Abstract
Shiga toxin–producing Escherichia coli (STEC) cause bloody diarrhea (BD), hemorrhagic colitis (HC), and even hemolytic uremic syndrome (HUS). In Nordic countries, STEC are widely spread and usually associated with gastrointestinal symptoms and HUS. The objective of this study was to investigate the occurrence of STEC in Swedish patients over 10 years of age from 2003 through 2015, and to analyze the correlation of critical STEC virulence factors with clinical symptoms and duration of stx shedding. Diarrheal stool samples were screened for presence of stx by real-time PCR. All STEC isolates were characterized by DNA microarray assay and PCR to determine serogenotypes, stx subtypes, and presence of intimin gene eae and enterohaemolysin gene ehxA. Multilocus sequencing typing (MLST) was used to assess phylogenetic relationships. Clinical features were collected and analyzed using data from the routine infection control measures in the county. A total of 14,550 samples were enrolled in this 12-years period study, and 175 (1.2%) stools were stx positive by real-time PCR. The overall incidence of STEC infection was 4.9 cases per 100,000 person-years during the project period. Seventy-five isolates, with one isolate per sample were recovered, among which 43 were from non-bloody stools, 32 from BD, and 3 out of the 75 STEC positive patients developed HUS. The presence of stx2 in both stools and isolates were associated with BD (p = 0.008, p = 0.05), and the presence of eae in isolates was related to BD (p = 0.008). The predominant serogenotypes associated with BD were O157:H7, O26:H11, O121:H19, and O103:H2. Isolates from HUS were O104:H4 and O98: H21 serotypes. Phylogenetic analysis revealed our strains were highly diverse, and showed close relatedness to HUS-associated STEC collection strains. In conclusion, the presence of stx2 in stool was related to BD already at the initial diagnostic procedure, thus could be used as risk predictor at an early stage. STEC isolates with stx2 and eae were significantly associated with BD. The predominant serotypes associated with BD were O157:H7, O26:H11, O121:H19, and O103:H2. Nevertheless, the pathogenic potential of other serotypes and genotypes should not be neglected.
Keywords: Shiga toxin-producing Escherichia coli, virulence factor, stx subtype, shedding, clinical symptoms
Introduction
Shiga toxin–producing Escherichia coli (STEC) cause diseases ranging in severity from asymptomatic infection, non-bloody diarrhea (NBD) to bloody diarrhea (BD), hemorrhagic colitis (HC), and even life-threatening hemolytic uremic syndrome (HUS) (Tarr et al., 2005). STEC are widely spread and associated with gastrointestinal symptoms and HUS in the Nordic countries (Haugum et al., 2014; Naseer et al., 2017; Pedersen et al., 2017). The number of STEC cases reported in Sweden has been rising ever since all STEC was notified in 2004 (Peter Nolskog and Cecilia Jernberg, 2017), with the largest O157 STEC outbreak occurring on the west coast of Sweden in 2005 (Söderström et al., 2008). In 2011, a large outbreak caused by a Shiga toxin 2–producing E. coli serotype O104:H4 resulted in 3,816 STEC cases in Germany, and subsequently spread throughout other European countries including Sweden (Guy et al., 2012, 2013). This highlighted the clinical significance of other STEC serotypes than O157 as a great threat to public health.
The production of Shiga toxin (Stx) is the primary virulence trait responsible for STEC disease, along with the presence of an outer membrane protein intimin (eae), which can lead to the formation of attaching and effacing (A/E) lesions (Elliott et al., 2000). Stx can be divided into two major types (Stx1 and Stx2), several subtypes and variants for each toxin type have been described (Scheutz et al., 2012). Shiga toxin subtypes have been found to differ in receptor preference and toxin potency (Fuller et al., 2011). Strains producing subtype Stx2a, Stx2c, or Stx2d which display close sequences relatedness, are often associated with development of HC and HUS (Orth et al., 2007; Kawano et al., 2008). Epidemiological studies suggest that stx2 along with the presence of eae is more often associated with severe disease and development of HUS (Orth et al., 2007). However, not all STEC infections with stx2 and eae positive strains develop severe clinical outcomes. The clinical significance of STEC for humans is further determined by the production and interplay of other virulence factors, such as the plasmid-encoded enterohaemolysin (ehxA). The prevalence of virulence factors in clinical strains and their role in disease development are not yet fully understood.
The duration of STEC carriage is of great importance since individuals who continue to excrete the pathogen may serve as a source of infection for secondary cases. In Sweden, there is a legal requirement that all individuals with HUS-associated STEC infection must have at least one stx negative control sample before returning to pre-school or work at risk (Peter Nolskog and Cecilia Jernberg, 2017). Our previous investigation of STEC in Swedish children revealed that stx genes were excreted for several months in some patients, with a maximal duration of 256 days (Matussek et al., 2016). However, the average duration of STEC shedding in adult patients is largely unknown.
The objective of this study was to depict the prevalence and molecular characteristics of STEC in patients over 10 years of age with diarrhea in Region Jönköping County, Sweden, and illustrate the correlation of several critical STEC virulence factors (stx subtypes, eae and ehxA) to clinical symptoms and duration of STEC shedding.
Materials and methods
Ethics approval statement
Formal consent is not required, as sample and data collection is part of the routine microbiological and contact tracing work in Region Jönköping County, Sweden.
Setting, isolation of STEC, and DNA extraction
Region Jönköping County has ~330,000 inhabitants, served by three hospitals and 46 health care centers and the clinical microbiology laboratory in Jönköping, Sweden, receives all microbiology samples in the region. The current study comprised all routine diarrheal stool samples where STEC analysis was requested by the clinician. In addition, STEC analysis was performed on stool samples submitted for routine bacterial culture where BD and/or HUS was mentioned on the referral as well as from individuals involved in contact tracing around an index case. All samples included were evaluated for the presence of stx by real-time PCR on suspensions of overnight cultures on blood agar plates at the clinical microbiology laboratory. Primers (MWG-Biotech, Ebersberg, Germany) and probes (TIB MOLBIOL, Berlin, Germany) used in the current study for detection of stx1 and stx2 were described previously (Bellin et al., 2001; Table S1). Real-time PCR was performed on LightCycler® 480 Instrument II (Roche Diagnostics GmbH, Mannheim, Germany). The amplification program included an initial denaturation step at 95°C for 120 s and 45 cycles of denaturation at 95°C for 1 s, annealing at 55°C for 5 s (reached with a touchdown from 60°C over the course of the first five cycles), and extension at 72°C for 20 s. E. coli EDL 933 was used as a positive control strain and nuclease-free water was used as negative control for real-time PCR method. PCR positive samples were then sent to the Karolinska University Laboratory, Stockholm, Sweden for confirmation and isolation of STEC strains on Sorbitol-MacConkey (SMAC) plates according to methods previously described (Svenungsson et al., 2000). Patients were sampled weekly until they were stx-PCR negative, and the duration of stx shedding was defined as the time from the first positive sample to the first negative sample (Matussek et al., 2016). Bacterial DNA was extracted from overnight cultures with EZ1 DNA Tissue Kit (Qiagen, Hilden, Germany) on EZ1 instrument (Qiagen).
Clinical data collection
Clinical data was collected from all patients who tested positive for stx by PCR over 10 years of age in Jönköping county from April 2003 through January 2015 through a questionnaire and by reviewing medical records as part of the routine infection control measures in the county. Clinical manifestations included were diarrhea, BD, abdominal pain, vomiting, fever and HUS. HUS was characterized by three primary symptoms: thrombocytopenia, microangiopathic hemolytic anemia, and acute renal injury according to Bryan et al. (2015).
Serotyping, stx subtyping and detection of eae and ehxA
In Sweden, all STEC isolates are submitted to The Public Health Agency of Sweden for confirmation and further typing as part of the national microbial surveillance program. This includes STEC O-type serogrouping by agglutination in micro titer plates using antisera (SSI Diagnostica, Copenhagen, Denmark).
All STEC isolates were further subjected to microarray based assays with the E. coli SeroGenoTyping AS-1 Kit, ShigaToxType AS-2 Kit (Alere Technologies GmbH, Germany) to determine the serogenotype (O:H) (Geue et al., 2014a) and stx allele/subtype (Scheutz et al., 2012; Geue et al., 2014b) according to manufacturer's instructions (http://alere-technologies.com/en/products/lab-solutions/e-coli/e-coli-serogenotyping-kit.html, http://alere-technologies.com/en/products/lab-solutions/shigatoxin.html). Detection of eae and ehxA was done by PCR (Bai et al., 2016).
MLST
Multilocus sequence typing (MLST) was used to characterize phylogenetic relationships of strains. Defined fragments of the seven housekeeping genes (i.e., adk, icd, fumC, recA, mdh, gyrB, and purA) were amplified and sequenced according to the E. coli MLST website (https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search). Sequences types (STs) were assigned based on the allelic profile of the seven housekeeping genes. STs of isolates from this study were compared with HUS-associated enterohemorrhagic E. coli (HUSEC) collection strains (www.ehec.org) (Mellmann et al., 2008). A minimum spanning tree (MST) based on these STs was generated using BioNumerics version 7.6 (Applied Maths NV, Sint-Martens-Latem, Belgium).
Statistical analyses
χ2 and Fisher's exact test were done for comparing categorical data using IBM® SPSS® Statistics 24.0 (IBM, US). Multiple testing correction by using the Benjamini-Hochberg method was conducted to correct the individual p-value for each gene. p < 0.05 was considered statistically significant.
Results
Patient data and the prevalence of STEC
A total of 14,550 specimens were enrolled in this 12-years period investigation. 175 (1.2%) stools were stx positive by real-time PCR on suspensions of overnight cultures on blood agar plates, and 85 of them were eae positive. The overall incidence of STEC infection was 4.9 cases per 100,000 person-years, with the annual incidence varying from 3.70 to 8.03 cases per 100,000 persons per year (Table 1). The presence of stx2 in stool samples was associated with BD (p = 0.008), while a higher positive rate of stx1+stx2 was detected in non-bloody stool (NBS) samples (p = 0.008; Table 2). Seventy-five stx positive stools yielded isolates on SMAC plates, with one isolate from one sample each, giving a culture positive rate of 42.9%. STEC patient data and clinical symptoms are presented in Table 3. The presence of stx2-only in isolates was associated with BD (p = 0.05; Table 2).
Table 1.
STEC incidence in adults over 10 years of age from April 2003 to January 2015, Jönköping county, Sweden.
| Sampling year | Population(>10 years old) | STEC cases | Annual incidence rate per 100,000 persons | No. of STEC isolates | Culture positive rate (%) |
|---|---|---|---|---|---|
| 2003 | 291,922 | 12 | 4.11 | 7 | 58.33 |
| 2004 | 293,045 | 14 | 4.78 | 8 | 57.14 |
| 2005 | 294,097 | 17 | 5.78 | 6 | 35.29 |
| 2006 | 295,187 | 11 | 3.73 | 4 | 36.36 |
| 2007 | 296,628 | 14 | 4.71 | 7 | 50.00 |
| 2008 | 297,835 | 11 | 3.70 | 3 | 27.27 |
| 2009 | 298,028 | 14 | 4.70 | 4 | 28.57 |
| 2010 | 298,272 | 14 | 4.70 | 2 | 14.29 |
| 2011 | 298,650 | 24 | 8.03 | 11 | 45.83 |
| 2012 | 299,367 | 14 | 4.68 | 10 | 71.43 |
| 2013 | 300,898 | 15 | 4.98 | 8 | 53.33 |
| 2014 | 303,195 | 15 | 4.95 | 5 | 33.33 |
| Total | 3567,124 | 175 | 4.91 | 75 | 42.86 |
Table 2.
Prevalence of stx and eae detected on stools and isolates in correlation with bloody diarrhea (BD) and non-bloody stools (NBS).
| Virulence genes | No. of stools (%) | p-value | No. of isolates (%) | p-value | ||
|---|---|---|---|---|---|---|
| BD (n = 53) | NBS (n = 122) | BD (n = 32) | NBS (n = 43) | |||
| stx1-only | 14(26.4) | 37(30.3) | 0.601 | 7(21.9) | 14(32.6) | 0.601 |
| stx2-only | 26(49.1) | 15(12.3) | 0.008* | 17(53.1) | 10(23.3) | 0.05* |
| stx1+stx2 | 13(24.5) | 70(57.4) | 0.008* | 8(25.0) | 19(44.2) | 0.271 |
| eae | 32(60.4) | 53(43.4) | 0.163 | 22(68.8) | 13(30.2) | 0.008* |
Statistically significant difference.
Table 3.
Patient data and clinical symptoms for all STEC infections yielding isolates.
| All cases | BD | NBS | |
|---|---|---|---|
| No. (%) | 75 | 32 (43) | 43 (57) |
| Median age of patients | 41 (10–87) | 49 (15–87) | 38 (10–82) |
| Median length of carriage, in days | 17 (0–294) | 17 (7–197) | 18 (0–294) |
| Aquired bacteria abroad (%) | 19 (25.3) | 7 (21.9) | 12 (27.9) |
| Diarrhea (%) | 54 (72.0) | 32 (100) | 22 (51.2) |
| Abdominal pain (%) | 39 (52.0) | 25 (78.1) | 14 (32.6) |
| Fever (%) | 10 (13.3) | 2 (6.3) | 8 (18.6) |
| Vomiting (%) | 3 (4.0) | 0 (0) | 3 (7.0) |
| HUS (%) | 3 (4) | 2 (6.3) | 1 (2.3) |
stx subtypes and presence of eae, ehxA
Overall, two stx1 subtypes (i.e., stx1a and stx1c) and six stx2 subtypes (i.e., stx2a, stx2b, stx2c, stx2d, stx2e, and stx2g) were detected, resulting in a total of 16 different stx subtype combinations (Table 4 and Table S2). BD cases were caused by stx1a-only, stx1a+stx2a, stx1a+stx2c, stx1c+stx2b, stx2a-only, stx2a+stx2c, stx2b-only, and stx2g-only positive isolates. Two of the three HUS isolates carried stx2a-only, and one HUS isolate possessed stx1a-only. stx2a only, stx2a+stx2c, stx1a+stx2a, and stx1a+stx2c were more often detected in isolates from BD cases, while no statistically significant difference was found (Table S2). In total, 35 isolates harbored eae, and 59 carried ehxA. Interestingly, almost all eae positive isolates, with one exception, carried ehxA. The presence of eae in isolates was found to be associated with BD (p = 0.008; Table 2). Notably, only one of three HUS isolates carried the ehxA gene, and none of them harbored eae.
Table 4.
Sequence types, serotypes, virulence genes in 75 STEC isolates and associated-clinical symptomsa.
| ST | No. | Serotypes | Virulence genes/stx subtypes | Clinical symptoms |
|---|---|---|---|---|
| 11 | 15 | O157:H7 (15) | stx2a+stx2c (11), stx1a+stx2a (1), stx1a+stx2c (3), eae (15), ehxA (15) | BD (11), NBS (4) |
| 21 | 9 | O26:H11 (9) | stx1a (8), stx2a (1), eae (3), ehxA (9) | BD (6), NBS (3) |
| 655 | 6 | O121:H19 (6) | stx2a (5), stx1a+stx2a (1), eae (6), ehxA (5) | BD (3), NBS (3) |
| 17 | 5 | O103:H2 (5) | stx1a (5), eae (5), ehxA (5) | BD (2), NBS (3) |
| 678 | 4 | O104:H4 (4) | stx2a (4) | HUS (2), BD (1), NBS (1) |
| 442 | 3 | O146:H21 (1), O91:H21 (2) | stx2b (1), stx2d (2), ehxA (3) | BD (1), NBS (2) |
| 504 | 3 | O117:H7 (2), O156:H7 (1) | stx1a (3) | NBS (3) |
| 10 | 2 | O113:H4 (1), O4:H16 (1) | stx1c (1), stx2d (1), ehxA (1) | NBS (2) |
| 13 | 2 | O117:H8 (2) | stx1c (2), ehxA (2) | NBS (2) |
| 33 | 2 | O91:H14 (2) | stx1a+stx2b (2), ehxA (2) | NBS (2) |
| 1724 | 2 | O150:H10 (2) | stx2b (2), ehxA (2) | NBS (2) |
| 25 | 1 | O128ab:H2 | stx1c+stx2b, ehxA | NBS |
| 58 | 1 | O126:H20 | stx2a, ehxA | NBS |
| 119 | 1 | O165:H25 | stx2a+stx2c, eae, ehxA | BD |
| 137 | 1 | O145:H28 | stx2a, eae, ehxA | NBS |
| 301 | 1 | O180:H2 | stx2a, eae, ehxA | NBS |
| 306 | 1 | O98:H21 | stx1a, ehxA | HUS |
| 325 | 1 | O15:H16 | stx2g | NBS |
| 329 | 1 | O136:H12 | stx2a, ehxA | NBS |
| 388 | 1 | O112ab:H2 | stx2b+stx2d | NBS |
| 410 | 1 | O8:H9 | stx2c | NBS |
| 447 | 1 | O5:H19 | stx1c, ehxA | NBS |
| 657 | 1 | O183:H18 | stx1a+stx2a, ehxA | BD |
| 658 | 1 | O103:H28 | stx1a+stx2a, eae, ehxA | BD |
| 679 | 1 | O163:H19 | stx1a+stx2d, ehxA | NBS |
| 738 | 1 | O146:H28 | stx2b | NBS |
| 811 | 1 | O128ac:H2 | stx1c+stx2b | BD |
| 1494 | 1 | O9a:H21 | stx2e | NBS |
| 1804 | 1 | O157:H7 | stx1a+stx2c, eae, ehxA | BD |
| 2388 | 1 | O15:H27 | stx1c+stx2b | NBS |
| 3101 | 1 | O78:H4 | stx1c+stx2b, ehxA | NBS |
| N1b | 1 | O91:H21 | stx2c+stx2d, eae, ehxA | NBS |
| N2b | 1 | O187:H28 | stx2g, ehxA | BD |
The number of isolates is indicated in parentheses if it contains more than one isolate.
Two new STs assigned in this study.
Serotypes
Thirty-four distinct serotypes were found in 75 isolates, which comprised of 29 distinct O serogroups and 17 H types. The most prevalent serotype was O157:H7 (n = 16), followed by O26:H11 (n = 9), O121:H19 (n = 6), O103:H2 (n = 5), O104:H4 (n = 4), and O91:H21 (n = 3) (Table 4). Serotypes causing 32 BD cases were O157:H7(n = 12), O26:H11(n = 6), O121:H19 (n = 3), O104:H4 (n = 3), O103:H2 (n = 2), O103:H28 (n = 1), O128ac:H2 (n = 1), O146:H21(n = 1), O165:H25 (n = 1), O183:H18 (n = 1), O187:H28 (n = 1). Isolates from individuals that developed HUS were assigned to O104:H4 (n = 2), O98:H21 (n = 1), which harbored stx2a-only and stx1a-only subtype, respectively.
Duration of stx shedding
Data on duration of stx shedding was available for 39 (52%) patients, ranging from 0 to 294 days. The median length of carriage was 17 days (Table 3), which was used to separate short (<2.5 weeks) and long (≥ 2.5 weeks) carriage. No statistically significant difference was found between the presence of genes tested and long duration of stx shedding in this study (Table S3).
MLST
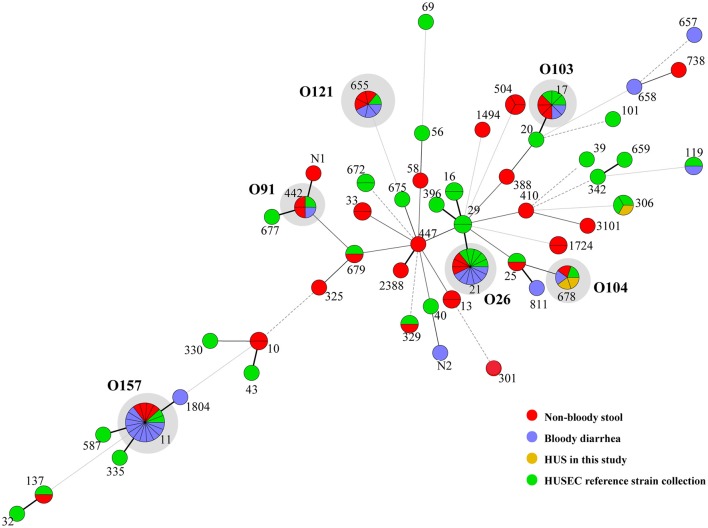
Thirty-three sequence types (STs) were obtained from 75 STEC isolates, including two novel STs (Table 4). It's noteworthy that isolates belonging to the same serotype (O157:H7, O26:H11, O121:H19, O103:H2, and O104:H4) were assigned as same ST (i.e., ST11, ST21, ST655, ST17, and ST678, respectively). 12 STs from BD cases and 26 STs from NBS in this study were scattered throughout the phylogenetic tree (Figure 1). Except the two HUS associated STs (ST678 and ST306), we found several STs from both NBS and BD in this study shared same STs (ST11, ST17, ST21, ST442, ST655, and ST679) or showed close relationship with HUSEC collection strains.
Figure 1.
Minimum spanning tree of 33 STs in this study and 31 STs from HUSEC collection. Each circle represents an ST with the size being proportional to the number of isolates. The colors for the slices of the pie represent the sources of isolates (see labeling in the lower right corner). The predominant STs comprising of strains in this study and HUSEC reference collection are indicated in gray shadows, with serogroups demonstrated aside.
Discussion
The incidence of reported human STEC cases varies geographically in Sweden. For example, 30.3 cases per 100,000 inhabitants were reported in Halland in 2016, while the reported incidence in Örebro County was 0 (Peter Nolskog and Cecilia Jernberg, 2017). In this report, the annual STEC incidence in individuals over 10 years of age in Jönköping county varied from 3.70 to 8.03 cases per 100,000 person-years during the project period. Sweden shows higher STEC incidence when compared with other Nordic countries and non-European countries (Pedersen et al., 2017; Vasant et al., 2017). The various STEC incidence might be effected by different local sampling and analysis routines (Peter Nolskog and Cecilia Jernberg, 2017). STEC isolation was failed in 57.1% of stx positive samples on SMAC agar in this study, including one HUS case, which could result in underestimation of clinical significance of emergent non-O157 seropathotypes. The low isolation rate might be due to shortcomings in current STEC screening protocols, for instance, the SMAC agar, which has been widely used for O157 isolation, is not sufficiently discriminatory to detect emergent non-O157 serotypes (Thomas et al., 2017). On the other hand, the transport conditions and time delay to culture in the reference laboratory, lack of culture enrichment broth in the current study could also be taken into account. Further studies are required to improve the diagnostic algorithm applied on clinical samples in combination with PCR diagnostics and culture based molecular serotyping as reported previously (de Boer et al., 2015). Remarkably, with increasing availability of whole-genome sequences of E. coli strains genome-scale metabolic models, whole genome sequencing (WGS) will contribute to greater understanding of STEC, aid to the development of improved culture-based detection methods in routine STEC diagnosis and provide a fast and accurate alternative to conventional identification and typing techniques (Sadiq et al., 2014; Lindsey et al., 2016; Parsons et al., 2016; Newell and La Ragione, 2018).
In this report, we found that stx2 was related to BD already at the initial PCR diagnosis on stools. Further characterization of STEC isolates enhanced that isolates harboring stx2 and eae were significantly associated with BD, therefore could be used as key genetic markers for risk assessment. It's noteworthy that we did not found association between stx1/stx2 subtypes and clinical symptoms, which is understandable as the number of isolates assigned as each subtype is limited in this study. Remarkably, three HUS isolates were all eae negative, no correlation between eae/stx2 subtypes and HUS was observed, which was also in agreement with a recent report from Denmark (Pedersen et al., 2017). This might partly be due to the low number of HUS cases in our study, and it is noteworthy that the two O104:H4 HUS isolates were from Swedish patients that visited Germany during the O104:H4 outbreak in 2011, exhibiting same serotype, stx2a subtype and absence of eae with the outbreak strain, thus they might be the same strain. In the O104:H4 outbreak strain, instead of intimin (eae), it's the aggregative adherence encoded by pAA plasmid genes (aggA, aggR) that anchored the bacterium to the intestinal mucosa (Bielaszewska et al., 2011; Navarro-Garcia, 2014). The absence of eae in HUS and BD cases implied that other adherence factors might contribute to severe disease outcomes. It's noteworthy that one HUS patient was infected with stx1a-only isolate, the same genotype was also observed in HUS case in Germany, suggesting that stx2 and eae could not reliably differentiate between HUS-associated and non-HUS-associated STEC strains. Hence, the potential of stx1 and other stx2 subtypes in development of HUS should not be neglected.
The predominant serotypes associated with BD cases in this study were in accordance with our previous survey in children (Matussek et al., 2016), and also similar to that reported in Norway (Naseer et al., 2017). Except O104:H4 isolates, one O98:H21 isolate from a HUS case, showed same sequence type (ST306), O serogroup and stx1 type as a HUSEC strain in Germany, highlighting the potential risk of non-O157 seropathotypes that were not predominantly reported. Interestingly, there is a good concordance observed between serotype and ST in this study. Isolates of same serotype exhibited higher tendency to be assigned as same ST, especially for predominant serotypes (O157:H7, O26:H11, O121:H19, O103:H2, and O104:H4). Thus, ST could be used to help molecularly assign a serotype in STEC genotyping. Moreover, these predominant STs were more often detected in BD cases, and all were found in HUSEC collection in Germany (Mellmann et al., 2008), implying that some STs showed higher pathogenic potential, could be used as high risk prediction in molecular typing.
The present study was confined to several limitations, for example, the low STEC isolation rate, which might result in the underestimation of clinical significance of non-O157 serotypes, and also insufficient association of the presence of virulence genes with symptomatology; limited number of virulence genes detected, HUS isolates and duration of shedding data in this study, which might prevent an adequate statistical association analysis between diverse genes/genotypes and clinical symptoms as well as duration of stx shedding. Hence, enhancement of STEC diagnosis of various serotypes in clinical samples, and whole genome-based analysis of molecular traits in correlation with clinical symptoms and duration of stx shedding is further needed.
In conclusion, here we report a large scale STEC investigation in Swedish patients over a 12-years period. The overall incidence of STEC infection was 4.9 cases per 100,000 person-years, 1.2% samples were stx PCR positive in investigated cases. By further characterization on serotypes, key virulence genes, stx subtypes, and MLST, we found the presence of stx2 in stool was related to BD already at the initial PCR diagnostic procedure performed directly on stool suspensions, thus could be used as risk predictor at an early stage. Further, isolates with stx2 and eae were significantly associated with BD. Isolates with high virulent stx2a/stx2c subtypes and serotypes (O157:H7, O26:H11, O121:H19, and O103:H2) are more prevalent in BD case, while strains with stx1a only and non-predominant serotype was associated with HUS. Thus, the pathogenic potential of other serotypes and genotypes should not be neglected.
Author contributions
AM and XB designed the experiments. SMe, CJ, SMo, and RE performed the experiments. I-ME and SMe collected clinical data. SMo and SL contributed analysis. XB and AM wrote the paper. SMe, CJ, SMo, RE, and SL polished the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by grants from Futurum, the Academy for Health and Care, Region Jönköping County (FUTURUM-425271).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00125/full#supplementary-material
References
- Bai X., Hu B., Xu Y., Sun H., Zhao A., Ba P., et al. (2016). Molecular and phylogenetic characterization of non-O157 shiga toxin-producing Escherichia coli strains in China. Front. Cell. Infect. Microbiol. 6:143. 10.3389/fcimb.2016.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin T., Pulz M., Matussek A., Hempen H. G., Gunzer F. (2001). Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J. Clin. Microbiol. 39, 370–374. 10.1128/JCM.39.1.370-374.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M., Mellmann A., Zhang W., Köck R., Fruth A., Bauwens A., et al. (2011). Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11, 671–676. 10.1016/S1473-3099(11)70165-7 [DOI] [PubMed] [Google Scholar]
- Bryan A., Youngster I., McAdam A. J. (2015). Shiga toxin producing Escherichia coli. Clin. Lab. Med. 35, 247–272. 10.1016/j.cll.2015.02.004 [DOI] [PubMed] [Google Scholar]
- de Boer R. F., Ferdous M., Ott A., Scheper H. R., Wisselink G. J., Heck M. E., et al. (2015). Assessing the public health risk of Shiga toxin-producing Escherichia coli by use of a rapid diagnostic screening algorithm. J. Clin. Microbiol. 53, 1588–1598. 10.1128/JCM.03590-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S. J., Sperandio V., Girón J. A., Shin S., Mellies J. L., Wainwright L., et al. (2000). The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68, 6115–6126. 10.1128/IAI.68.11.6115-6126.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C. A., Pellino C. A., Flagler M. J., Strasser J. E., Weiss A. A. (2011). Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79, 1329–1337. 10.1128/IAI.01182-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geue L., Monecke S., Engelmann I., Braun S., Slickers P., Ehricht R. (2014a). Rapid microarray-based DNA genoserotyping of Escherichia coli. Microbiol. Immunol. 58, 77–86. 10.1111/1348-0421.12120 [DOI] [PubMed] [Google Scholar]
- Geue L., Stieber B., Monecke S., Engelmann I., Gunzer F., Slickers P., et al. (2014b). Development of a rapid microarray-based DNA subtyping assay for the alleles of Shiga toxins 1 and 2 of Escherichia coli. J. Clin. Microbiol. 52, 2898–2904. 10.1128/JCM.01048-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L., Jernberg C., Arvén Norling J., Ivarsson S., Hedenstrom I., Melefors O., et al. (2013). Adaptive mutations and replacements of virulence traits in the Escherichia coli O104:H4 outbreak population. PLoS ONE 8:e63027. 10.1371/journal.pone.0063027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L., Jernberg C., Ivarsson S., Hedenström I., Engstrand L., Andersson S. G. (2012). Genomic diversity of the 2011 European outbreaks of Escherichia coli O104:H4. Proc. Natl. Acad. Sci. U.S.A. 109, E3627–E3628. 10.1073/pnas.1206246110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugum K., Brandal L. T., Lindstedt B. A., Wester A. L., Bergh K., Afset J. E. (2014). PCR-based detection and molecular characterization of shiga toxin-producing Escherichia coli strains in a routine microbiology laboratory over 16 years. J. Clin. Microbiol. 52, 3156–3163. 10.1128/JCM.00453-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano K., Okada M., Haga T., Maeda K., Goto Y. (2008). Relationship between pathogenicity for humans and stx genotype in Shiga toxin-producing Escherichia coli serotype O157. Eur. J. Clin. Microbiol. Infect. Dis. 27, 227–232. 10.1007/s10096-007-0420-3 [DOI] [PubMed] [Google Scholar]
- Lindsey R. L., Pouseele H., Chen J. C., Strockbine N. A., Carleton H. A. (2016). Implementation of Whole Genome Sequencing (WGS) for identification and characterization of shiga toxin-producing Escherichia coli (STEC) in the United States. Front. Microbiol. 7:766. 10.3389/fmicb.2016.00766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matussek A., Einemo I. M., Jogenfors A., Löfdahl S., Löfgren S. (2016). Shiga toxin-producing Escherichia coli in diarrheal stool of swedish children: evaluation of polymerase chain reaction screening and duration of shiga toxin shedding. J. Pediatric Infect. Dis. Soc. 5, 147–151. 10.1093/jpids/piv003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A., Bielaszewska M., Köck R., Friedrich A. W., Fruth A., Middendorf B., et al. (2008). Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerging Infect. Dis. 14, 1287–1290. 10.3201/eid1408.071082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer U., Lobersli I., Hindrum M., Bruvik T., Brandal L. T. (2017). Virulence factors of Shiga toxin-producing Escherichia coli and the risk of developing haemolytic uraemic syndrome in Norway, 1992–2013. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1613–1620. 10.1007/s10096-017-2974-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F. (2014). Escherichia coli O104:H4 pathogenesis: an enteroaggregative, E. coli/shiga toxin-producing, E. coli explosive cocktail of high virulence. Microbiol. Spectr. 2 10.1128/microbiolspec.EHEC-0008-2013 [DOI] [PubMed] [Google Scholar]
- Newell D. G., La Ragione R. M. (2018). Enterohaemorrhagic and other Shiga toxin-producing Escherichia coli (STEC): Where are we now regarding diagnostics and control strategies? Transbound. Emerg. Dis. [Epub ahead of print]. 10.1111/tbed.12789 [DOI] [PubMed] [Google Scholar]
- Orth D., Grif K., Khan A. B., Naim A., Dierich M. P., Wurzner R. (2007). The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn. Microbiol. Infect. Dis. 59, 235–242. 10.1016/j.diagmicrobio.2007.04.013 [DOI] [PubMed] [Google Scholar]
- Parsons B. D., Zelyas N., Berenger B. M., Chui L. (2016). Detection, characterization, and typing of shiga toxin-producing Escherichia coli. Front. Microbiol. 7:478. 10.3389/fmicb.2016.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen R. M., Nielsen M. T. K., Möller S., Ethelberg S., Skov M. N., Kolmos H. J., et al. (2017). Shiga toxin-producing Escherichia coli: incidence and clinical features in a setting with complete screening of patients with suspected infective diarrhoea. Clin. Microbiol. Infect. [Epub ahead of print]. 10.1016/j.cmi.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Peter Nolskog B. S., Cecilia Jernberg. (2017). New Procedures for Contraceptive Measures in EHEC Infection. pdf. Medical Comment. Läkartidningen. Available online at: http://www.lakartidningen.se/Klinik-och-vetenskap/Kommentar/2017/07/Nya-rutiner-for-smittskyddsatgarder-vid-EHEC-infektion/
- Sadiq S. M., Hazen T. H., Rasko D. A., Eppinger M. (2014). EHEC genomics: past, present, and future. Microbiol. Spectr. 2:EHEC-0020-2013. 10.1128/microbiolspec.EHEC-0020-2013 [DOI] [PubMed] [Google Scholar]
- Scheutz F., Teel L. D., Beutin L., Piérard D., Buvens G., Karch H., et al. (2012). Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50, 2951–2963. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderström A., Osterberg P., Lindqvist A., Jönsson B., Lindberg A., Blide Ulander S., et al. (2008). A large Escherichia coli O157 outbreak in Sweden associated with locally produced lettuce. Foodborne Pathog. Dis. 5, 339–349. 10.1089/fpd.2007.0065 [DOI] [PubMed] [Google Scholar]
- Svenungsson B., Lagergren A., Ekwall E., Evengard B., Hedlund K. O., Kärnell A., et al. (2000). Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin. Infect. Dis. 30, 770–778. 10.1086/313770 [DOI] [PubMed] [Google Scholar]
- Tarr P. I., Gordon C. A., Chandler W. L. (2005). Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- Thomas R. R., Brooks H. J., O'Brien R. (2017). Prevalence of Shiga toxin-producing and enteropathogenic Escherichia coli marker genes in diarrhoeic stools in a New Zealand catchment area. J. Clin. Pathol. 70, 81–84. 10.1136/jclinpath-2016-203882 [DOI] [PubMed] [Google Scholar]
- Vasant B. R., Stafford R. J., Jennison A. V., Bennett S. M., Bell R. J., Doyle C. J., et al. (2017). Mild illness during outbreak of shiga toxin-producing Escherichia coli O157 infections associated with agricultural show, Australia. Emerging Infect. Dis. 23, 1686–1689. 10.3201/eid2310.161836 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.