we identified the spines and, then, quantified them using the cell counter tool in ImageJ (http://rsb.info.nih.gov/ij). The average thickness of the postsynaptic density (PSD) was measured as described previously (Dosemeci et al. 2001). Results in Scribf/f and mutant animals are presented as the mean ± SEM. The measurements were all performed by experimenter's blind to the genotype.
Yeast Two-Hybrid Screening
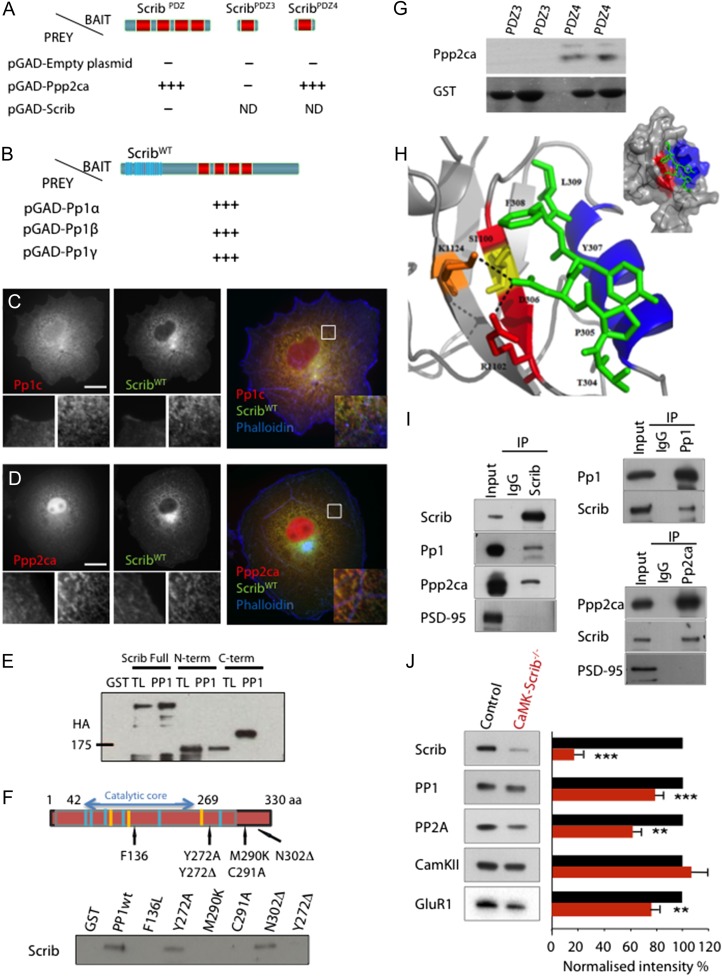
The PDZ3 and PDZ4 portions of rat Scrib (aa 990–1070 and 1086–1180, respectively) were subcloned into pGBTK7 vector (Clontech) in-frame with the DNA-binding domain of GAL4 and used as a bait for the screening as previously described (Sans et al. 2003). Yeast two-hybrid screening and assays were performed accordingly to the Matchmaker™ Gold Yeast Two-Hybrid System protocol (Clontech). AH109 cells expressing ScribPDZ3 and ScribPDZ4 were combined with Y187 cells expressing a P10 mouse brain cDNA library (Yi et al. 2007). The mating mixture was plated on SD/Ade−/Trp−/Leu−/His− plates. From 1.5 × 103 colonies obtained 5 days after transformation, 56 passed high stringency conditions. Library plasmids from those colonies were rescued, amplified by PCR and sequenced, including Ppp2ca. Interactions between Ppp1c and Ppp2ca phosphatases and Scrib constructs transfected into the haploid yeast strain AH109 were tested through mating of the two yeast strains.
For additional yeast two-hybrid screens, baits were cloned in pBTM116 to express proteins fused to LexA-BD, which carries Trp1. A human mammary gland epithelial library cloned in pACT2 (Clontech), which carries Leu2 as a selection marker, was screened with Scrib, Erbin or LANO as baits. L40 Trp+Leu+ cotransformants were grown on plates with supplemented minimum medium that lacked tryptophan, leucine, histidine and contained 10 mM 3-aminotriazole (3-AT) and then tested for the β-galactosidase activity by the filter method.
Glutathione S-Transferase Binding Assays
For GST pull-down assays, full or truncated HA-tagged versions (NH2- and carboxy-terminal) of hScrib or PP2A were expressed in COS7 or HEK cells and assayed for binding to recombinant GST-PP1 fusion proteins or GST-PDZ3 or PDZ4 of Scrib (Montcouquiol et al. 2006b) expressed in BL21 using pGEX-4T-1 and purified directly from bacterial extract on glutathione–Sepharose-4B beads.
Cells were washed twice with cold PBS and lysed in lysis buffer (50 mM HEPES [pH 7.5], 10% glycerol, 150 mM NaCl, 1% Triton X-100, 1.5 mM MgCl2, and 1 mM ethyleneglycol-bis(2-aminoethylether)-N,N,N′,N′-tetra acetic acid, EGTA) supplemented with 1 mM phenylmethylsulphonylfluoride [PMSF], 10 μg/ml aprotinin, and 10 μg/ml leupeptin. After preclearing, cellular lysates were incubated with the appropriate GST-tagged proteins bound to agarose beads for 2 h. Protein complexes bound to beads were recovered and washed once with lysis buffer and twice with HNTG buffer (same as lysis buffer with 0.1% Triton X-100 final). Beads containing complexes were boiled in 2× sample buffer, separated on 7.5% or 10 % SDS-PAGE and transferred on nitrocellulose for western blot analysis with the appropriated antibody. GST pull down performed with wild type and different mutant PP1 were done as mentioned above using Caco-2 cell lysate. Anti-HA 3F10 mouse monoclonal and anti-Scrib (C20) rabbit antibodies are from Roche Molecular Biochemicals and Santa Cruz Biotechnology, respectively. The hScrib constructions are described in Audebert et al. (2004). PP1 mutations were done with the Quick Change Kit (Stratagene) according to the manufacturer's instructions.
Coimmunoprecipitation
For coimmunoprecipitation experiments, hippocampi from 1 month old Sprague Dawley rats were used. Soluble extracts of 0.5–1 ml were incubated with 25 µl of PPI or 10 µl of anti-Scrib rabbit antibody (Montcouquiol et al. 2006b). Samples were immobilized on Protein A/G agarose beads and beads were then pelleted by centrifugation and resuspended in 2× SDS sample buffer (Sans et al. 2003, 2005). Samples were finally analyzed by SDS/PAGE and immunoblotted using 1/500 anti-Scrib rabbit polyclonal antibody; 1/100 anti-PP2A (clone 46, BD Biosciences), 1/400 anti-PP1 (sc-7482, Santa Cruz Biotechnology).
PSD Preparation
At least 15 male mice of each genotype were used for the preparation of subcellular and PSD fractions. The detailed fractionation procedure is presented in Moreau et al. (2010) and Western blots were performed essentially as described previously (Sans et al. 2000). Briefly, pellets (synaptosomes) were resuspended in buffer (32 mM sucrose in 12 mM Tris, pH 8.1) solubilized in ice-cold 0.5% Triton X-100, then centrifuged and the pellet was resuspended in 6 mM Tris, pH 8.1, and flash frozen. BCA Protein Assay was first used to estimate Protein concentration, then the concentration was adjusted and verified using infrared (IR)-based protein quantitation (Direct Detect® Infrared Spectrometer, Millipore). An equal amount of protein was loaded for both genotype and the quantification of immunoblots was performed using Quantity One software (Biorad). Rabbit polyclonal antibody anti-Scrib was diluted at 1/500, mouse monoclonal anti-CaMKII (C265, Sigma-Aldrich) at 1/5000, PP2A at 1/250, and PP1 at 1/400. Each experiment was repeated three times, and representative blots are shown.
COS-7 Cell Culture, Transfection, and Immunocytochemistry
Cells were cultured in DMEM, supplemented with 10% (v/v) fetal calf serum (Life Technologies), 2 mM L-glutamine, and penicillin/streptomycin (50 U/ml) and transfected as previously described (Sans et al. 2005). hScrib-GFP (green fluorescent protein) and HA-tagged PPpp1c or Ppp2ca cDNA samples were generously provided by I. Macara and D. Brautigan (University of Virginia, Charlottesville, VA, USA), respectively. About 48 h after transfection, cells were fixed with 4% PFA in 0.1 M PBS at pH 7.4, washed and permeabilized with 0.25% Triton X-100 in PBS. After blocking, cells were incubated with the following primary antibodies: mouse anti-GFP (1/1000; JL-8, Clontech), mouse anti-HA (1/1000; MMS-101P, Covance). Following washout, cells were incubated with Alexa Fluor 488, 568 conjugated anti mouse antibodies and Alexa Fluor 647-phalloidin (Life Technologies). Images were obtained on a Zeiss AxioImager Z1 microscope.
Interaction Model
The PDZ domains are small domains whose structure is highly conserved. There are many structures in the PDB showing an interaction between a PDZ domain and the C-terminal peptide of a partner. In these known structures, the C-terminal of the target peptide comes to be fixed to a hydrophobic pocket. To build an interaction model between residues 1082 to 1179 of Scrib (PDZ4 domain) and 304 to 309 of Ppp2ca, we used the known structure of the Scrib PDZ4 domain (PDB: 1UJU). We searched the structure of a PDZ domain in complex whose protein sequence is most similar to Scrib PDZ4 domain, and found the PDB 1MFG (Birrane et al. 2003). We aligned the two PDZ domain structures one on the other, and residues 304–309 of the peptide Ppp2ca on the peptide of the PDB 1MFG. The C-terminal residues of Ppp2ca are thus placed in the hydrophobic pocket of Scrib PDZ4 domain. This new PDB file was used as the basis for this interaction model. We used Coot to build the model and MolProbity to test its quality (Piguel et al. 2014). The final model did not show clashes between Ppp2ca peptide and the Scrib PDZ4 domain. We used PyMOL to prepare the figures.
Electrophysiological Recordings
Littermate male mice between postnatal Day 29 and P34 were used for all electrophysiological recordings. To record activity of CA3-CA1 synapses, we electrically stimulated Schaffer collateral fibers and recorded CA1 field excitatory postsynaptic potentials (fEPSPs). Basal transmission was evaluated by increasing the intensity of stimulation from 0 to 10 mV in 1 mV steps. Paired-pulse ratio (PPR) was examined by delivering two stimulations at the same intensity separated by 25, 50, 100, or 200 ms. LTP protocols consisted of 2 or 3 trains at 100 Hz for 1 s and LTD protocol consisted of 900 pulses delivered every second at 1 Hz during 15 min. Averages of the last 10 min were compared between Scribf/f and CaMK-Scrib−/− mice, unless mentioned otherwise. During extracellular field recordings, at least 20 min of stable fEPSP slopes (baseline) were recorded before starting any experiment. aCSF contained the following drugs: 10 µM strychnine, 100 µM picrotoxin, and 100 µM RS-MCPG ((RS)-α-Methyl-4-carboxyphenylglycine). D-AP5 was used when indicated to test LTP and LTD dependence on NMDARs. For AMPAR-mediated miniature EPSCs experiments, CA1 cells were voltage clamped at −70 mV in the presence of TTX (0.5 µM). Amplitude and frequency of at least 200 continuous mEPSC events per cell were analyzed. Recorded data were amplified by Multiclamp 700B (Axon instruments) and recorded on the hard disk using pClamp9 (Axon instruments). All drugs were purchased from Tocris Bioscience. All experiments were performed without prior knowledge of mice genotype.
Behavioral Testing
Behavioral experiments were conducted on adult Scribf/f and CaMK-Scrib−/− male mice littermates.
Mice were tested in activity cages for their locomotor activity, in the Plus maze test to measure anxiety-like behavior as well as in the Y-maze to test spontaneous alternation (Moreau et al. 2010). All behavioral tracking images were analyzed with Viewpoint video tracking. During the Plus maze test, mice were placed in the center of the maze and allowed free access to all arms for 5 min (60 lux in the center). The percentage time spent in the open arms was computed. During the Y-maze test, mice were placed at the end of one of the arms of the maze and allowed free access to three arms for 5 min (60 lux in the center). The sequence of arms entries, the total number of arm entries, and the number of triads are recorded in order to calculate the percentage of alternation.
Morris water maze testing took place in a circular pool (diameter, 150 cm) filled with water (19–20°C) rendered opaque by a nontoxic white cosmetic adjuvant. Mice were trained to locate a submerged platform to escape from the water (14 cm diameter, 1.5 cm below the water surface) using spatial cues placed on the walls. Each subject was placed by the tail into the water, immediately facing the perimeter, at one of the cardinal compass points. Mice were released from a different starting point at each trial, and sequence of these starting points was randomized from day to day. Visible platform task was also performed in the same water maze using a 15-min inter-trial interval and a total of 6 trials. Three daily trials with 5 min inter-trial interval and a cutoff at 60 s were conducted during training phase. One or 24 h after the last session of acquisition, a probe test for spatial discrimination was conducted. For that, the hidden platform was removed and each subject was placed into the water diagonally opposite to the target quadrant. Time spent in the target quadrant (% of total time, chance level = 25%) in addition to the number of the platform location crossings (where the platform was located during training) were measured over 60 s.
Enriched Environment
Environment enrichment started during prenatal period and continued postnatally for at least 4 weeks before the onset of experiments. Enriched cages consisted of large (46 × 37 × 21 cm) cages that contained at least five mice per cage and equipped with running wheels, small houses, and several toys. Position of running wheels and small houses changed on a daily basis whereas toys were daily rearranged and some replaced with new ones of different shapes, textures, and colors in order to stimulate animals exploratory behavior. All mice received standard lab chow and water ad libitum. The experimenters were blind to genotype throughout electron microscopy studies, electrophysiological, and behavioral testing.
Statistical Analysis
Statistical analyses performed for each experiment are summarized in Supplementary Table S1 that indicates chosen statistical test, n and P values, as well as degree of freedom and F/t values. Data were tested for normality using D'Agostino & Pearson omnibus normality test where appropriate and all graphs represent mean ± SEM. GraphPad Prism software was used for statistical analysis and P < 0.05 was considered as statistically significant.
Results
Postnatal Scrib Deficiency Causes no Gross Defect in Hippocampus Morphology
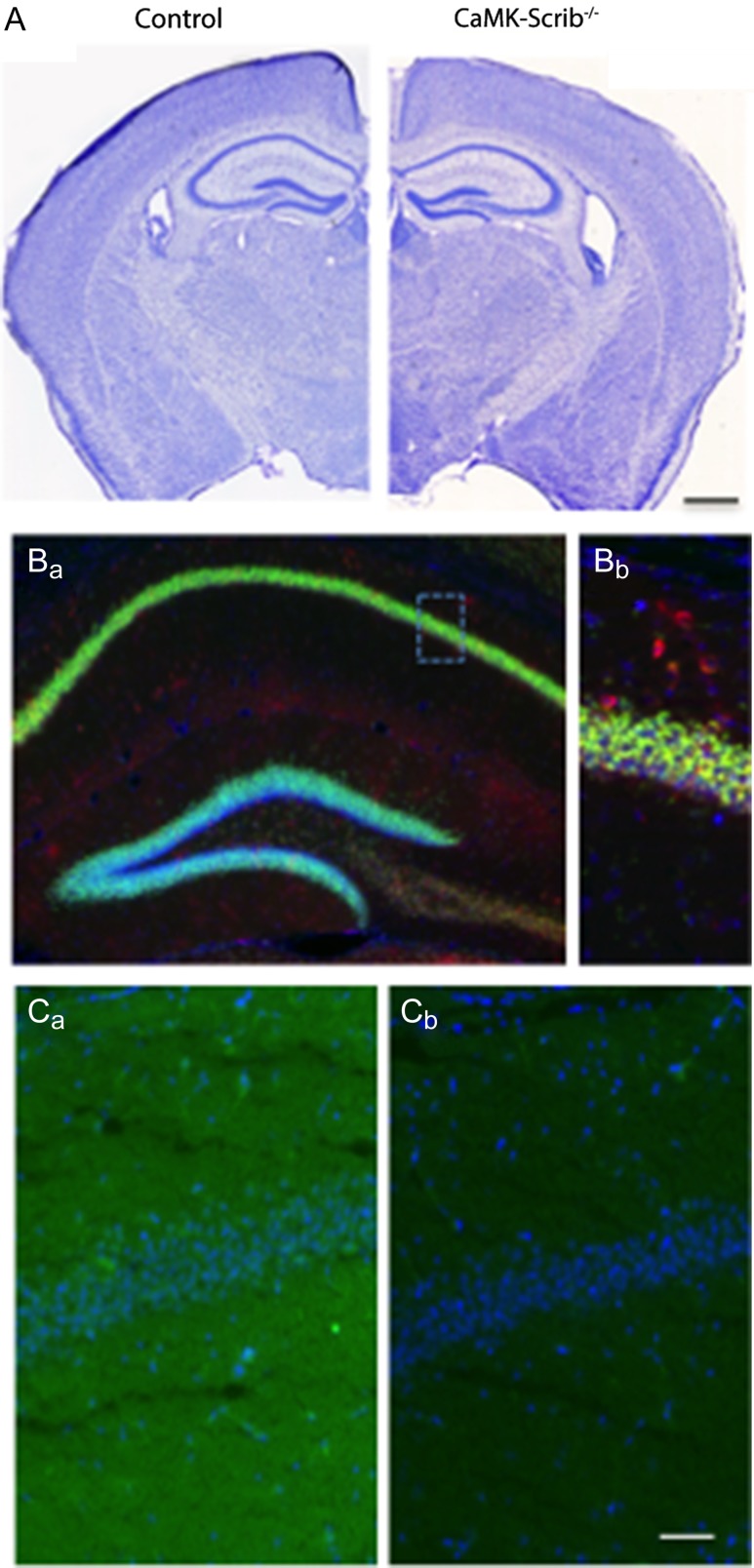
To investigate whether Scrib plays a role in hippocampal structure and function during the postnatal stages, we crossed mice carrying a conditional allele of the scrib gene (Scribf/f) (Yamben et al. 2013), with mice carrying the CaMKIIα-Cre-recombinase allele (Casanova et al. 2001) and engineered Scribf/f,CaMKII-cre conditional knock-out mice (here referred to as CaMK-Scrib−/−). CaMKIIα promoter restricted the excision of the scrib gene to CaMKII-expressing neurons of the postnatal forebrain (Supplementary Fig. S1A). Littermate Scribf/f mice with intact scrib gene were used as control (here called Control). Both genotypes developed normally and were able to breed. CaMK-Scrib−/− mice did not exhibit any gross abnormalities in the morphology of their adult brains stained with cresyl violet, although a slight lateral ventricle enlargement was observed along the rostrocaudal axis (Fig. 1A). The Scribf/f gene was detected by PCR at all examined ages (2, 5, and 10 weeks) and in all examined tissues (tail, cortex, and hippocampus) of Control and CaMK-Scrib−/− mice (Supplementary Fig. S1B). In contrast, cre-recombinase and scribΔf/f genes were detected only in CaMK-Scrib−/− forebrain structures as early as 2 weeks postnatally. As expected, both intact scribf/f and excised scribΔf/f were present in CaMK-Scrib−/− hippocampus and cortex because excision of scrib takes place only in CaMKII-positive neurons.
Figure 1.
Generation of Scrib conditional knock-out mouse: CaMK-Scrib−/−. (A) Coronal sections stained with cresyl violet showing normal brain gross anatomy in control and CaMK-Scrib−/− mice. Scale bar = 1 mm. (B) NeuroTrace 530/615 (red) and DAPI (blue) staining showing expression in the hippocampus of 10-week-old Ai6f/+/CaMKIIα-Cre/+ double-transgenic mouse (Ba), and a higher magnification of CA1 pyramidal layer (Bb). Scale bar = 40 µm. (C) Scrib expression (green) in dorsal hippocampus of adult Control (Ca) and CaMK-Scrib−/− (Cb) mice and DAPI staining (blue). Scale bar = 50 µm.
The following figure supplement is available for Figure 1: Supplementary Fig. S1. Generation of Scrib conditional knock-out mouse.
The spatial pattern of Cre/loxP recombination in the CaMK-Scrib−/− mouse line was examined by crossing this line with an Ai6 ZsGreen1 reporter mouse (Madisen et al. 2010) and brain sections from the progeny were stained with DAPI and NeuroTrace fluorescent Nissl stains. At 10 weeks of age, strong green fluorescence certifying the recombination occurred in virtually all pyramidal cells in the hippocampus (Fig. 1B). As a result, Scrib protein levels decreased in the hippocampus of CaMK-Scrib−/− mice as early as 2 weeks postnatally (n = 4, P < 0.05) (Supplementary Fig. S1C). At 5 weeks old (5 w), Scrib levels in CaMK-Scrib−/− mice were reduced by 70% compared to their Control littermates, and by up to 80% at 10 weeks (P < 0.05). The residual Scrib (~20%) detected in the hippocampus was mainly due to the expression of Scrib in interneurons, glia and endothelial cells in which Scrib downregulation does not occur. In regions outside the forebrain, such as the cerebellum where the CaMK-Cre-recombinase is not expressed, no decrease in Scrib was observed (n = 3, P > 0.10) (Supplementary Fig. S1D). Finally, immunohistochemistry labeling with an antibody against Scrib (Montcouquiol et al. 2006b) confirmed the selective deletion of scrib in the hippocampus of the CaMK-Scrib−/− mice resulting in a loss of Scrib protein (Fig. 1C). In summary, we generated a mouse model in which Scrib is preserved during embryonic development and is specifically eliminated in the excitatory neurons of the hippocampus at postnatal stages.
Slower Spatial Learning and Impaired Memory Consolidation in CaMK-Scrib−/− Mice
To determine whether the loss of postnatal Scrib affects hippocampal-dependent memory, we first investigated the behavioral phenotype of CaMK-Scrib−/− mice to assess possible alterations that could bias interpretations of their performances during memory tests. In tests for locomotor activity (Supplementary Fig. S2A), anxiety-like behaviors (Supplementary Fig. S2B) and spontaneous alternations in a Y-maze (Supplementary Fig. S2C), we found no differences between the two genotypes, indicating that locomotor activity, emotionality, and spontaneous navigation are preserved in CaMK-Scrib−/− mice.
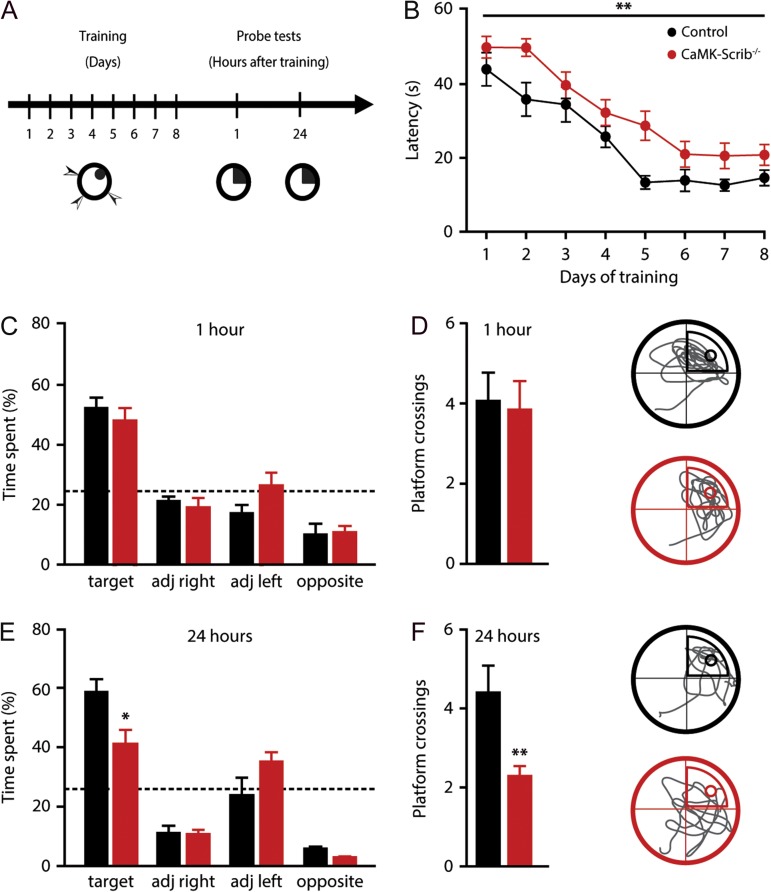
We then proceeded to evaluate hippocampus-dependent spatial learning and reference memory in CaMK-Scrib−/− mice using the Morris water maze test (Fig. 2A). During training, Control and CaMK-Scrib−/− groups improved their performance over time (n = 12–17, P < 0.001) (Fig. 2B). However, CaMK-Scrib−/− mice showed a slower learning rate compared to Control mice, as revealed by their significantly higher latencies (P < 0.01) and distance traveled (P = 0.01, Supplementary Fig. S3A) to find the hidden platform over the 8-day acquisition phase, although swim speed was similar in both groups (P = 0.84) (Supplementary Fig. S3B). Noticeably, by the end of the acquisition phase, the performances of both groups were equivalent (D7; P = 0.15 and D8; P = 0.41) and they also performed equally well in the visible platform task that requires the comprehension of the water-maze rule (find a refuge platform) as well as normal sensorimotor abilities (P = 0.84) (Supplementary Fig. S3C).
Figure 2.
Impaired long-term spatial memory in CaMK-Scrib−/− mice. (A) Experimental timeline of hidden platform water maze test showing an 8-day training phase followed by two probe tests at 1 and 24 h after the last training session. (B) Slower spatial learning in CaMK-Scrib−/− (red; n = 17) compared to Control mice (black; n = 12) revealed by mean latency to reach the hidden platform in Morris water maze over 8 days of training. Data were compared using two-way analysis of variance (ANOVA) test, genotype effect, P < 0.01. (C) Equivalent time spent searching in each quadrant during a 60-s probe trial performed without the platform at 1 h (red; n = 6 and black; n = 9). Data were compared using unpaired t-test. (D) Same number of platform crossings and representative swim trace patterns during probe test at 1 h for Control (top) and CaMK-Scrib−/− (bottom); a circle marks target location. Data were compared using unpaired t-test. (E) Time spent searching in target quadrant during probe test at 24 h was decreased in CaMK-Scrib−/− (red; n = 8) compared to Control mice (black; n = 6). Data were compared using unpaired t-test, P < 0.05. (F) Fewer platform crossings were counted for CaMK-Scrib−/− mice indicating imprecise localization of the hidden platform and representative swim trace patterns during probe test at 24 h for Control (top) and CaMK-Scrib−/− (bottom); a circle marks target location. Data were compared using unpaired t-test, P < 0.01. Data are represented as mean ± SEM.
The following figure supplements are available for Figure 2: Supplementary Fig. S2. Normal locomotor activity, anxiety levels, and spatial working memory in CaMK-Scrib−/−mice;Supplementary Fig. S3. CaMK-Scrib−/−mice exhibit normal short-term but impaired long-term spatial memory.
Memory retention was assessed at two time points, 1 and 24 h after training. First, to evaluate short-term memory, a retrieval probe test was conducted in half of the animals from each genotype group 1 h after the last acquisition session. Control and CaMK-Scrib−/− mice spent equivalent amounts of time in the target quadrant (49.8 ± 3.7%, n = 6, and 45.9 ± 3.7%, n = 9, respectively, P = 0.27) (Fig. 2C) and had a similar number of platform crossings, reflecting precise spatial localization of the platform in both groups (Control: 4 ± 0.7 and CaMK-Scrib−/−: 3.8 ± 0.7, P = 0.84) (Fig. 2D). However, when these same animals were tested with a probe test at 24 h post-training, CaMK-Scrib−/− mice displayed impaired long-term memory. This was apparent in the significantly lower amount of time spent in the target quadrant (CaMK-Scrib−/− mice: 40.2 ± 5%, n = 8; Control 58.4 ± 4.3%, n = 6, P < 0.05) (Fig. 2E) and the reduced number of platform crossings (CaMK-Scrib−/−: 2.1 ± 0.2; Control: 4.3 ± 0.7, P < 0.01) (Fig. 2F) in CaMK-Scrib−/− mice compared to Control mice. Given that the absence of the platform during the probe test at 1 h can induce extinction learning that could have interfered with the measurement of long-term memory recall at the 24 h probe test, we tested the second half of trained animals only at 24 h post-training, which were not tested at the 1 h post-training time-point. Again, we found that the CaMK-Scrib−/− mice exhibited a similar decrease in the amount of time spent in the target quadrant (44.8 ± 7.4%, n = 8) compared to Control mice (67.5 ± 3.7%, n = 6, P < 0.05) (Supplementary Fig. S3D). A similar difference was observed in the number of platform crossings (CaMK-Scrib−/−: 1.9 ± 0.4 and Control: 5.8 ± 0.6, P < 0.001) (Supplementary Fig. S3E). These data reveal impairments in long-term, but not short-term, spatial memory in CaMK-Scrib−/− mice suggesting a deficit in spatial memory consolidation.
Reduced Number of Active CA3-CA1 Synapses in CaMK-Scrib−/− Mice
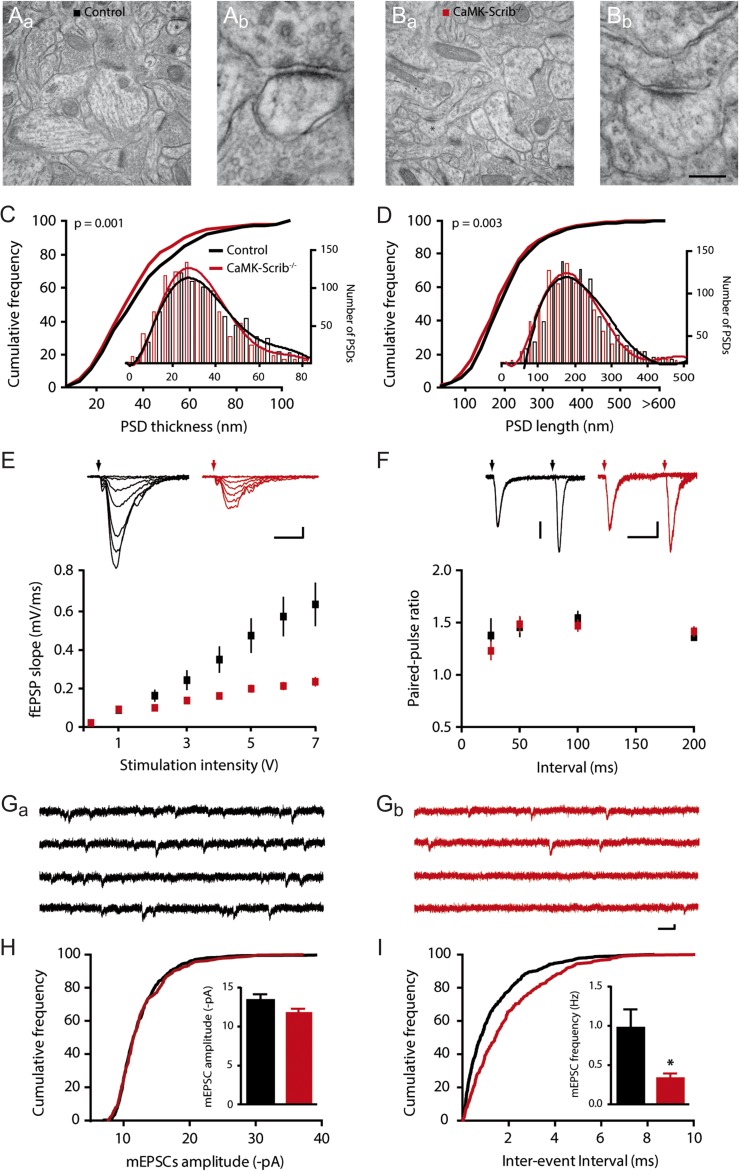
To analyze the potential cellular mechanisms underlying the observed spatial memory impairment, we examined whether the deletion of Scrib in pyramidal neurons had altered synaptic morphology and function. We focused on the CA1 region given its key role in the acquisition and retention of spatial reference memory (Martin and Clark 2007). An analysis of electron microscopy images from the CA1 stratum radiatum yielded a similar number of spines/µm² in Control (0.54 ± 0.02 spines/µm2) and CaMK-Scrib−/− neurons (0.61 ± 0.02 spines/µm2) (n = 1892/4, P = 0.06) (Fig. 3A and C). An assessment of the PSD size indicated a shifted cumulative probability curve toward smaller PSD thicknessess (Control: 41.95 ± 0.90 nm and CaMK-Scrib−/−: 41.05 ± 1.14 nm, n = 1892/4, P = 0.001) and PSD lengths (Control: 216.14 ± 3.02 nm and CaMK-Scrib−/−: 204.02 ± 2.82 nm, n = 1892/4, P < 0.01) (Fig. 3C and D). Specifically, PSDs from CaMK-Scrib−/− CA1 neurons showed an increased probability (38.9%) of being shorter than 170 nm compared to those from Control (33.6%). These data indicate that although the spine density is similar in both genotypes, stratum radiatum CA1 neurons from CaMK-Scrib−/− mice display a larger portion of small immature PSDs.
Figure 3.
Reduced number of active CA3-CA1 synapses in CaMK-Scrib−/− mice. (A and B) Representative low magnification electron micrographs of hippocampal CA1 stratum radiatum region of Control (Aa) and CaMK-Scrib−/− (Ba) mice and a higher magnification of a spinous synapse, marked by an asterisks in the respective low magnification panels (Ab and Bb). Scale bar = 588 nm (Aa, Ba) and 88 nm (Ab, Bb). (C and D) Shifted distribution of PSD thickness (C) and PSD length (D) towards smaller PSDs (shorter than 200 nm) in the stratum radiatum of CaMK-Scrib−/− (red) compared to Control (black) (n = 1892/4 mice). Data were compared using Mann–Whitney, P = 0.001 (C) and P < 0.01 (D). (E) Input-output relationship was reduced in CaMK-Scrib−/− slices (red) with respect to Control (black) (n = 12/6, P < 0.0001). Scale bars = 10 ms; 0.2 mV. Data were compared using two-way ANOVA, genotype effect, P < 0.001. (F) Similar PPRs at 25, 50, 100, or 200 ms in Control and CaMK-Scrib−/− slices (n = 6). Scale bars = 50 ms; 50 (Control) and 20 (CaMK-Scrib−/−) µV. Data were compared using Mann–Whitney. (G) Representative mEPSCs traces from Control (Ga) and CaMK-Scrib−/− (Gb) CA1 neurons. Scale bars = 75 ms; 5 pA. (H) Similar average amplitudes of mEPSCs at Control and CaMK-Scrib−/− CA3-CA1 synapses (n = 5). Data were compared using Mann–Whitney. (I) Average frequency of mEPSCs was decreased in CaMK-Scrib−/− compared to Control CA3-CA1 synapses (n = 5). Data were compared using Mann–Whitney, P < 0.05. Data are represented as mean ± SEM.
As small PSDs (less than 170 nm) appear to be devoid of AMPARs (Takumi et al. 1999), our findings suggest that CaMK-Scrib−/− might have a larger population of CA1 synapses that are non-functional during basal synaptic transmission. If this is the case, both basal synaptic transmission at CA3-CA1 synapses and the number of active synapses during basal synaptic transmission should be reduced in CaMK-Scrib−/− CA1 neurons. To test this possibility, field excitatory postsynaptic potentials (fEPSPs) were measured to evaluate CA3-CA1 basal glutamatergic transmission. Input-output relationship was found to be altered in CaMK-Scrib−/− slices in comparison to Control littermates, with a maximal slope of fEPSPs ~65% smaller (n = 12/6 mice; P < 0.001) (Fig. 3E). This finding, reflecting a severe decrease in glutamatergic transmission, could be due to distinct pre- and/or postsynaptic mechanisms, including a reduction in quantal content (n.P) and/or in quantal size (q). To test whether a change in the probability of glutamate release (P) could account for the reduction in synaptic transmission, we examined short-term facilitation at CA3-CA1 synapses at four different intervals (25, 50, 100, and 200 ms). For all these intervals, the PPR was found to be similar in CaMK-Scrib−/− and Control acute slices indicating normal probability of glutamate release (P) (n = 6, P > 0.35 for all four intervals) (Fig. 3F). Thus, the reduced input-output relation could be due to a decrease in quantal size (q) or quantal content (n.P). We hence measured AMPARs-mediated miniature excitatory postsynaptic currents (mEPSCs) using whole-cell patch clamp recordings in the presence of TTX (Fig. 3G). The average amplitudes of mEPSCs in CA1 neurons were similar in the Control and CaMK-Scrib−/− mice indicating normal quantal size (13.38 ± 0.76 and 11.88 ± 0.42 pA, respectively, n = 5, P = 0.22) (Fig. 3H). Importantly, mEPSC frequency was reduced by more than 70% in CaMK-Scrib−/− mice (0.33 ± 0.06 Hz) compared to their Control littermates (0.97 ± 0.23 Hz, n = 5, P < 0.05) (Fig. 3I). The decrease in quantal content shown by the mEPSCs frequency could be due to a change in the probability of release (P) and/or the number of active synapses (n). A PPR analysis revealed (P) the PPR to be unaltered in CaMK-Scrib−/− synapses, indicating that the number of active synapses (n) must be decreased in CaMK-Scrib−/− CA1 neurons. Because the spine density was similar in both genotypes, we conclude that the decrease in (n) is due to an increase in the number of silent synapses in CaMK-Scrib−/− neurons. This is in agreement with our morphological analysis revealing an increased population of immature synapses in the stratum radiatum of CaMK-Scrib−/− CA1 neurons (Fig. 3A–D).
Altered NMDAR-Dependent Bidirectional Plasticity in CA3-CA1 CaMK-Scrib−/− Synapses
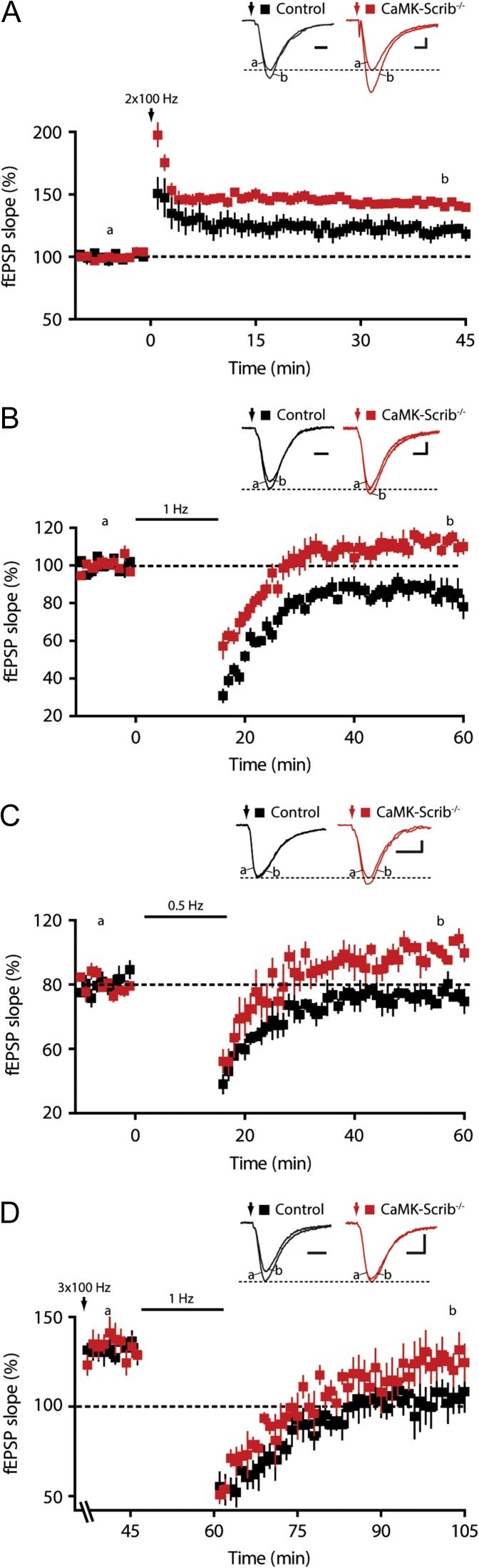
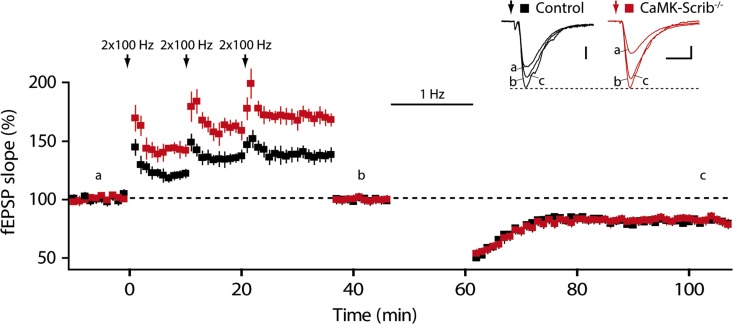
The decrease in synaptic transmission due to the loss of Scrib can impact long-term synaptic plasticity in the hippocampus. To examine this, NMDAR-dependent long-term potentiation (LTP) at the Schaffer collaterals was induced using a high frequency stimulation (HFS) consisting of 2 trains at 100 Hz. This yielded a significant increase in LTP amplitude at CaMK-Scrib−/− synapses (145 ± 0.6%, n = 6/5 mice) compared to Control synapses (121.1 ± 0.7%, n = 5) (P = 0.01) (Fig. 4A). The application of D-AP5, a specific NMDAR antagonist, completely blocked LTP induction, indicating that it was NMDAR-dependent (102 ± 3.7%, n = 6, P = 0.44). Subsequently, NMDAR-dependent long-term depression (LTD) was also investigated using a low frequency stimulation (LFS) protocol of 15 min at 1 Hz. Synaptic strength was significantly depressed to 85.6 ± 1.1% of initial baseline following LTD induction in Control slices (n = 6/5 mice, P < 0.05) (Fig. 4B). This LTD was NMDAR-dependent, as it was completely blocked by D-AP5 (102.5 ± 0.85%, n = 4, P = 0.12). Surprisingly, rather than generating LTD, LFS of Schaffer collaterals generated a significant LTP at CaMK-Scrib−/− CA3-CA1 synapses (112.2 ± 1.5%; n = 7/6 mice, P = 0.01). A weaker stimulation protocol at 0.5 Hz did not induce any change in Control or CaMK-Scrib−/− mice (103.4 ± 0.6%, n = 5, P = 0.31 and 110 ± 0.8%, n = 4, P = 0.13, respectively) (Fig. 4C). Long-term depotentiation, which is a form of LTD induced by LFS at potentiated synapses, was also impaired in CaMK-Scrib−/− compared to Control synapses (Fig. 4D). Indeed, in response to LFS, potentiated Control synapses were depotentiated (101.6 ± 7.5%, n = 6, P < 0.05), whereas CaMK-Scrib−/− synapses remained potentiated (128.9 ± 8.7%, n = 6, P = 0.84). In conclusion, while LTP was enhanced, LTD and depotentiation were both abolished at CaMK-Scrib−/− CA3-CA1 synapses.
Figure 4.
Enhanced LTP and abolished LTD in CaMK-Scrib−/− synapses. (A) Enhanced LTP induced by HFS consisting of 2 trains at 100 Hz at CA3-CA1 CaMK-Scrib−/− synapses (red; n = 6/5) compared to Control synapses (black; n = 5, P = 0.01). Scale bars = 5 (Control) and 4 (CaMK-Scrib−/−) ms; 0.1 mV. Data were compared using Mann–Whitney, P = 0.01. (B) Abolished LTD in CA3-CA1 CaMK-Scrib−/− synapses (n = 6/5) that generated LTP following LFS at 1 Hz stimulation in contrast to Control synapses (n = 7/6) that induced LTD. Scale bars = 5 (Control) and 4 (CaMK-Scrib−/−) ms; 0.1 mV. Data were compared using Wilcoxon matched-pairs, P < 0.05 (Control) and P = 0.01 (CaMK-Scrib−/−). (C) A weaker stimulation protocol at 0.5 Hz did not induce any change in Control (n = 5, P = 0.31) or CaMK-Scrib−/− mice (n = 4, P = 0.13). Scale bars = 10 ms; 0.1 mV. Data were compared using Wilcoxon matched-pairs. (D) Long-term depotentiation at CA3-CA1 synapses, potentiated with HFS consisting of 3 trains at 100 Hz followed by LFS at 1 Hz delivery, was impaired in CaMK-Scrib−/− (n = 5) compared to Control mice (n = 6). Scale bars = 10 (Control) and 8 (CaMK-Scrib−/−) ms; 0.1 mV. Data were compared using Wilcoxon matched-pairs, P < 0.05 (Control).
Scrib Scaffolds PP1/PP2A at the PSD of Excitatory Hippocampal Synapses
CaMK-Scrib−/− synapses not only did not express LTD but, instead, exhibited LTP following the LTD-inducing protocol (LFS), as shown in Fig. 4B. This indicates inappropriate intracellular signaling due to the absence of Scrib and its potential downstream partners. To identify interacting proteins of Scrib that could participate in LTD induction, different domains of Scrib were used as baits to screen with a P10 brain library using yeast two-hybrid screening. In our screen using the PDZ4 domain (aa 1086–1180 of Q80U72–3), one of the 53 clones identified as interacting with Scrib encoded a portion of the PP2A catalytic subunit alpha (Ppp2ca). Because phosphatases are important for LTD expression (Mulkey et al. 1994; Morishita et al. 2001), we proceeded to investigate potential interactions between Scrib and members of this phosphoprotein phosphatase family. We analyzed the binding properties of Ppp2ca in a yeast two-hybrid assay and found that it indeed interacted specifically with PDZ4 (Fig. 5A). In addition, independent yeast two-hybrid screens of a human mammary gland epithelial library using Scrib as bait identified protein phosphatase 1 (Pp1) alpha, beta, and gamma as partners (Fig. 5B). Interestingly, in a secondary screening we observed that the interaction between Scrib and Pp1 proteins was conserved during evolution, as Scribble from Drosophila or LET-413 (Erbin homolog) from Caenorhabditis elegans were able to interact with PP1 (Borg and Santoni, unpublished). When GFP-tagged Scrib and a Myc-tagged Pp1γ or Ppp2ca were expressed in heterologous cells, colocalization of both phosphatases with Scrib at the membrane and in intracellular structures was observed (Fig. 5C and D). Further biochemical approaches were undertaken to ascertain the Pp1-Scrib interaction. First, we produced recombinant GST-Pp1 fusion proteins and performed GST pull-down assays. We expressed either full or truncated HA-tagged versions ([aa 1–724] NH2- and [aa 717–1630] carboxy-terminal) of Scrib and assayed their binding to recombinant GST-Pp1. The NH2-terminal fusion protein contains the LRR domain and the C-terminal contains the PDZ domains. Full-length Scrib interacted with GST-Pp1, but with neither GST alone nor the GST-C-terminal. We identified the NH2-terminal region of Scrib as responsible for the interaction with PP1 (Fig. 5E), and therefore, PP1 does not interact with PDZ domains of Scrib. Second, we decided to map the regions within Pp1 proteins involved in this interaction. PP1 was truncated or mutated within its catalytic domain at sites shown to decrease or abrogate its phosphatase activity (Y272A), or deletion of the tyrosine in position 272 (Y272Δ) or to be involved in the binding to the Pp1 inhibitor I2 (M290K-C291 A) or implicated in the binding to two Pp1 binding partners PNUTS and spinophilin (Y272Δ) (Watanabe et al. 2001). We also introduced a point mutation analogous to a mutation known to affect the role of glc-7–10 (the yeast Pp1 homolog) in cell polarity processes (mutant F136L) (Fig. 5F). A loss of binding was obvious with the mutants affecting the catalytic activity of Pp1 while a small carboxy-terminal truncation (N302Δ) had no effect on PP1 binding to Scrib. The Y272A mutation that decreased the phosphatase activity of PP1 did not affect the interaction with hScrib (Fig. 5F). For Ppp2ca, based on these results, the Ppp2ca-Scrib interaction was narrowed down to a PDZ-binding domain-PDZ4 domain interaction (Fig. 5G). We used modeling to assess important amino-acids involved in this interaction. In the Scrib-Ppp2ca complex model, the C-terminus of Ppp2ca is fixed in the hydrophobic pocket formed between helix B and strand B of the Scrib PDZ4 domain (Fig. 5H), as is commonly the case between PDZ domains and their targets (Lee and Zheng 2010). Additionally, the residue D306 of the Ppp2ca C-terminal is favorably positioned to form a salt bridge with R1102 and/or K1124 residues of Scrib. Hence, both hydrophobic interactions and a salt bridge should allow for the interaction between the two partners. The generated model with the NetPhos 2.0 server showed the presence of two serines at the contact zone, one of which (S1100) can potentially be phosphorylated, although this remains to be confirmed. This serine, S1100, is ideally positioned to disrupt the interaction between Scrib1 and Ppp2ca, as the steric bulk of the added phosphate could prevent the formation of a salt bridge between D306 (Ppp2ca) and R1102/K1124 (Scrib1). We finally demonstrated that both endogenous PP1 and Ppp2ca, but not PSD-95, interacted with Scrib in hippocampal lysates by coimmunoprecipitation using an antibody against Scrib or PP1/Ppp2ca (Fig. 5I).
Figure 5.
Scrib scaffolds PP1 and PP2A phosphatases. (A) Directed yeast two-hybrid assays with Scrib and PP2A catalytic subunit (Ppp2ca) constructs. Schematic domain structures of ScribPDZ, ScribPDZ3, and ScribPDZ4 used as baits. Ppp2ca binds ScribPDZ and ScribPDZ4, but not ScribPDZ3. (B) Directed yeast two-hybrid assays with Scrib and Pp1 constructs. Pp1α,β,γ can bind Scrib. (C and D) ScribWT colocalizes with Pp1c (C) and Ppp2ca (D) catalytic subunits. COS-7 cells transiently co-transfected with hScrib-GFP (green) and HA-tagged Pp1c (red) (C) or hScrib-GFP (green) and Ppp2ca (red) (D). Phalloidin is shown in blue. Bottom white panels show higher magnification of individual or merged staining at the plasma membrane and cytoplasm level. Scale bars = 15 µm. (E) Three N-terminal HA-tagged constructs that contain either the full-length (Full) or N-terminal/LRR domain (N-term) or C-terminal/PDZ domains (C-term) truncated form of Scrib were expressed in COS cells. Lysates were pulled down with GST alone (GST) or GST-PP1 (PP1) and submitted to SDS-PAGE and western blot analysis with an anti-HA antibody. Input lanes (TL) represent 10% of total extract used for each pull down; (F) Schematic representation of PP1: the catalytic core domain span from amino-acid residue 42–269 and the binding domain from 270 to 330. Blue and Yellow stripes represent GDxHG, GDxVDRG or GNHE sequences. The different point mutations (F136L, Y272 A, M290K, C291A) and truncations of the tyrosine in position 272 [Y272Δ] or asparagine in position 302 [N302Δ] are mentioned with their corresponding positions. Resulting complexes were analyzed by anti-Scrib antibody western blot. (G) GST pull downs indicate that Ppp2ca can interact with PDZ4 in a dose-dependent manner but not with PDZ3. (H) Model of the interaction of Scrib-PDZ4 K1124 (orange) and R1102 (red) residues with the T304 to L309 residues from Ppp2ca C-terminus (green) and overall view of the model of the interaction (top right) between the alpha helix B (blue) and beta strand B (red) of Scrib-PDZ4 (gray) and Ppp2ca C-terminus (green). (I) Endogenous coimmunoprecipitation of Scrib, Pp1 and Ppp2ca from the hippocampus. Supernatants were immunoprecipitated with Scrib antibodies. The precipitates show positive immunoblotting for Pp1 and Ppp2ca subunits. (J) Immunoblot analysis of PSD fractions from Control and CaMK-Scrib−/− mice, and quantification of Scrib, CaMKII, PP1 and PP2 proteins. The blots were normalized to WT (100%; black histograms). Data were compared using Mann–Whitney, P < 0.01. Data are represented as mean ± SEM.
The following figure supplement is available for Figure 5: Supplementary Fig. S4. CaMK-Scrib1−/− mice exhibit normal level of PP1, PP2A, GluR1, and PSD95 in cell lysates.
These data suggest that Scrib acts as a scaffold protein for PP1 and PP2A through an interaction with Ppp2ca, its catalytic subunit alpha, at the synapse and led us to investigate the protein expression levels of these phosphatases at the PSDs of CaMK-Scrib−/− synapses lacking Scrib. Pp1 and Ppp2ca levels were found to be decreased by ~20 and 35%, respectively, at CaMK-Scrib−/− PSDs compared to Control synapses (n = 16, P < 0.01) (Fig. 5J), while they were unchanged in the total cell lysates (Supplementary Fig. S4). The levels of GluR1 were also decreased in the PSD fraction but not in homogenates and this is consistent with the fact that PSDs are smaller in the CamK-Scrib−/− mice. On the other hand, the levels of the CaMKII kinase were unchanged in the CaMK-Scrib−/− mice compared to Control mice (n = 4, P = 0.63) (Fig. 5J). This shows that Scrib interacts directly with PP1/PP2A phosphatases and strongly indicates that it ensures their proper localization at the synapse.
Saturating the LTP Signaling Pathway Rescues LTD Expression in CA3-CA1 CaMK-Scrib−/− Synapses
In the absence of Scrib, reduced levels of PP1/PP2A can entail a decrease in the LTD signaling pathway following LFS, which can favor the competing pathway downstream of CaMKII, resulting in LTP induction instead of LTD. Previous studies in mutant mice have reported that high (saturated) levels of active CaMKII (phosphorylated T286) prevent LTP and favor LTD (Bejar et al. 2002). Hence, we attempted to rescue the CaMK-Scrib−/− phenotype by saturating the competing LTP pathway to enhance the probability of recruiting the PP1/PP2A-dependent pathway during LTD. To saturate the LTP signaling pathway, we induced three consecutive HFS at 2 × 100 Hz and then delivered the LTD-inducing protocol (LFS) at 1 Hz for 15 min. Under these conditions, saturated CaMK-Scrib−/− CA3-CA1 synapses were able to express a form of LTD similarly to Control synapses (81.2 ± 0.4% and 82.5 ± 0.6%, respectively, n = 5, P = 0.94) (Fig. 6). Hence, by saturating the LTP signaling pathway, we were able to rescue LTD in the CaMK-Scrib−/− mice.
Figure 6.
Saturation of LTP pathway rescues long-term depotentiation in CaMK-Scrib−/− synapses.Three strong protocols of LTP consisting of 2 trains at 100 Hz were delivered to saturate synapses and followed by LFS to induce long-term depotentiation, which was similar in Control and CaMK-Scrib−/− slices (n = 5). Scale bars = 10 ms; 100 (Control) and 50 (CaMK-Scrib−/−) µV. Data were compared using Mann–Whitney. Data are represented as mean ± SEM.
Exposure to EE Rescues Synaptic Function and Memory Formation in CaMK-Scrib−/− Mice
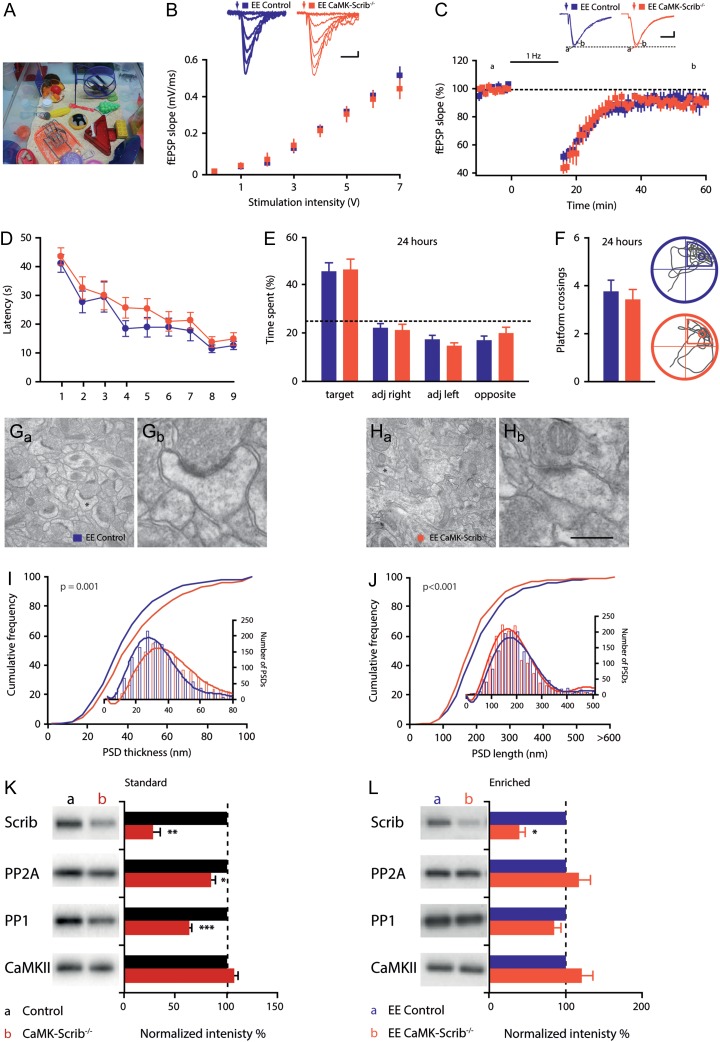
Finally, we aimed at achieving a more physiological rescue of LTD and spatial memory by exposure to an EE (Fig. 7A). Indeed, EE has been documented to increase synaptic transmission, enhance synaptic plasticity, and improve spatial memory (van Praag et al. 2000; Foster and Dumas 2001; Chancey et al. 2013). Importantly, EE exposure restored basal glutamatergic transmission, illustrated by a normal input-output relationship in EE CaMK-Scrib−/− mice compared to EE Control mice (n = 5 and n = 6, respectively, P = 0.51) (Fig. 7B). Furthermore, under these conditions it was possible to induce LTD using LFS in EE CaMK-Scrib−/− CA3-CA1 synapses (90.4 ± 3%, n = 7/6 mice, P < 0.05) that was not different from LTD induced in EE Control synapses (93.5 ± 5.8%, n = 7/6 mice, P = 0.60) (Fig. 7C).
Figure 7.
Exposure to EE rescues synaptic function and memory consolidation in CaMK-Scrib−/− mice. (A) Example of an EE cage. (B) Input-output relationship was restored in EE CaMK-Scrib−/− (n = 5) with respect to EE Control slices (n = 6). Scale bars = 10 ms; 100 µV. Data were compared using two-way ANOVA. (C) Normal LTD expression following LFS at 1 Hz stimulation in both EE CaMK-Scrib−/− (orange) and EE Control CA3-CA1 synapses (purple) (n = 7/6). Scale bars = 10 ms; 100 µV. Data were compared using Mann–Whitney and Wilcoxon matched-pairs (P < 0.05 for EE CaMK-Scrib−/−). (D) Spatial learning was normal in EE Control and EE CaMK-Scrib−/− mice revealed by mean latency to reach the hidden platform in Morris water maze over 9 days of training. Data were compared using two-way ANOVA. (E) Equivalent time spent searching in each quadrant during long-term probe test performed at 24 h after the last session of training (n = 15). Data were compared using unpaired t-test. (F) Same number of platform crossings and representative swim trace patterns during probe test at 24 h for EE Control (top) and EE CaMK-Scrib−/− mice (bottom); a circle marks target location. Data were compared using unpaired t-test. (G and H) Representative low magnification electron micrographs of hippocampal CA1 stratum radiatum region of EE Control (G) and EE CaMK-Scrib1−/− (H) mice and a higher magnification of a spinous synapse, marked by an asterisks in the respective low magnification panels (Gb and Hb). Scale bar = 1.1 µm (Ga, Ha) and 310 nm (Gb, Hb). (I and J) Shifted distribution of PSD thickness (I) towards thicker PSDs and smaller PSD length (J) (shorter than 200 nm) in the stratum radiatum of EE CaMK-Scrib1−/− (orange) compared to EE Control (purple) (n = 2838/6 mice). Data were compared using Mann–Whitney, P = 0.001 (I) and P < 0.001 (J). (K and L) Immunoblot analysis of PSD fractions from Control and CaMK-Scrib−/− mice in standard (K) and enriched (L) environments, and quantification of Scrib1, CaMKII, PP1, and PP2 proteins in each condition. The blots were normalized to WT (100%; black histograms). Data are represented as mean ± SEM.
The following figure supplements are available for Figure 7: Supplementary Fig. S5. Exposure to EE rescues spatial learning in CaMK-Scrib−/−mice.
These electrophysiological findings correlated with a cognitive rescue of spatial learning in the Morris water maze, as revealed by equivalent latencies to find the hidden platform observed in EE Control and EE CaMK-Scrib−/− mice during the acquisition phase (n = 15, genotype effect P = 0.08) (Supplementary Fig. S5A). Following EE, both genotypes improved their performances in terms of reduced latency to find the platform (P < 0.001) and reduced distance traveled to reach the platform (P < 0.0001) (Supplementary Fig. S5B). As expected, during the short-term (1 h) probe test, no difference was observed in the time spent in the target quadrant (P = 0.25) (Supplementary Fig. S5C) or in the number of platform crossings (P = 0.76) (Supplementary Fig. S5D) between EE Control mice (41.3 ± 2.8% and 2.7 ± 0.25, respectively) and EE CaMK-Scrib−/− mice (46.6 ± 3.5% and 2.8 ± 0.35, respectively). More strikingly, the long-term memory impairment observed during the 24-h probe test was prevented by EE in the CaMK-Scrib−/− mice. Indeed, both genotypes submitted to EE spent similar amounts of time in the target quadrant (EE Control: 45.1 ± 3.8% and EE CaMK-Scrib−/−: 45.9 ± 4.5%, P = 0.90) (Fig. 7E) and exhibited an equal number of platform crossings (EE Control: 3.7 ± 0.5 and EE CaMK-Scrib−/−: 3.4 ± 0.45, P = 0.63) (Fig. 7F). These data indicate that exposure to EE prevents impairments in synaptic transmission, LTD expression and spatial learning and memory in CaMK-Scrib−/− mice.
Next, we investigated the impact of EE exposure on the morphological features of CaMK-Scrib−/− mice in the CA1 stratum radiatum (Fig. 7G and H). We found a significantly higher spine density in CaMK-Scrib−/− EE mice (0.67 ± 0.02 spines/µm2) compared to EE Control mice (0.60 ± 0.01 spines/µm2, n = 2838/6, P = 0.023). In addition, PSDs from EE CaMK-Scrib−/− mice displayed a shifted cumulative probability curve toward higher thickness (EE Control: 41.24 ± 0.51 nm and EE CaMK-Scrib−/−: 46.389 ± 0.61 nm, n = 2838/6, P = 0.001) and lower length values (EE Control: 224.40 ± 2.58 nm and EE CaMK-Scrib−/−: 203.70 ± 2.19 nm, n = 2838/6, P < 0.001) (Fig. 7I and J, respectively). Specifically, PSDs of EE CaMK-Scrib−/− showed an increased probability (33.6%) of being shorter than 170 nm compared to those of EE Control (31.2%). These data indicate that although EE CaMK-Scrib−/− CA1 neurons still displayed a larger portion of spines with small immature PSDs compared to EE Control, a higher spine density and PSD thickness induced by EE exposure was found in EE CaMK-Scrib−/− mice.
Finally, we evaluated the levels of the PP1/2A phosphatases in standard and EE-exposed mice and found reduced levels of PP2A and PP1 in the synaptosomes in standard mice but not in the EE-exposed CaMK-Scrib−/− mice. In summary, these data showed that EE preserves phosphatase levels in EE CaMK-Scrib−/− mice and is associated with a recovery of CA1 plasticity and spatial learning and memory deficits induced by the absence of Scrib.
Discussion
In this study, we identified a novel role of the PCP protein Scrib during postnatal stages, which makes Scrib key in the synaptic maturation and plasticity underlying spatial learning and memory consolidation in the hippocampus. Our major findings show that (1) Scrib in postnatal excitatory neurons is necessary for hippocampus-dependent spatial learning and memory consolidation, (2) the functional maturation and NMDAR-dependent bidirectional plasticity of glutamatergic CA3-CA1 synapses depend on Scrib, (3) the PCP protein Scrib regulates at the PSD the levels of PP1/PP2A phosphatases, which are required for synaptic plasticity, and finally (4) exposure to an EE re-establishes spatial learning and memory consolidation in mice lacking Scrib and recovers synaptic function in the hippocampus, showing that the genetic loss of Scrib can be overcome by environmental manipulations based on a combination of cognitive, motor and social stimulations.
Functional Maturation of Hippocampal Synapses Depends on the PCP Protein Scrib
Our results show that the PCP protein Scrib at postnatal stages plays an important role in basal synaptic transmission by regulating the number of immature or inactive synapses that do not participate in basal synaptic transmission (Isaac et al. 1995; Liao et al. 1995). The novelty of our results resides in the demonstration of the key role of a PCP protein during postnatal brain stages, as PCP proteins are well documented to play a role in prenatal brain development but not during postnatal stages. A study with an embryonic mutation of the core PCP protein Prickle2 showed that Prickle2 affects the frequency but not the amplitude of miniature excitatory currents in hippocampal slices (Sowers et al. 2013). However, the decrease in the frequency observed in Prickle2 mutants seems to result from a deficiency in synaptogenesis rather than synaptic maturation. Downregulation of the core PCP protein Vangl2 through shRNA constructs also affects synaptogenesis in rat cultured hippocampal neurons (Nagaoka et al. 2014). These studies suggest different functions for each PCP protein that engage different pathways regulating synaptic transmission in the hippocampus. Interestingly, we recently revealed that Scrib controls the number of NMDARs at the plasma membrane and is involved in an NMDAR-subunit switch that is associated with D-serine stimulation prior to NMDA/D-serine activation (Piguel et al. 2014). The scaffold protein PSD-95, whose mutation in mice leads to a deficiency in the NMDAR-subunit switch, has been reported to convert immature silent synapses into functional synapses (Stein et al. 2003; Béîque et al. 2006; Sanz-Clemente et al. 2013). Hence, we suggest that the PCP protein Scrib, similarly to the scaffold protein PSD-95, is necessary for morpho-functional maturation of glutamatergic synapses through a mechanism that involves both cytoskeleton remodeling and an NMDAR-subunit switch. Finally, new silent synapses formed following synaptic inactivity in CA1 neurons have been shown to be recruited during LTP induction, leading to enhanced potentiation (Arendt et al. 2013). Therefore, we hypothesize that the observed enhanced LTP at CaMK-Scrib−/− CA3-CA1 synapses might be due to the increased immature population of synapses that is potentially recruited during LTP induction. These results, together with our previous studies, highlight the importance of regulating the PCP protein Scrib in mature neurons for normal functional synapses.
NMDAR-Dependent Bidirectional Plasticity at Hippocampal Synapses Requires the PCP Protein Scrib
We show that the loss of the PCP protein Scrib in postnatal neurons enhanced NMDAR-dependent LTP and completely abolished LTD at hippocampal synapses. Only the core PCP protein Celsr3 has been reported to affect synaptic plasticity, specifically LTP, in the hippocampus of conditional mutant mice (Feng et al. 2012b). As this study used embryonic mutations of Celsr3, it is not possible to conclude whether the LTP impairment observed in these mice is due to developmental problems that can result in impaired LTP or a direct role of postnatal Celsr3 in regulating LTP in the hippocampus. In our study, we demonstrate a role for a key PCP protein in regulating NMDAR-dependent synaptic plasticity and suggest a possible mechanism for this regulation in postnatal hippocampal neurons. Indeed, our search for interacting partners of Scrib that could help to explain the abolished LTD in conditional mutants led to the identification of PP1/PP2A phosphatases. We notably showed that Scrib directly interacts with PP1 and PP2A and that their levels are strongly reduced at synapses in the absence of Scrib. This original interaction in neurons is in agreement with previous work demonstrating that Scrib functions as a PP1-regulatory protein by interacting directly with PP1 in heterologous or cancer cells (Nagasaka et al. 2013; Young et al. 2013). Interestingly, widerborst, a B′ regulatory subunit of PP2A, was previously implicated in PCP mechanisms in both Drosophila and in Zebrafish (Hannus et al. 2002). In mice, PP1/PP2A has been reported to be necessary for both LTD and long-term depotentiation expression at CA3-CA1 synapses, and partial inhibition of PP1 alters bidirectional plasticity by inducing a shift that favors potentiation in the hippocampus (Jouvenceau et al. 2003). In addition, loss of Scrib not only abolished LTD expression but generated significant LTP following LTD-inducing protocol at CA3-CA1 synapses. Interestingly, altered bidirectional plasticity similar to the one observed in CaMK-Scrib−/− mice was obtained in Neurogranin knock-out mice (Krucker et al. 2002). Neurogranin regulates the equilibrium between kinase and phosphatase activities by binding available Ca2+/calmodulin at basal levels. In normal conditions, a modest increase in Ca2+ preferentially activates calcineurin instead of CaMKII, and consequently, PP1/PP2A initiates a series of substrate-specific dephosphorylations and triggers LTD. If PP1/PP2A levels are reduced at synapses, as is the case in our mutants, the resulting dephosphorylation process is slowed down compared to the competing phosphorylation process by CaMKII (Lisman and Zhabotinsky 2001; Xia and Storm 2005). This leads to the activation of a LTP signaling pathway instead of LTD at modest Ca2+ levels, as observed in CaMK-Scrib−/− mice. Consistent with this model, we were able to rescue long-term depotentiation expression at CaMK-Scrib−/− CA3-CA1 synapses by in vitro saturation of the LTP signaling pathway. Furthermore, recent data from our group demonstrated a direct interaction between Scrib and NMDAR-subunits in hippocampal neurons (Piguel et al. 2014). We hence suggest that Scrib is a scaffold protein that localizes PP1/PP2A phosphatases in the immediate vicinity of NMDARs, hence bridging the calcium influx of NMDARs to specific downstream signaling phosphatases necessary for bidirectional plasticity.
Memory Consolidation Requires Scrib
Loss of Scrib in postnatal excitatory hippocampal neurons led to delayed spatial learning and impaired long-term, but not short-term, spatial memory. These results reveal that the PCP protein Scrib plays a mild role in spatial learning but is indispensable for hippocampus-dependent spatial memory consolidation. Interestingly, studies using embryonic mutations have shown that the PCP proteins Prickle2, Celsr3, and Scrib may be involved in spatial learning (Moreau et al. 2010; Feng et al. 2012b; Sowers et al. 2013). However, the alterations in spatial learning observed in these studies could be due to modifications that occur during brain development that have been shown to require PCP proteins (Shima et al. 2007; Goodrich 2008; Tissir and Goffinet 2010; Feng et al. 2012a; Sowers et al. 2013; Hagiwara et al. 2014; Liu et al. 2014; Nagaoka et al. 2014). A possible mechanism that occurs during postnatal stages by which Scrib regulate learning and memory at the molecular level might involve PP1/2A phosphatases. We showed that Scrib directly interacts with PP1 and PP2A and that their levels are strongly reduced at synapses in the absence of Scrib. Learning and memory processes have been reported to be regulated by PP1 and PP2A phosphatases (Mansuy and Shenolikar 2006). PP2A has been shown to be necessary specifically in the consolidation phase, starting 50 min after a learning session in chickens (Bennett et al. 2001). Conditional knock-out of a regulator of the PP2A catalytic subunit (α4) leads to impairment in learning and long-term memory in the water maze paradigm (Yamashita et al. 2006). Pharmacological administration of an inhibitor of PP1/PP2A results in impaired NMADR-dependent LTD and spatial reference memory in rodents (He et al. 2001). Moreover, inhibiting LTD expression by blocking AMPAR endocytosis results in impairments in early consolidation (>1 h post-training) of spatial reference memory (Ge et al. 2010). Additionally, NMDAR-dependent LTD has been recently shown to be required for consolidation, but not acquisition, of hippocampus-dependent fear memory (Liu et al. 2014). Finally, downregulation of protein phosphatases, as observed in CaMK-Scrib−/− mice, is often associated with cognitive decline and dementia, such as in Alzheimer's disease (Tian and Wang 2002). These observations suggest that deficits in spatial memory consolidation in CaMK-Scrib−/− mice might involve NMDAR-dependent LTD impairment due to an alteration in Scrib-PP1/PP2A interactions and their downstream targets. In favor of this hypothesis, rescue of LTD expression at CaMK-Scrib−/− CA3-CA1 synapses by EE exposure led to the recovery of normal spatial memory consolidation.
Beneficial Effect of Environmental Enrichment Despite Genetic Mutation
We were able to overcome the absence of Scrib at hippocampal synapses using EE exposure, which yielded the rescue of synaptic basal transmission and LTD expression at CA3-CA1 synapses. EE has been documented to increase synaptic transmission, enhance synaptic plasticity, improve spatial memory (van Praag et al. 2000; Foster and Dumas 2001; Chancey et al. 2013), and even reduce the severity of seizures in epileptic mice (Morelli et al. 2014). In CaMK-Scrib−/− mice, EE exposure that restored glutamatergic synaptic function also prevented memory impairment, suggesting a causal link between altered synaptic plasticity and memory deficits observed in the absence of Scrib. Other studies have also reported reversed learning and memory impairments by EE exposure in different memory-deficient animal models (Frick and Fernandez 2003; Need et al. 2003; Dahlqvist et al. 2004) and in children with developmental disabilities such as autism (Kozulin et al. 2010). EE rescue in CaMK-Scrib−/− mice is independent of Scrib expression and likely involves compensatory phenomena to overcome synaptic dysfunction and memory impairment. Indeed, EE exposure was unable to convert immature synapses into mature synapses in the absence of Scrib, as a larger portion of spines with small immature PSDs in the stratum radiatum was still observed in CaMK-Scrib−/− EE. However, EE exposure in CaMK-Scrib−/− EE CA1 neurons increased spinogenesis and PSD thickness, which together were sufficient to overcome the increase in the number of immature synapses in the absence of Scrib. The increase in PSD thickness could indicate recruitment of other scaffold proteins and/or better coupling between glutamate receptors and downstream effectors and the cytoskeleton. For instance, EE exposure has been reported to increase the expression of the scaffold protein PSD-95 following 2 weeks of EE exposure (Nithianantharajah et al. 2004). Hence, a likely mechanism for the rescue by EE exposure in CaMK-Scrib−/− mice is the up-regulation of other scaffold proteins, such as PSD-95, and an increase in synaptogenesis to compensate for the loss of Scrib. Our study, along with others, demonstrates that EE exposure is a powerful tool that is sufficient to prevent and treat synaptic dysfunction associated with memory impairment caused by a genetic mutation.
PCP Proteins and Mental Disorders
Our results indicate that the PCP protein Scrib is one of the important PSD proteins whose mutations, which disrupt synapse development and function, are associated with cognitive defects or mental disorders in humans (Bayés et al. 2011). Recent large studies designed to identify genes with rare copy number variants or de novo mutations implicated in autism spectrum disorder (ASD) have identified three autistic patients: one with a 540-kb deleted region in 8q24.3 encompassing five genes, including SCRIB (the human orthologue of scrib) (Hu et al. 2015) and two with a missense mutation in SCRIB (c.1114C>T, c.1774C>T) (Pinto et al. 2010; Neale et al. 2012; Iossifov et al. 2014). Additionally, Scrib spontaneous mutation in Scribcrc mice leads to emotional and cognitive phenotypes related to ASD (Moreau et al. 2010). Moreover, transcriptional and splicing dysregulation of scrib were found in the brains of individuals with ASD (Voigeneagu et al. 2011; Irimia et al. 2014). Interestingly, mutations in other PCP genes, such as FAT1, Dvl2 or Prickle2, have also been reported in ASD patients, strongly implicating the PCP signaling pathway in neurodevelopmental pathologies (Cukier et al. 2014; review in Sans et al. 2016). In this study, we report specific alterations in spatial memory formation in Scrib conditional knock-out mice that might be considered as a mouse model for the intellectual disabilities present in certain patients with ASDs.
Conclusion
Our results identify a key PCP protein, Scrib, as one of the fundamental scaffold proteins that enable hippocampal glutamatergic synapses to mature and to express bidirectional plasticity required for memory formation. Also, this study offers a unique insight into the mechanism by which the PCP protein Scrib can regulate memory formation at postnatal stages. Overall, this work highlights the fundamental role of PCP proteins and signaling, beyond developmental stages, in ensuring normal higher-order brain function such as learning and memory.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Supplementary Material
Notes
We thank Drs N.G. Copeland and N.A. Jenkins for the Scrib floxed mutant (NIH, USA), Prof. Dr G. Schütz and colleagues for the CaMKII Cre mice (DKFZ, Germany), and Prof. D.L. Brautigan for the PP1 and PP2A constructs (University of Virginia School of Medicine, USA). We thank M.-C. Donat, E. Richard, and F. Loll for technical assistance. We thank Dr D.N. Abrous for kindly providing access to her water maze tracking and Neurolucida systems and Drs M. Koehl and S. Tronel for their help and advice. We also thank the “animal and genotyping facilities” members of the Neurocentre for technical assistance, notably S. Laumond, D. Gonzales, and co-workers, the “Biochemistry and Biophysics Facility” of Bordeaux Neurocampus funded by the Labex B.R.A.I.N., the Electron Microscopy Research Services, Faculty of Medical Sciences, Newcastle University. We also would like to thank Drs Carmen Sandi, Jerome Ezan, and Andy Trevelyan for helpful comments and discussion. Conflict of Interest: None declared.
Funding
This work was supported byINSERM and INSERM AVENIR grants (NS and MM), Conseil Régional d'Aquitaine (NS/MM), Neurocampus program (MM), La Fondation pour la Recherche Médicale (NS, MM), ANR NeuroScrib ANR-07-NEUR-031-01 (NS, MM), ANR MossyPCP ANR-12-BSV4-0016-01 (NS), Royal Society International Joint Project grant (CR, NS) and the Wellcome Trust Institutional Strategic Support Fund Project, Faculty of Medical Sciences, Newcastle University (CR). J.P. Borg's lab is funded by la Ligue Contre le Cancer (Label Ligue), INSERM, and Institut Paoli-Calmettes. MLH was an AXA funded student of the University of Bordeaux graduate programme and also received funding for a 4th year from the Fondation pour la Recherche Medicale (FRM; FDT20120925405). VLP was supported by a Ph.D. fellowship from the 7th Framework Program (FP7) Marie Curie ITN SyMBaD, the FRM “4eme année de these” (FDT20130928124) and the Schizo Oui Foundation. SDSC is supported by an ENC Neurasmus Ph.D. fellowship. NHP was supported by the Conseil Régional d'Aquitaine/INSERM during his Ph.D. and the FRM “Bourse de soudure” fellowship.JP Borg is a scholar of Institut Universitaire de France. SHRO is supported by INSERM, Université de Bordeaux, and FRM (Equipe FRM; DEQ 201 303 26519). The Montcouquiol/Sans & Oliet labsare members of the Labex B.R.A.I.N.
References
- Arendt KL, Sarti F, Chen L. 2013. Chronic inactivation of a neural circuit enhances LTP by inducing silent synapse formation. J Neurosci. 33:2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Lecine P, Bellaiche Y, Dupont JL, Premont RT, Sempere C, Strub JM, et al. . 2004. Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr Biol. 14:987–995. [DOI] [PubMed] [Google Scholar]
- Bayés A, van de Lagemaat LN, Collins MO, Croning MDR, Whittle IR, Choudhary JS, Grant SGN. 2011. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 14:19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R, Axelrod JD. 2011. Pointing in the right direction: new developments in the field of planar cell polarity. Nat Rev Genet. 12:385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béîque J, Lin D, Kang M, Aizawa H, Takamiya K, Huganir RL. 2006. Synapse-specific regulation of AMPA receptor function by PSD-95 Results. Proc Natl Acad Sci USA. 103:19535–19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Yasuda R, Krugers H, Hood K, Mayford M. 2002. Transgenic calmodulin-dependent protein kinase II activation: dose-dependent effects on synaptic plasticity, learning, and memory. J Neurosci. 22:5719–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett PC, Zhao W, Ng KT. 2001. Concentration-dependent effects of protein phosphatase (PP) inhibitors implicate PP1 and PP2A in different stages of memory formation. Neurobiol Learn Mem. 75:91–110. [DOI] [PubMed] [Google Scholar]
- Bilder D, Birnbaum D, Borg J, Bryant P, Huigbretse J, Jansen E, Kennedy MB, Labouesse M, Legouis R, Mechler B, et al. . 2000. Collective nomenclature for LAP proteins. Nat Cell Biol. 2:2000. [DOI] [PubMed] [Google Scholar]
- Birrane G, Chung J, Ladias JA. 2003. Novel mode of ligand recognition by the Erbin PDZ domain. J Biol Chem. 278:1399–1402. [DOI] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, Schutz G. 2001. A CamKIIα iCre BAC allows brain-specific gene inactivation. Genesis. 31:37–42. [DOI] [PubMed] [Google Scholar]
- Chancey JH, Adlaf EW, Sapp MC, Pugh PC, Wadiche JI, Overstreet-Wadiche LS. 2013. GABA depolarization is required for experience-dependent synapse unsilencing in adult-born neurons. J Neurosci. 33:6614–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbard J-R, Djiane A, Wu J, Mlodzik M. 2010. The apical/basal-polarity determinant Scribble cooperates with the PCP core factor Stbm/Vang and functions as one of its effectors. Dev Biol. 333:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier HN, Dueker ND, Slifer SH, Lee JM, Whitehead PL, Lalanne E, Leyva N, Konidari I, Gentry RC, Hulme WF, et al. . 2014. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist P, Ronnback ÃA, Bergstrom S-A, Soderstrom I, Olsson T. 2004. Environmental enrichment reverses learning impairment in the Morris water maze after focal cerebral ischemia in rats. Eur J Neurosci. 19:2288–2298. [DOI] [PubMed] [Google Scholar]
- Dosemeci A, Vinade L, Winters CA, Pozzo-miller L, Reese TS. 2001. Glutamate-induced transient modification of the postsynaptic density. Proc Natl Acad Sci USA. 98:10428–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Kauffman JS, Caddy J, Zarbalis K, Peterson AS, Jane SM, Russell SM, Humbert PO. 2007. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 26:2272–2282. [DOI] [PubMed] [Google Scholar]
- Ezan J, Montcouquiol M. 2013. Revisiting planar cell polarity in the inner ear. Semin Cell Dev Biol. 24:499–506. [DOI] [PubMed] [Google Scholar]
- Feng J, Han Q, Zhou L. 2012. a. Planar cell polarity genes, Celsr1-3, in neural development. Neurosci Bull. 28:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Xu Y, Wang M, Ruan Y, So K-F, Tissir F, Goffinet A, Zhou L. 2012. b. A role for atypical cadherin Celsr3 in hippocampal maturation and connectivity. J Neurosci. 32:13729–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TC, Dumas TC. 2001. Mechanism for increased hippocampal synaptic strength following differential experience. J Neurophysiol. 85:1377–1383. [DOI] [PubMed] [Google Scholar]
- Frick KM, Fernandez SM. 2003. Enrichment enhances spatial memory and increases synaptophysin levels in aged female mice. Neurobiol Aging. 24:615–626. [DOI] [PubMed] [Google Scholar]
- Ge Y, Dong Z, Bagot RC, Howland JG, Phillips AG, Wong TP, Wang YT. 2010. Hippocampal long-term depression is required for the consolidation of spatial memory. Proc Natl Acad Sci USA. 107:16697–16702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV. 2008. The plane facts of PCP in the CNS. Neuron. 60:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Yasumura M, Hida Y, Inoue E, Ohtsuka T. 2014. The planar cell polarity protein Vangl2 bidirectionally regulates dendritic branching in cultured hippocampal neurons. Mol Brain. 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannus M, Feiguin F, Heisenberg C, Eaton S. 2002. Planar cell polarization requires Widerborst, a B′ regulatory subunit of protein phosphatase 2A. Development. 129:3493–3503. [DOI] [PubMed] [Google Scholar]
- He J, Yamada K, Zou LB, Nabeshima T. 2001. Spatial memory deficit and neurodegeneration induced by the direct injection of okadaic acid into the hippocampus in rats. J Neural Transm. 108:1435–1443. [DOI] [PubMed] [Google Scholar]
- Hu J, Sathanoori M, Kochmar S, Azage M, Mann S, Madan-Khetarpal S, Goldstein A, Surti U. 2015. A novel maternally inherited 8q24.3 and a rare paternally inherited 14q23.3 CNVs in a family with neurodevelopmental disorders. Am J Med Genet. A167A:1921–1926. [DOI] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O'Hanlon D, et al. . 2014. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 159:1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac JT, Nicoll RA, Malenka RC. 1995. Evidence for silent synapses: implications for the expression of LTP. Neuron. 15:427–434. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. . 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 515:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenceau A, Billard J-M, Haditsch U, Mansuy IM, Dutar P. 2003. Different phosphatase-dependent mechanisms mediate long-term depression and depotentiation of long-term potentiation in mouse hippocampal CA1 area. Eur J Neurosci. 18:1279–1285. [DOI] [PubMed] [Google Scholar]
- Kozulin A, Lebeer J, Madella-Noja A, Gonzalez F, Jeffrey I, Rosenthal N, Koslowsky M. 2010. Cognitive modifiability of children with developmental disabilities: a multicentre study using Feuerstein's Instrumental Enrichment-Basic program. Res Dev Disabil. 31:551–559. [DOI] [PubMed] [Google Scholar]
- Krucker T, Siggins GR, McNamara RK, Lindsley Ka, Dao A, Allison DW, De Lecea L, Lovenberg TW, Sutcliffe JG, Gerendasy DD. 2002. Targeted disruption of RC3 reveals a calmodulin-based mechanism for regulating metaplasticity in the hippocampus. J Neurosci. 22:5525–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zheng JJ. 2010. PDZ domains and their binding partners : structure, specificity, and modification. Cell Commun Signal. 28(8):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Hessler NA, Malinow R. 1995. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 375:400–404. [DOI] [PubMed] [Google Scholar]
- Lisman JE, Zhabotinsky AM. 2001. A model of synaptic memory : viewpoint A CaMKII/PP1 switch that potentiates transmission by organizing an AMPA receptor anchoring assembly. Neuron. 31:191–201. [DOI] [PubMed] [Google Scholar]
- Liu X, Gu Q-H, Duan K, Li Z. 2014. NMDA receptor-dependent LTD is required for consolidation but not acquisition of fear memory. J Neurosci. 34:8741–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Hatim A, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Allan R, et al. . 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy IM, Shenolikar S. 2006. Protein serine/threonine phosphatases in neuronal plasticity and disorders of learning and memory. Trends Neurosci. 29:679–686. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Clark RE. 2007. The rodent hippocampus and spatial memory: from synapses to systems. Cell Mol life Sci. 64:401–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Crenshaw EB, Kelley MW. 2006. a. Noncanonical Wnt signaling and neural polarity. Annu Rev Neurosci. 29:363–386. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Rachel RA, Lanford PJ, Copeland NG, Jenkins NA, Kelley MW. 2003. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 423:173–177. [DOI] [PubMed] [Google Scholar]
- Montcouquiol M, Sans N, Huss D, Kach J, Dickman JD, Forge A, Rachel RA, Copeland NG, Jenkins NA, Bogani D, et al. . 2006. b. Asymmetric localization of Vangl2 and Fz3 indicate novel mechanisms for planar cell polarity in mammals. J Neurosci. 26:5265–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MM, Piguel N, Papouin T, Koehl M, Durand CM, Rubio ME, Loll F, Richard EM, Mazzocco C, Racca C, et al. . 2010. The planar polarity protein Scribble1 is essential for neuronal plasticity and brain function. J Neurosci. 30:9738–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli E, Ghiglieri V, Pendolino V, Bagetta V, Pignataro A, Fejtova A, Costa C, Ammassari-Teule M, Gundelfinger ED, Picconi B, et al. . 2014. Environmental enrichment restores CA1 hippocampal LTP and reduces severity of seizures in epileptic mice. Exp Neurol. 261:320–327. [DOI] [PubMed] [Google Scholar]
- Morishita W, Connor JH, Xia H, Quinlan EM, Shenolikar S, Malenka RC, Carolina N. 2001. Regulation of Synaptic strength by protein phosphatase 1. Neuron. 32:1133–1148. [DOI] [PubMed] [Google Scholar]
- Mulkey RM, Endo S, Shenolikar S, Malenka RC. 1994. Involvment of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature. 369:486–488. [DOI] [PubMed] [Google Scholar]
- Nagaoka T, Ohashi R, Inutsuka A, Sakai S, Fujisawa N, Yokoyama M, Huang YH, Igarashi M, Kishi M. 2014. The Wnt/planar cell polarity pathway component Vangl2 induces synapse formation through direct control of N-cadherin. Cell Rep. 6:916–927. [DOI] [PubMed] [Google Scholar]
- Nagasaka K, Seiki T, Yamashita A, Massimi P, Subbaiah VK, Thomas M, Kranjec C, Kawana K, Nakagawa S, Yano T, et al. . 2013. A novel interaction between hScrib and PP1γ downregulates ERK signaling and suppresses oncogene-induced cell transformation. PLoS One. 8:e53752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. . 2012. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 485:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Irvine EE, Giese KP. 2003. Learning and memory impairments in Kvbeta1.1-null mutants are rescued by environmental enrichment or ageing. Eur J Neurosci. 18:1640–1644. [DOI] [PubMed] [Google Scholar]
- Nithianantharajah J, Levis H, Murphy M. 2004. Environmental enrichment results in cortical and subcortical changes in levels of synaptophysin and PSD-95 proteins. Neurobiol Learn Mem. 81:200–210. [DOI] [PubMed] [Google Scholar]
- Piguel NH, Fievre S, Blanc JM, Carta M, Moreau MM, Moutin E, Pinheiro VL, Medina C, Ezan J, Lasvaux L, et al. . 2014. Scribble1/AP2 complex coordinates NMDA receptor endocytic recycling. Cell Rep. 9:712–727. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. . 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richier L, Williton K, Clattenburg L, Colwill K, O'Brien M, Tsang C, Kolar A, Zinck N, Metalnikov P, Trimble WS, et al. . 2010. NOS1AP associates with Scribble and regulates dendritic spine development. J Neurosci. 30:4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Ezan J, Moreau M, Montcouquiol M. 2016. Planar cell polarity (PCP) gene mutations in Autism Spectrum Disorder, Intellectual Disabilities and Related Deletion/Duplication Syndromes In: Sala C, Verpelli C, editors. Neuronal and Synaptic Dysfunction in Autism Spectrum Disorder and Intellectual Disability. San Diego: Academic Press; p. 189–219. [Google Scholar]
- Sans N, Petralia RS, Wang Y, Ii JB, Hell JW, Wenthold RJ. 2000. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 20:1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Prybylowski K, Petralia RS, Chang K, Wang Y-X, Racca C, Vicini S, Wenthold RJ. 2003. NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat Cell Biol. 5:520–530. [DOI] [PubMed] [Google Scholar]
- Sans N, Wang PY, Du Q, Petralia RS, Wang Y-X, Nakka S, Blumer JB, Macara IG, Wenthold RJ. 2005. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol. 7:1179–1190. [DOI] [PubMed] [Google Scholar]
- Sanz-Clemente A, Nicoll RA, Roche KW. 2013. Diversity in NMDA receptor composition: many regulators, many consequences. Neuroscience. 19:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Kawaguchi S, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, Hirano T, Uemura T. 2007. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci. 10:963–969. [DOI] [PubMed] [Google Scholar]
- Sowers LP, Loo L, Wu Y, Campbell E, Ulrich JD, Wu S, Paemka L, Wassink T, Meyer K, Bing X, et al. . 2013. Disruption of the non-canonical Wnt gene PRICKLE2 leads to autism-like behaviors with evidence for hippocampal synaptic dysfunction. Mol Psychiatry. 18:1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein V, House DRC, Bredt DS, Nicoll RA. 2003. Postsynaptic density-95 mimics and occludes hippocampal long-term potentiation and enhances long-term depression. J Neurosci. 23:5503–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi Y, Ramírez-León V, Laake P, Rinvik E, Ottersen OP. 1999. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 2:618–624. [DOI] [PubMed] [Google Scholar]
- Tian Q, Wang J. 2002. Role of serine/threonine protein phosphatase in Alzheimer's disease. Neurosignals. 11:262–269. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. 2010. Planar cell polarity signaling in neural development. Curr Opin Neurobiol. 20:572–577. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. 2013. Shaping the nervous system: role of the core planar cell polarity genes. Nat Rev Neurosci. 4:525–535. [DOI] [PubMed] [Google Scholar]
- Van Praag H, Kempermann G, Gage FH. 2000. Neural consequences of environmental enrichment. Nat Rev Neurosci. 1:191–198. [DOI] [PubMed] [Google Scholar]
- Verghese S, Waghmare I, Kwon H, Hanes K, Kango-Singh M. 2012. Scribble acts in the Drosophila fat-hippo pathway to regulate warts activity. PLoS One. 7:e47173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB. 2012. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu Rev Cell Dev Biol. 28:627–653. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nathans J. 2007. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development. 134:647–658. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Huang H, Horiuchi A, da Cruze Silva EF, Hsieh-wilson L, Allen PB, Shenolikar S, Greengard P, Nairn AC. 2001. Protein phosphatase 1 regulation by inhibitors and targeting subunits. Proc Natl Acad Sci. 98:3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Storm DR. 2005. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 6:267–276. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Inui S, Maeda K, Rong D, Takagi K, Fukunaga K, Sakaguchi N. 2006. Regulation of CaMKII by α 4/PP2Ac contributes to learning and memory. Brain Res. 82:1–10. [DOI] [PubMed] [Google Scholar]
- Yamben IF, Rachel RA, Shatadal S, Copeland NG, Jenkins NA, Warming S, Griep AE. 2013. Scrib is required for epithelial cell identity and prevents epithelial to mesenchymal transition in the mouse. Dev Biol. 384:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z, Petralia RS, Fu Z, Swanwick CC, Wang Y-X, Prybylowski K, Sans N, Vicini S, Wenthold RJ. 2007. The role of the PDZ protein GIPC in regulating NMDA receptor trafficking. J Neurosci. 27:11663–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LC, Hartig N, Muñoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, Vijayakumar V, Vietri Rudan M, Gewinner C, Henderson S, et al. . 2013. An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol Cell. 52:679–692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.