Abstract
Tissue engineering has been recognized as a translational approach to replace damaged tissue or whole organs. Engineering tissue, however, faces an outstanding knowledge gap in the challenge to fully recapitulate complex organ-specific features. Major components, such as cells, matrix, and architecture, must each be carefully controlled to engineer tissue-specific structure and function that mimics what is found in vivo. Here we review different methods to engineer tissue, and discuss critical challenges in recapitulating the unique features and functional units in four major organs—the kidney, liver, heart, and lung, which are also the top four candidates for organ transplantation in the USA. We highlight advances in tissue engineering approaches to enable the regeneration of complex tissue and organ substitutes, and provide tissue-specific models for drug testing and disease modeling. We discuss the current challenges and future perspectives toward engineering human tissue models.
Tissue engineering has emerged as a promising approach with two major goals: (1) to develop tissue and organ substitutes for clinical transplantation to replace damaged regions and restore organ function and (2) to build human tissue chips and replace animal models for drug screening and disease modeling. To date, multiple levels of complexity have been achieved with existing technologies to precisely position cells at scales from the single cell to whole tissue-level architectures. Clinical success has been achieved in simple flat tissue transplantation, such as skin and bladder,[1,2] which contain few cell types and require simpler engineering designs. Multiple human organs-on-a-chip have been actively developed for the study of drug response and pharmaceutical kinetics.[3–7] Engineering complex metabolically-demanding tissues, however, requires higher-order organization across interacting functional compartments (e.g., parenchyma and vasculature; cells, and matrix), at molecular, cellular and tissue scales, in addition to adequate mass to generate physiological tissue function.[8,9]
Each organ varies in its unique structural components—namely different cell types, matrix, and architecture among them, biophysical environment—pressure and flow, and biochemical stimuli—oxygen tension, cytokines, and growth factors, to support the specific organ function. In this review, we discuss the unique features and critical engineering challenges to tissue engineering in four major organs, focusing on the organs that top the organ transplant waiting list in the USA: the kidney, the liver, the heart, and the lung.[10,11] We will review advanced tissue engineering techniques that enable engineering organ-specific functional units for drug testing and reconstruction of thick tissue constructs for transplant. We will discuss organ-specific cells, matrix sources, and architectures, and highlight the bottlenecks and prospective for organ-specific tissue engineering.
Defining features of the kidney, liver, heart, and lung
The human body has levels of organization that build on each other: cells and matrices make up tissues, tissues make up organs, and organs make up systems to support the various bodily functions. At each level of organization, structure is closely related to function. The structure of kidneys, liver, heart, and lungs, therefore, reflects their specialized functions. Kidneys filter toxins and waste out of the body and produce urine. The liver produces and regulates many chemicals, substances, and proteins in the body. The heart pumps blood throughout the body, and lungs take up oxygen and release carbon dioxide. These functions are achieved via either repeating functional units, such as in the lungs, liver, and kidneys, or adequate mass, such as in the heart. It is important to recognize the basic structural units or mass in each organ to achieve tissue or organ-level functions.
Kidney
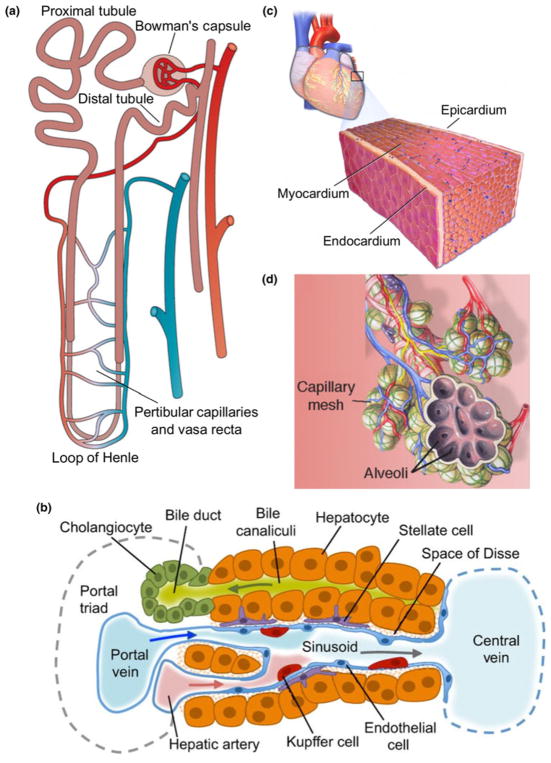
The fundamental functional unit of the kidney is the nephron [Fig. 1(a)], with each human kidney containing between 6 × 105 and 1.4 × 106 nephrons.[12] A nephron is composed of multiple segments, beginning with Bowman’s capsule, followed by the proximal tubule in the cortex, loop of Henle in the medulla, distal tubule in the cortex, and collecting ducts toward the ureter. These segments selectively filter, secrete, or reabsorb solutes, regulate the composition and volume of extracellular fluid, and maintain blood pressure. The kidney capillaries wrap closely around the nephron, providing nutrient support as well as actively participating in solute exchange. In fact, the kidney is highly vascular, receives about 25% of cardiac output, and is particularly susceptible to injury due to its dual dynamic functions.[13–16] After injury, these segments along with capillaries have limited regenerative capacity, which may contribute to tissue ischemia, tubular dysfunction, inflammation, fibrosis, and the development of chronic kidney diseases.[16,17]
Figure 1.
Functional units of the kidney, liver, heart, and lung. The nephron (a), liver sinusoid (b), and alveolus (d), are considered the functional units of the kidney, liver, and lung, respectively. The heart contains distinct epicardial, myocardial, and endocardial layers that each contribute to its function (c). Figures adapted from Bussolati and Camussi (a),[18] Gordillo et al. (b),[19] Laizzo (c),[20] and Desai et al. (d).[21]
The functional structural unit at the exchange interface of the kidney consists of three components: the tubular lumen, the vessel lumen, and a thin layer of basement membrane in between. Both sides of lumen are lined with specialized cells. Fenestrated endothelial cells with a rich glycocalyx along the capillary lumen, and epithelial cells with various signatures corresponding to different nephron segments in the tubular lumen.[22] For example, proximal tubular epithelial cells have a densely-packed bed of straight microvilli on the apical surface (brush border), narrow tight junctions, lateral cell surface folds with Na+/K+-ATPase transporters, and transmembrane water channels such as aquaporin-1 to control absorption and fluid transport.[23] Specialized epithelial cells called podocytes line the visceral layer of Bowman’s space, and have unique foot processes that interdigitate extensively while leaving small gaps in between (~25 nm wide). These gaps are spanned by a filtration slit membrane to control filtration, although its chemical composition is still a subject of investigation.[24,25] The basement membrane matrix is <1 μm thick, and rich in collagen IV and laminin.[26,27] Other cell types such as mesangial cells and perivascular cells may not directly participate in exchange, but rather provide structural support and secrete signals during injury and may even regulate blood flow by their contractility.[28] These specialized cells and matrix control the transport efficiency between blood and urine. Therefore, the engineering challenges for the kidney exchange interface rely on the recapitulating the close proximity of vessels and tubules with appropriate cell phenotypes and matrix to ensure the proper transport and accurate recreation of renal physiology and pathology.
Liver
The liver has a highly-organized architecture to serve its function, which has provided a daunting challenge to liver tissue engineering to reconstruct the tissue structure and function at large scales. For example, hepatocytes are known to require organized stromal support for proper function.[29–33] Liver function is supported by four major structural components: the hepatocytes that perform the metabolic reactions, the connective tissue stroma, the sinusoidal capillaries that deliver vascular flow to the hepatocytes, and the perisinusoidal space between the capillaries and the hepatocytes (also known as the space of Disse).[34] In addition, the classic hepatic lobule architecture describes a hexagonal mass of tissue that surrounds a central vein, and whose corners are the portal canals containing the triad of hepatic portal vein, hepatic artery, and bile duct.[35] Two distinct fluid flows travel in opposite directions in the liver lobule to provide a mix of oxygenated and deoxygenated blood from hepatic artery and hepatic portal veins, that travels through the sinusoid and drains into the central vein. This unique architecture provides the hierarchy for efficient cell–cell interactions and complex transport.
One unique structure in the liver that supports its special function is the hepatic sinusoids [Fig 1(b)]. The sinusoids have a discontinuous endothelium, containing both large fenestrae without diaphragms and large gaps between adjacent endothelial cells.[36,37] Part of the endothelial lining are stellate sinusoidal macrophages, also known as Kupffer cells, that play an important role in innate immunity in the liver.[38] In between the basal side of the hepatocytes and the sinusoids is the perisinusoidal space that allows for significant exchange between blood plasma and the hepatocytes. The gaps in the sinusoidal endothelium and the lack of a continuous basal lamina, as well as microvilli projections on the basal side of the hepatocytes, combine to increase the material exchange efficiency in this space. Within the perisinusoidal space, there also exist hepatic stellate cells, which are the primary sites for vitamin A storage.[39] These cells contain contractile elements that can constrict the sinusoids to increase vascular resistance, and they are also involved in extracellular matrix remodeling in response to liver injury.[40] Engineering liver-specific tissue requires not only these specialized cells, but also biomimetic architecture among cells in addition to adequate mass for physiological function, which further requires a hierarchical vasculature and perfusion support.
Heart
The heart is a muscular pump located in the chest cavity that requires high metabolic support to drive unidirectional blood flow into the pulmonary and systemic circulations during each contraction cycle. This delicate and powerful pump requires a highly organized layered architecture to generate contractile force efficiently and rhythmically, and a highly organized vasculature to provide sufficient nutrient support.
There are three layers in the heart muscle: epicardium, myocardium, and endocardium, from the outermost to the innermost, respectively [Fig. 1(c)]. The epicardium contains the coronary blood vessels and nerves to support the heart’s metabolic demand and rhythmic electrical propagation in the myocardium. The myocardium is a thick muscle layer with a helical architecture such that contraction propagates in an asynchronous manner, leading to both shortening and twisting of the ventricle during pumping[41] and maximized contraction and pumping efficiency.[42–44] In addition to cardiomyocytes, the myocardium also contains a large amount of cardiac fibroblasts and supporting vasculature. Nearly every myocardial cell is within 20 μm of a perfused capillary to facilitate the delivery of nutrients and oxygen and the removal of waste to support their high metabolic demand.[45] Along the epicardial surface, two coronary arteries branch off and divide into smaller arterioles and capillaries, which then penetrate deeply into the myocardium. The coronary blood supply can adapt to varying mechanical and metabolic circumstances by dynamically altering coronary vascular resistance. In the innermost layer, the endocardium makes direct contact with blood and is continuous with the circulatory system. It encompasses a specialized endothelial layer, its underlying layer of connective tissue and smooth muscle, and a deeper layer of connective tissue that contains the conductive fibers to initiate the rhythmic pumping activity.
The heart valves that control the flow between heart chambers are another unique cardiac structure. Valve disorders are common, and there is significant medical need to replace damaged or diseased valves.[46,47] These valves are rather thin, but are subjected to significant mechanical forces during normal function. Cardiac tissue engineering has focused on these two major goals: to remuscularize the heart via either cell injection or thick myocardium transplant, and to recreate live mechanically sound heart valves. Though much progress has been made, state-of-the-art engineered cardiac tissue remains functionally inferior to the mature normal myocardium found in vivo. The hierarchical organization of cardiomyocytes, vascularization, and multi-cellular proximity in three-dimensional (3D) thick constructs still pose a critical challenge, and the interactions between vasculature and perfusion with cardiomyocyte maturation remain elusive.
Lung
The fundamental functional unit of the lungs is the air–blood exchange interface in the respiratory zone, called alveoli [Fig. 1(d)]. The lungs have highly branched hierarchical airways in which there are more than 20 generations of airway branching, each resulting in narrower, shorter, and more numerous tubes. At the end of the airways, dense clusters form, known as alveolar sacs, each with diameter about 0.2 mm.[48,49] The number of alveoli increases along the bronchioles. Neighboring alveoli in these clusters are separated by a very thin layer of connective tissue, mostly composed of collagen and elastin fibers, known as the alveolar septum.[50] The alveolar space, septum, and surrounding capillaries form an air–blood barrier as a double-layer wall system.[51] These barriers contain both a thin and a thick portion.[52,53] The thin portion only consists of a surfactant layer, a type I pneumocyte with its basal lamina, and the capillary endothelial cells with its basal lamina. In the thick portion, some connective tissue and fibers can be found between the two layers of basal laminae that alter the thickness of the membrane, and is the site for fluid accumulation that drains into the lymphatic system. The architecture of the lung is particularly challenging to replicate using modern engineering approaches due to its hierarchical structure in three dimensions and biomechanical properties that permit breathing. Not only are the vascular and alveolar compartments organized in complex trees, but also the whole network resides in a flexible and dynamic matrix constantly in motion. In light of these challenges, lung tissue engineering has focused on either whole organ development or two cell layer interfaces, with limited structural and functional recapitulation.
Engineering organ-specific tissue and functional units
Complex organ structure may be recapitulated to different degrees by controlling cells, matrix, and architectures at the molecular, cellular, and tissue scales. In general, the engineering approaches balance two competing principles, top-down, and bottom-up engineering. Top-down engineering may be driven by the cells or scaffolds. Macroscopic scaffolds, or even whole organ scaffolds can be repopulated with cells to allow for cellular remodeling and self-assembly, and take the shape of a complex engineered tissue.[54–61] In contrast, bottom-up approaches involve engineering the smallest component elements of a tissue and directly assembling them into a larger construct.[62] Like building a house brick-by-brick, bottom-up engineering aims to explicitly specify fundamental units and organizations of cell–cell and cell–matrix interactions. Stem cell engineering has also made it possible to generate organoids, and organ-specific parenchyma from a single-cell source—human pluripotent stem cells (hPSC).[63] Here we focus on reviewing the state of the art of engineering approaches to allow for the control of geometry, cell–matrix architecture, matrix patterning, and their application toward engineering organs or organ-specific functional units.
Whole-organ scaffolds
The most advanced top-down method uses whole organs as the scaffold. With donor or cadaveric organs as the source, whole organs can be decellularized and then re-seeded with a patient’s own cells.[56,57] This method has the major advantage of native matrix composition and complex architecture to regenerate functional whole organs that may be used in therapeutic approaches for organ transplantation. Decellularization is commonly achieved by perfusion of detergents, often including SDS and Triton X-100.[64] The process yields a scaffold with both native structure and a complex, cell-produced matrix that maintains the spatial distribution of extracellular matrix proteins.[65] These cell-free organs can be seeded with cells from another individual or organism. Once seeded, material and microenvironmental cues are thought to direct the organization of cells within the scaffold. Success has been made for all four organs.[58–61,66,67] After recellularization and culture under perfusion, rat kidneys were shown to produce dilute urine,[61] hearts could weakly pump at physiological loads,[58] and lungs could oxygenate blood.[59,60] These successes have drawn considerable interest and follow-up in whole organ decellularization as a potential therapeutic approach for organ transplantation. Effective strategies to mature the cells seeded on the organ, as well as reliable sources for the large number of cells necessary to repopulate whole organs remain hurdles to the progression of this technology.
Advances in stem cell technology could provide reliable cell sources to address this problem. Induced pluripotent stem cells (iPSC), which can be formed by treatment of adult somatic cells with reprogramming factors, can differentiate into all cell types in the body.[68,69] Through this technology, it is possible to generate many patient-specific cells that may be useful in seeding decellularized organs. Their regenerative potential and ability to differentiate into multiple cell lineages to further promote recellularization may lead to higher levels of organ-specific function.[70–72]
Organ-specific functional units
As whole organ scaffolds continue to improve, engineered organ-specific tissue has drawn considerable attention as a method to recapitulate organ functional units to understand organ microphysiology, mechanisms of injury and the regeneration response, drug toxicity, and disease progression. These so-called microphysiological systems (MPS) have pushed forward the frontiers of engineering, fundamental biology, and pharmaceutical applications.[3,4,73–75] These successes will drive a shift in the drug development process from animal models toward high-throughput screening in human MPS to benefit healthcare. The common features of these MPS include miniaturization to minimize the requirement for cells and reagents, controlled exposure to biophysical (i.e., flow, pressure, oxygen tension) and biochemical stimuli (organ-specific growth factors and cytokines), and spatially controlled cell–cell and cell–matrix interactions. The engineering platforms range from simple microfluidic flow chambers,[7] to complex 3D tissues with controlled interactions between parenchyma and vasculature.[76,77] Many MPS may be connected to attempt to capture the diverse interactions among tissues throughout the body. The following techniques have been applied to achieve various levels of complexity.
Soft lithography
In the past 15 years, soft lithography, as a bottom-up engineering approach, has been developed and exploited extensively to benefit the understanding of biology and medicine. Using light patterning, small features, ranging from the hundreds of nanometers to hundreds of microns, can be created in various photoresists and then transferred to a “soft” material, like polydimethylsiloxane (PDMS).[78–81] This scale covers the microscale features found in many organs and allows the creation of biomimetic structures. Once these structures have been patterned into PDMS, cells can be seeded directly after modifying the surface of the PDMS or a second round of patterning can transfer the features to a hydrogel for 3D culture. Soft lithography has been used in the study of cell monolayers under flow,[82–84] cell–cell layer interactions,[85–87] microcontact printing,[88] and a wide variety of other engineering applications.[89,90] In these approaches, soft lithography provides very high resolution for engineered features. This enables the creation of geometries on the same scale as those found in organs that are generally durable and easy to work with. The limitation of this approach is that due to the transfer step from wafer to PDMS, designs must be confined to planar patterns that do not contain undercuts or overhangs. This fact, combined with the inability of cells to interact with and remodel the PDMS itself, make luminal structures one of the primary applications of soft lithography.
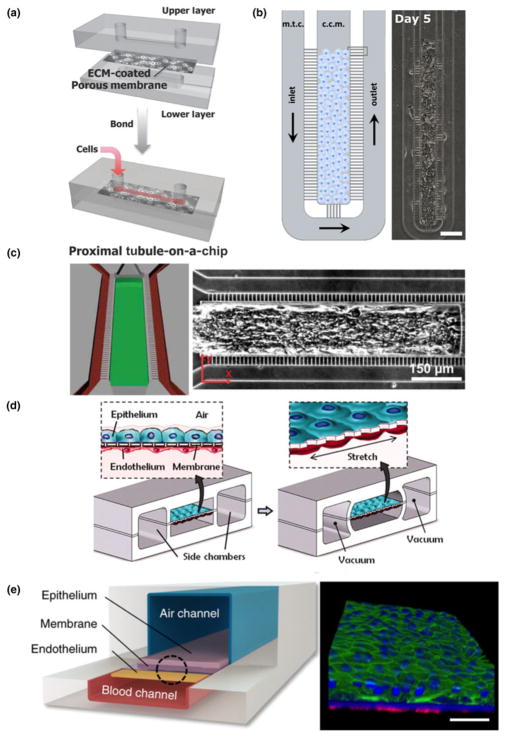
Nonetheless, due to its simplicity and high-throughput fabrication, PDMS-based soft lithography has been applied to generate the kidney endothelial–epithelial interface to evaluate drug-induced nephrotoxicity. Representative studies include those by the Ingber group[91] and the Takayama group,[92] where multilayered devices were made in PDMS, which include two channels separated by a porous membrane. Cells coat the top channel and the bottom channel is left unseeded to mimic the interstitial space [Fig. 2(a)]. Perfusion of both channels allows drug uptake and clearance to be evaluated under physiologic conditions. Results from their studies highlighted the need for culture under biomimetic conditions, as culture under flow altered the cytotoxicity of applied drugs and reflected a shortcoming of static culture techniques. In particular, the Takayama group[92] showed a different drug response to continuous low-concentration perfusion or a bolus style delivery. Continuous delivery of the drug was more damaging to the cells, resulting in greater loss of junctional proteins and an increase in permeability. These studies demonstrate the possibility of these models to explore the dynamics of renal drug toxicity and provide more than a simple binary readout for a drug’s effect.
Figure 2.
Soft lithography in organ-specific engineering. Jang et al. engineered a proximal tubule model through soft lithography with an upper layer where cells are under fluid flow and a lower layer modeling the interstitial space in the kidney (a, figure adapted from Jang et al.[91]). Soft lithographic approaches by Gori et al. expose hepatocyte clusters to physiological mass transport (b, figure adapted from Gori et al.[93]). Mathur et al. created 3D microtissues using cardiomyocytes in a mold with controlled transport properties (c, figure adapted from Mathur et al.[7]). The Ingber group has developed multiple soft lithographic platforms that mimic the mechanical forces of the lung (d, figure adapted from Huh et al.[87]) and its unique air and blood interfaces (e, figure adapted from Benam et al.[94]).
PDMS based soft lithography has also been used in engineering microenvironments for liver, heart and lungs. Lee et al. have generated a cord of hepatocytes in a PDMS-based microfluidic chamber to mimic the liver sinusoid in its mass transport properties.[95] Although this system simplifies the sinusoid by culturing purely hepatocytes, it applies consistent physiologic transport conditions matched to healthy tissue, and allows researchers to understand the response of hepatocytes under controlled hydrodynamic conditions. A modified version of this device was recently used by Gori et al. to model nonalcoholic fatty liver disease [Fig. 2(b)].[93] Their work showed that hepatocytes cultured on this platform had triglyceride accumulation closer to in vivo conditions than the two-dimensional (2D) culture. Similarly PDMS-based microfluidics have also been explored for hepatocyte metabolism. The Leclerc group developed a platform to culture primary hepatocytes under dynamic flow conditions mimicking those found in vivo, and showed enhanced recapitulation of an in vivo like phenotype.[96]
Soft lithography in heart tissue engineering studies has been generally limited to creating cardiomyocyte monolayers or cell sheets for high-throughput drug testing[97,98] For example, the Kim group has shown that nanopatterned cues can induce the alignment and organization of iPS-derived cardiomyocytes.[99] These sheets can also be made using cells from a diseased background (e.g., muscular dystrophy) to explore their unique biology and investigate possible therapeutic approaches.[100] Mathur et al. further expanded the usage of soft lithography to create aligned 3D microtissues with iPSC-derived cardiomyocytes [Fig. 2(c)].[7] In this model, cardiomyocytes are exposed to drugs in a way that more closely mimics that of healthy cardiac tissue. By creating an artificial endothelial-like barrier, the mass transport to the tissue is controlled and more physiological. As a result, this device was more predictive of the effectiveness of a clinically available cardiac therapeutic than conventional 2D platforms.
The greatest tissue complexity achieved with PDMS-based soft lithography is in the recreation of the lung exchange barrier with a device with controlled motion to mimic the breathing rhythm and mechanical tension. The Ingber group has used this to make important strides in modeling the mechanics of lung tissue.[87] In a multilayered PDMS device, two channels serve to mimic the lung [Fig. 2(d)]. The lower chamber contains an endothelial layer and is separated from the upper chamber by a porous PDMS layer. An epithelial layer lines the upper chamber. Two additional chambers flank the cell containing channels on either side. By pulling a vacuum through these additional chambers, the epithelial and endothelial layers can be stretched, mimicking the mechanical forces experienced in the lung. When cultured with immune cells and stimulated by tumor necrosis factor-α, neutrophils could transmigrate across the endothelium into the epithelium. Bacteria in the airway channel could drive neutrophils across the endothelium to consume bacteria on the epithelial surface. Additionally, this model provided insight into the role of mechanical strain in the uptake of nanoparticulates. The lung on chip model was further refined to include a differentiated and beating mucociliary bronchiolar epithelium [Fig. 2(e)].[94] Using primary cells isolated from healthy and diseased patients, this small-airway model was used to investigate asthma and chronic obstructive pulmonary disease (COPD). This allowed the analysis of both the effectiveness of some therapeutic agents as well as provided insight into their mechanism of action.
Hydrogel-based casting, molding, and stamping
Advances in biomaterials, particularly hydrogels, have made it possible to provide important matrix support and modulate cell–matrix interactions, to achieve long-term culture and recapitulate in vivo microphysiology.[101,102] A long-standing technique is to embed cells into bulk gel constructs, so that cells can readily communicate with the surrounding matrix and other cells to reorganize and remodel the microenvironment toward functional constructs.[103] This approach is relatively simple, however, often taking on an amorphous macroscopic structure. Engineering constructs with controllable features need to combine the advances in hydrogel development with different molding and casting engineering techniques to more closely mimic the essential structural architecture of cells, tissues, and organs. There are techniques to create shapes and structures by subtracting or adding materials. Tissue can be formed by step-wise casting and layered construction or through macroscopic casting or automatic printing. Each technique, however, has different limits and criteria for material selection.
Subtractive casting
Subtractive casting is often used to create a void space or hollow tube in bulk hydrogels. Simple geometries such as a straight tube can be achieved by crosslinking a bulk hydrogel around a straight rod (e.g., an acupuncture needle).[104,105] Once the gel has set, the rod is removed, and an open lumen is formed. Taking advantage of this simple setup, this technique has been used to make endothelialized tubes for the endothelial response to biophysical and biochemical stimuli and their interactions with surrounding cells.[105] This approach benefits from its simple setup, but is also limited by its simple geometry. Multiple rods can be positioned in a spatially distributed manner with macroscopic control to generate multiple tubes in one hydrogel device.[106] These complexities provide an opportunity to control the biophysical flow and transport among tubes to mimic the vascular–lymphatic communication, as well as the exchange between the vasculature and surrounding tissue, or tubules, as in the kidneys for example.
Sacrificial material molding
Complex structures can also be made with subtractive casting, using sacrificial materials via multi-step casting and molding techniques. To create complex patterns in gel, networks must first be created in a dissolvable or removable material that can be embedded in a bulk gel. Once surrounded by cross-linked gel, the network pattern is then dissolved or evacuated from the matrix. Engineered patterns have been created in multiple materials, including gelatin[107] and sugar.[108] For very complex geometries, the entire vasculature of an organ has been converted into a sacrificial material that could then be encased and dissolved in a hydrogel.[109] The selection of materials for sacrificial molding, however, are limited. For cells to survive in the bulk matrix, the sacrificial step must be gentle, avoiding cytotoxic solvents and extreme temperatures.
Layered stamping
Rather than encasing a pattern within a gel, patterns may be transferred plane-by-plane through repeated steps of stamping and layering. Stamps containing negative or positive features made through lithography, milling, or other processes are covered in a hydrogel solution that is then set. Once the stamp is removed, these open features can then be used directly,[32] or this process can be repeated and the stamped hydrogel layers stacked to create complex 3D tissues.[110,111] The process of stacking and aligning many independent and often fragile layers can be challenging, although automation or refinements to the stacking process can address this limitation.
Macroscopic molding
For even larger constructs, molds from the millimeter to centimeter scale can also be created. These molds often sacrifice control of the microscale features of tissues in the name of a more functional or easier to manipulate macroscopic construct.[55,112] While the external geometry of these constructs can be tightly controlled, the localization of cells and matrix within the tissue, as well as the inclusion of an embedded vascular network is very limited. Other techniques are poised to bridge the gaps between these micro- and macro-scale approaches.[113] These approaches have pushed the frontier of organ-specific functional units to allow for cell and matrix patterning and remodeling simultaneously for both short- and long-term culture, and to better mimic the cellular, biochemical, and biophysical microphysiology found in vivo. Extensive applications have been made in this direction.
Subtractive casting has been applied to generate a single vessel or tubular structure to mimic kidney tubules. Kelly et al. have used single-tube-based devices (Nortis, Inc.) to recreate a microphysiological model of human kidney proximal tubule [Fig. 3(a)]. They demonstrated the appropriate polarity and marker expression of the proximal tubule in this 3D flow-directed MPS.[5] More importantly, these tubules exhibited biochemical and synthetic activity, as well as secretory and reabsorptive features associated with proximal tubule function in vivo. The same devices have also been used to generate a human four-cell sequentially layered self-assembled liver MPS.[114] In this system, human hepatocytes, endothelial cells, and Kupffer cells are all sequentially layered as cell suspensions into the device, followed by the addition of a layer of collagen gel containing encapsulated hepatic stellate cells.[114] Using this setup, they were able to demonstrate reasonable response to drugs in terms of toxicity and fibrotic response, and validated it as a liver MPS model for drug development. Furthermore, these individual MPS can be connected and coupled together to study inter-organ interactions (i.e., kidney proximal tubule model with intestine, liver, blood–brain barrier, and skeletal muscle).[76]
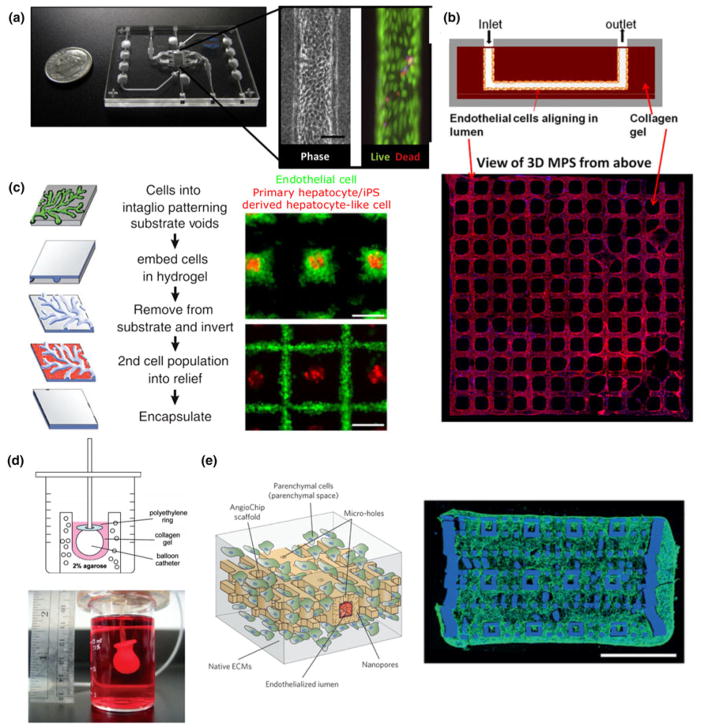
Figure 3.
Hydrogel casting and molding of organ functional units. By subtractive casting, Weber et al. formed linear channels in a collagen I gel lined with human proximal tubule epithelial cells with high viability (a, figure adapted from Weber et al.[5]). Our group has utilized stamping to form a 3D peritubular vascular network in collagen I containing human kidney microvascular endothelial cells (b, adapted from Ligresti et al.[115]). The InVERT molding strategy developed by Stevens et al. allows multiple hepatic subtypes to be cultured with spatial and geometric control (c, figure adapted from Stevens et al.[32]). By bulk molding, Lee et al. created centimeter-scale cardiac organoid chambers that had low levels of pump function (d, figure adapted from Lee et al.[112]). Highly vascularized 3D cardiac tissues were created by the Radisic group using a stamping and layering technique (e, figure adapted from Zhang et al.[111]).
A more complex subtractive casting process was recently applied in kidney by Huling et al. to recreate the kidney vasculature through several dissolving and casting steps.[109] First, whole rat kidneys were perfused with polycaprolactone (PCL) to create a cast of the renal vasculature. Kidney tissue was then dissolved away, and resulting PCL casting was coated with a layer of collagen I matrix. In a second digestion step, the PCL was then dissolved in acetone, leaving behind a perfusable copy of the kidney vasculature. Finally, this network was embedded in a bulk collagen gel, forming a densely vascularized engineered tissue containing glomerular structures. This multi-step casting technique recreated the vascular networks at the level of complexity advanced engineering techniques have yet to accomplish de novo. The system, however, may be too complex for understanding the cellular response to biophysical and biochemical cues at mechanistic level. A more controlled geometry with appropriate perfusion dynamics would allow for an understanding of drug-induced toxicity and the injury-regeneration capacities of kidney vascular networks. Our group has combined injection molding, lithography, and layered stamping to create 3D human kidney peritubular microvessel networks in collagen [Fig. 3(b)].[115] A major advantage of these devices is that cells may interact with and remodel the surrounding matrix, which can also be embedded with additional cell types such as pericytes. Our study showed for the first time that fenestrated human kidney microvessels can be created and maintained under flow. The advantage of this technique is that it allowed for the addition and combination of multiple layers, which can mimic the filtration barrier or endothelial–tubular interface for mechanistic studies of kidney exchange functions.
Similarly, more complex models of the liver have been achieved through molding and stamping for improved functional outcomes. For example, Stevens et al. developed a multistep casting process and applied it to liver tissue [Fig. 3(c)].[32] In this approach, cells are first patterned into the negative features of a mold (intaglio) and encapsulated in a hydrogel. Once set, the hydrogel is removed and inverted from the mold. The raised features in this hydrogel contain the first-cell population and a second-cell population can be patterned into the recesses. Using this technique, primary or iPSC-derived hepatic aggregates could be surrounded by endothelial or stromal cells, which were found to alter tissue function once implanted in mice. In more recent work, they have shown that engineered cords of endothelial cells and hepatocytes could be micropatterned to create a vascularized liver tissue when implanted in vivo.[116]
Hydrogel-based molding and casting have also been explored extensively in cardiac tissue engineering. Nunes et al. cast differentiated cells in a collagen gel around a surgical suture to promote the alignment of encapsulated PSC-derived cardiomyocytes.[55] Over the course of 1 week, the cells compacted the matrix, aligned along the length of the suture, and beat synchronously. Providing the cells with a surface to compact against is a common theme in engineered cardiac tissues, and has been achieved using other geometries. For example, Lee et al. cast primary rat cardiac cells in a collagen and Matrigel® matrix around an inflated balloon catheter [Fig. 3(d)].[112] Over 7–10 days, the cells compacted around this surface to form a model ventricle that beat and produced pressure. These hydrogel-based constructs may also be supplemented with mechanical loads (i.e., constant or cyclic stress and strain) that mimic the physiological conditions in vivo. On an absorbable gelatin sponge scaffold, Mihic et al. seeded human embryonic stem cell (hESC)-derived cardiomyocytes and applied cyclic uniaxial stretch.[117] Through this loading regime meant to mimic the mechanical load on cardiomyocytes in the first few months of post-natal life, cardiomyocytes attained a more mature phenotype and were implantable in ischemic rat hearts. Uniaxial stretch can also be applied directly to bulk hydrogels containing cardiomyocytes, as in constructs prepared by Tulloch et al.[118] In these conditions, stretch also promoted maturation and alignment of cardiomyocytes, and could be combined with endothelial cells to promote the self-assembly of vascular structures in the tissue.
A major thrust in hydrogel-based thick tissue engineering is to engineer perfusable vessel networks that support the high metabolic demands of thick tissues. Our group has used collagen gel-based soft lithography to pattern vessel networks in cardiac tissues.[54] When cardiomyocytes are combined with stromal cells in the bulk collagen and endothelial cells in the vessels, synchronized electrical activity was observed along with angiogenesis into avascular regions of the construct. Zhang et al. reported the development of a vascularized platform, which could support multiple tissues, including cardiac cells [Fig. 3(e)].[111] By repeated steps of casting and stamping from molds created by soft lithography, a complex 3D vascular network was created in a citric-acid-based elastomer. Around this network, hESC-derived cardiomyocytes could be cast in fibrin gel or Matrigel and resulted in contractile vascularized constructs that could be directly anastomosed to the femoral arteries and veins of rat hind limbs.
Bioprinting
More complex 3D structures can be achieved by bioprinting, a technique that has recently exploded in popularity and rapidly developed. Drawing inspiration from conventional 3D printing of plastics and metals, bioprinting may construct tissues in full complexity from the bottom up, by directly controlling the placement of cells and matrix.[119–121] With a range of natural and synthetic materials to choose from, hydrogels have become an essential component in many bioprinting applications.[120,122] Materials, including polymers such as collagen, and polyethylene glycol, can often be easily modified to tune features like their mechanical properties, degradation rate, and gelation kinetics.[123] These modifications enable tighter control over both the printability of a gel and its ability to match the properties of the tissue being printed in the pursuit of more biomimetic constructs. Composite hydrogels[124] and decellularized extracellular matrix hydrogels[121,125] further expand the range of material and biochemical properties available for bioprinting. The designs for bioprinted tissues may be a carefully engineered shape, or a personalized 3D reconstruction extracted from a patient’s imaging data (e.g., computed tomography and magnetic resonance imaging). Either through selective deposition, or selective cross-linking, bioprinting comes in several flavors, each with their own advantages and disadvantages.[113,126] The simplest version of bioprinting has two core components, a moveable stage and extruder. Together the stage and extruder have three axes of freedom (e.g., the stage may move in ±Z, and the extruder in ±X and ±Y ). The extruder deposits the bio-material onto the stage and builds up the tissue layer by layer. Each layer is like a single 2D print, with the extruder tracing out a single planar geometry. Once one layer is complete and has crosslinked, the stage is lowered (or the extruder is raised) by a set amount, and the geometry of the next layer is extruded. This process repeats until the whole 3D structure has been printed. Extruders come in several varieties and often eject material by air pressure or thermal actuation.[127]
Soft lithography bioprinters cut out a few of the moving parts and trade-in an extruder for a projector and photocrosslinkable biomaterials. Rather than tracing out each layer, the projector projects a photomask onto the bed, crosslinking a single layer of material and attaching it to the stage. This process is repeated layer by layer as the stage moves in the Z to create the tissue.[128] Advances in bioprinting provide the potential for the automated creation of complex geometries that is required toward engineering thick complex tissue.
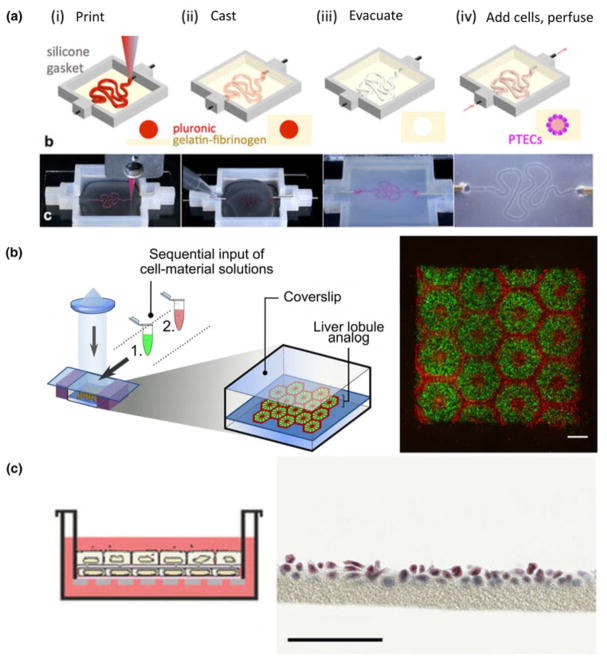
To date, bioprinting has been used to replicate parts of nephron, liver, heart, and lung architectures. The Lewis group recently reported a model of the kidney proximal tubule [Fig. 4(a)].[129] In this system, the proximal tubule geometry is first printed in a sacrificial layer of Pluronic®. This tubule is then encapsulated within a bulk gelatin–fibrin matrix and evacuated to leave behind a perfusable proximal tubule model. After seeding with proximal tubule epithelial cells, this model appeared to have improved functionality when compared with cells cultured in 2D alone and showed structural and functional nephrotoxicity when injured by cyclosporine.
Figure 4.
Bioprinting organ-specific tissues. Homan et al. created a model proximal tubule on chip by printing the tubule geometry in a fugitive material that is encapsulated in gel, and then evacuated leaving an open lumen. Proximal tubule epithelial cells could then be cultured in the tubule and further characterized (a, figure adapted from Homan et al.[129]). Ma et al. produced complex geometries that mimicked the hexagonal lobule structure in normal liver tissue through 3D bioprinting (b, figure adapted from Ma et al.[130]). To recreate the lung tissue interface in gel, Horváth et al. used 3D printing to print endothelial and epithelial cell layers separated by a thin layer of matrix (c, figure adapted from Horváth et al.[131]).
Faulkner-Jones et al. showed that both hiPSC and hESC could be printed directly in an alginate gel and subsequently differentiated into albumin producing hepatocyte-like cells.[132] Primary cells have been used by Nguyen et al., who modeled drug-induced liver injury in bioprinted tissues.[133] To create a high throughput liver-on-chip model, Bhise et al. bioprinted constructs containing hepatic spheroids.[134] By combining spheroids with bioprinting, constructs that better mimic the native oxygen gradient in tissues can be rapidly formed. Placed under continuous flow these constructs could be used to model hepatoxicity, and illustrate the utility of bioprinting as a technique for high-throughput screening.
While resolution limitations have generally been prohibitive for researchers attempting to mimic the microarchitecture of the liver, Ma et al. recently made strides in this approach.[130] Using a DLP-based bioprinter, hexagonal liver lobules matched with human dimensions were printed in two layers [Fig. 4(b)]. The first contained a parenchymal mixture of iPSC-derived hepatic progenitor cells, and the second a non-parenchymal combination of cells of endothelial and mesenchymal origin. Through this process, constructs with higher expression of several hepatic genes suggested that printing and patterning progenitor cells lead to more mature cells than 2D culture or 3D encapsulation alone.
Gaetani et al. used extrusion based bioprinting to show that human cardiac progenitor cells could be printed in an alginate-based matrix.[135] These cells showed good viability after printing and could migrate into an adjacent, non-printed matrix. In a later study, this group enhanced the viability of printed cells by altering the matrix to a hyaluronic acid and gelatin-based material.[136] Printed progenitor cells could be differentiated in vitro, and constructs implanted into a mouse myocardial infarct contained cardiomyocyte and endothelial populations. While functional improvements were limited, these printed patches reduced fibrosis in the infarct region. In a more direct attempt at mimicking the native cardiac structure, Zhang et al. directly printed endothelial cells and then filled in voids of the scaffold with iPSC-derived cardiomyocytes.[137] Endothelial cells were printed in a mesh-like microfibrous structure that the authors hypothesize could remodel to form open lumen. Around this structure cardiomyocytes were seeded and spontaneous beating of the construct was observed. By altering the structural anisotropy of the network, cardiomyocyte alignment and contraction amplitude could be tuned. Models like this could prove to be useful for both cardiac drug screening, and as therapeutics.
The complex air-interface of lung tissue is difficult to replicate, but has been a target for bioprinting. Horváth et al. printed endothelial and epithelial layers of cells separated by a thin layer of matrix that more closely matches what is found in vivo than in existing models of this interface [Fig. 4(c)].[131] While this approach is still in its infancy, it may represent a high-throughput technique to study and engineer the alveolar barrier. While bioprinting has not yet been widely applied to lung tissue engineering, it remains a promising approach as the technology matures.
Other techniques
Several other powerful techniques exist to create organ-specific tissues. Organoid cultures for the kidney,[138–140] liver,[141,142] heart,[143–145] and lung,[146,147] in which cells self-assemble into structures with organ-like hierarchy and features, provide insight into developmental and functional components of these organs. When combined with hPSC technology they may also ultimately serve as a source of therapeutic tissue. In addition, scaffold-free techniques, such as cell sheet engineering, also provide alternative methods to control 2D architecture at the molecular and cellular level,[97] which can further be assembled to form more complex tissues using multiple techniques.[148,149]
Perspectives and future directions
The goals, techniques, and approaches for organ-specific engineering are broad. Whether they are used to develop model biologic systems, to design high throughput drug discovery platforms, or to create therapeutic tissues, constructs built in the laboratory better replicate real biologic systems than ever before. The unique features of each organ lend themselves to certain approaches, and as a result progress toward each of these ends is heterogeneous.
In the kidney, a variety of strategies have led to success. While complete recreation of the nephron is beyond the limits of current synthetic techniques, piecemeal reconstruction of renal components is ongoing. Future advances in engineered kidney functional units are in three major directions: (1) recapitulating multiple-segments of a nephron with differentiated function such as the glomeruli, loop-of-Henle, distal tubule, and connecting ducts; (2) integration of kidney capillaries with tubular segments for the study of the delicate transport properties in health and disease; and (3) modeling patient-specific structure and function toward the study of nephrotoxicity and injury-regeneration within the scope of precision medicine. Outside of these methods for creating structure de novo, decellularized, and recellularized whole human kidneys would provide the most relevant matrix scaffold for the support of cellular function. Further progress would benefit the drug development industry and could represent a promising organ transplantation alternative with minimal immune response and rejection by incorporating patient-specific cells.
Liver tissue engineering likely has the longest history of these four organs. The scale of the lobule, regular structure, and clear cellular organization of the liver make simplified models of the liver feasible. Nonetheless, complex 3D liver tissue organization and function has mostly been demonstrated in animal models. The functional units of a liver, or a sustainable 3D liver tissue model have not been achieved. Future work is expected to advance the field of liver tissue engineering by incorporating appropriate liver sinusoidal vascular and stromal cells, identifying the important signals or cellular/structural support and fully recapitulating the cellular, biochemical, and biophysical signals in vivo.
Strategies for the heart have been similarly diverse, with many techniques leading to the production of contractile, but functionally immature, cardiac constructs. Although cardiomyocytes are the crux of these tissues, a focus on incorporating stromal and endothelial populations have increased the complexity of engineered cardiac tissues from all techniques. Vascularization remains as the most immediate need in the field, in both functional tissue and whole-organ scaffolding. Engineering perfusable heart tissue with proper architecture has advanced with recent progress. Future work remains to mature the tissue toward efficiently performing work in long-term culture, and rapidly integrating when implanted to improve perfusion and pump function in damaged cardiac tissue. Whole heart decellularization, though promising like other organs, requires further development in efficient cell seeding, retention, and maturation.
The lung remains an extraordinary challenge to engineer from scratch. Model systems produced through soft lithography remain a gold standard for in vitro approaches and continue to expand our understanding of pulmonary biology. The interactions between pulmonary endothelium and epithelium, and the unique properties of the liquid–air interface in the lung are all targets for these platforms. On the therapeutic side, synthetic alternatives that meet the unique vascular and mechanical demands of the lung have not yet emerged, positioning decellularized lungs as the de facto option for whole lung engineering. Early reports have hinted at their therapeutic efficacy, with transplant studies in rodents both showing promise and highlighting the difficulty of forming a non-leaky vasculature.
In addition to the continued development of engineering techniques, it is important to improve cell and matrix sources for both enhancing tissue function and promoting clinical translation. In particular, vascularization efforts in tissue engineering have mostly relied on human umbilical vein endothelial cells (HUVECs). HUVEC-formed vessels, unfortunately, are known for poor survival and immunogenicity in vivo,[150] and do not recapitulate organ-specific vessel characteristics.[151] It is therefore important to find alternative endothelial cell sources, ideally a common autologous patient cell source for all parenchymal cells in engineered tissues or organ functional units. While isolating primary cells from patient-specific biopsy samples is possible, advances in stem cell engineering provide the potential to create multiple cell lineages from patient-specific pluripotent stem cells.[152] It is possible that cells derived from a single-cell source could provide structural and functional support to each other and further enhance the functionality of the engineered tissues.
Recapitulating the complexity of native tissue matrices also remains an open challenge. The matrix can communicate essential cues related to cell fate[153] and function,[154] and is a critical component in the design of engineered tissues. To meet these design and fabrication needs, many advances have been made in the biomaterials available for engineered tissues, ranging from natural and synthetic biomaterials to decellularized matrices.[120,155] Despite these advances, an understanding and ability to engineer matrices that mimic the biochemical composition and ultrastructure of native tissue is limited. Decellularization protocols preserve many of these features, but still cause some changes to the matrix. Depending on the decellularization method used, the matrix’s structural, mechanical, and biochemical properties can change,[156] which may ultimately alter the characteristics of seeded cells. Hydrogels can also be derived from decellularized matrices enabling their use in a wider variety of tissue engineering techniques.[121] Forming these hydrogels is not without compromise, however, as some of the ultrastructural features of the native matrix are lost in the process of solubilization.[157] Advances in matrices that can guide the performance of encapsulated cells will lead to more complex and advanced constructs that better integrate with host tissue.
In general, the tissue-engineering approach has significantly advanced the tools available to study the physiology and pathology of many organs. The discussed approaches benefit not only the kidney, liver, heart, and lungs, but the engineering principles, biomaterials, and selection criteria can also be used to model any other organ. Toward next-level function of engineered tissue, one ignored, but unachieved topic is the integration of the nervous system, which will require the combination of neural cell biology and tissue engineering knowledge. While we move forward, it is important to keep in mind the basic anatomy and physiology of the body, learn from its organization, and continue to improve our tissue engineering models to better benefit healthcare.
Acknowledgments
We acknowledge the financial support of National Institute of Health Grants DP2DK102258, UH2/UH3 TR000504, UH2DK107343, and RO1HL130488.
References
- 1.MacNeil S. Progress and opportunities for tissue-engineered skin. Nature. 2007;445:874. doi: 10.1038/nature05664. [DOI] [PubMed] [Google Scholar]
- 2.Atala A, Bauer SB, Soker S, Yoo JJ, Retik AB. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet. 2006;367:1241. doi: 10.1016/S0140-6736(06)68438-9. [DOI] [PubMed] [Google Scholar]
- 3.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 4.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med (Maywood) 2014;239:1061. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, Wang Z, Yamaura Y, Hailey DW, Neumann T, Shen DD, Thummel KE, Muczynski KA, Himmelfarb J, Kelly EJ. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016;90:627. doi: 10.1016/j.kint.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez CE, Yen RW, Perez SM, Bedell HW, Povsic TJ, Reichert WM, Truskey GA. Human vascular microphysiological system for in vitro drug screening. Sci Rep. 2016;6:21579. doi: 10.1038/srep21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Healy KE. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atala A, Kasper FK, Mikos AG. Engineering complex tissues. Sci Transl Med. 2012;4:160rv12. doi: 10.1126/scitranslmed.3004890. [DOI] [PubMed] [Google Scholar]
- 9.Mikos AG, Herring SW, Ochareon P, Elisseeff J, Lu HH, Kandel R, Schoen FJ, Toner M, Mooney D, Atala A, Van Dyke ME, Kaplan D, Vunjak-Novakovic G. Engineering complex tissues. Tissue Eng. 2006;12:3307. doi: 10.1089/ten.2006.12.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellar CA. Solid organ transplantation overview and delection criteria. Am J Manag Care. 2015;21:S4. [PubMed] [Google Scholar]
- 11.Magee JC, Barr ML, Basadonna GP, Johnson MR, Mahadevan S, McBride MA, Schaubel DE, Leichtman AB. Repeat organ transplantation in the United States, 1996–2005. Am J Transplant. 2007;7:1424. doi: 10.1111/j.1600-6143.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 12.Pocock G, Richards CD, Richards DA. Human Physiology. Oxford University Press; 2013. [Google Scholar]
- 13.Jen KY, Haragsim L, Laszik ZG. Kidney microvasculature in health and disease. Exp Model Ren Dis Pathog Diagn. 2011;169:51. doi: 10.1159/000313945. [DOI] [PubMed] [Google Scholar]
- 14.Basile DP. The endothelial cell in ischemic acute kidney injury: implications for acute and chronic function. Kidney Int. 2007;72:151. doi: 10.1038/sj.ki.5002312. [DOI] [PubMed] [Google Scholar]
- 15.Basile DP, Friedrich JL, Spahic J, Knipe N, Mang H, Leonard EC, Changizi-Ashtiyani S, Bacallao RL, Molitoris BA, Sutton TA. Impaired endothelial proliferation and mesenchymal transition contribute to vascular rarefaction following acute kidney injury. Am J Physiol Physiol. 2011;300:F721. doi: 10.1152/ajprenal.00546.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basile DP. Rarefaction of peritubular capillaries following ischemic acute renal failure: a potential factor predisposing to progressive nephropathy. Curr Opin Nephrol Hypertens. 2004;13:1. doi: 10.1097/00041552-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med. 2014;371:58. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bussolati B, Camussi G. Therapeutic use of human renal progenitor cells for kidney regeneration. Nat Rev Nephrol. 2015;11:695. doi: 10.1038/nrneph.2015.126. [DOI] [PubMed] [Google Scholar]
- 19.Gordillo M, Evans T, Gouon-Evans V. Orchestrating liver development. Development. 2015;142:2094. doi: 10.1242/dev.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laizzo PA. Handbook of Cardiac Anatomy, Physiology, and Devices. Springer International Publishing; 2009. [Google Scholar]
- 21.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulger RE, Dobyan DC. Recent structure-function relationships in normal and injured mammalian kidneys. Anat Rec. 1983;205:1. doi: 10.1002/ar.1092050102. [DOI] [PubMed] [Google Scholar]
- 23.Furriols M, Chillarón J, Mora C, Castelló A, Bertran J, Camps M, Testar X, Vilaró S, Zorzano A, Palacín M. rBAT, related to L-cysteine transport, is localized to the microvilli of proximal straight tubules, and its expression is regulated in kidney by development. J Biol Chem. 1993;268:27060. [PubMed] [Google Scholar]
- 24.Greka A, Mundel P. Cell biology and pathology of podocytes. Annu Rev Physiol. 2012;74:299. doi: 10.1146/annurev-physiol-020911-153238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavenstädt H. Roles of the podocyte in glomerular function. Am J Physiol Renal Physiol. 2000;278:F173. doi: 10.1152/ajprenal.2000.278.2.F173. [DOI] [PubMed] [Google Scholar]
- 26.Salmon AHJ, Neal CR, Harper SJ. New aspects of glomerular filtration barrier structure and function: five layers (at least) not three. Curr Opin Nephrol Hypertens. 2009;18:197. doi: 10.1097/MNH.0b013e328329f837. [DOI] [PubMed] [Google Scholar]
- 27.Shirato I, Tomino Y, Koide H, Sakai T. Fine structure of the glomerular basement membrane of the rat kidney visualized by high-resolution scanning electron microscopy. Cell Tissue Res. 1991;266:1. doi: 10.1007/BF00678705. [DOI] [PubMed] [Google Scholar]
- 28.Cortes P, Méndez M, Riser BL, Guérin CJ, Rodríguez-Barbero A, Hassett C, Yee J. F-actin fiber distribution in glomerular cells: structural and functional implications. Kidney Int. 2000;58:2452. doi: 10.1046/j.1523-1755.2000.00428.x. [DOI] [PubMed] [Google Scholar]
- 29.Hui EE, Bhatia SN. Micromechanical control of cell–cell interactions. Proc Natl Acad Sci U S A. 2007;104:5722. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Probing the role of multicellular organization in three-dimensional microenvironments. Nat Methods. 2006;3:369. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 31.Bhatia SN, Underhill GH, Zaret KS, Fox IJ. Cell and tissue engineering for liver disease. Sci Transl Med. 2014;6:245sr2. doi: 10.1126/scitranslmed.3005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens KR, Ungrin MD, Schwartz RE, Ng S, Carvalho B, Christine KS, Chaturvedi RR, Li CY, Zandstra PW, Chen CS, Bhatia SN. InVERT molding for scalable control of tissue microarchitecture. Nat Commun. 2013;4:1847. doi: 10.1038/ncomms2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan J, Logan DJ, Root DE, Carpenter AE, Bhatia SN. High-throughput platform for identifying molecular factors involved in phenotypic stabilization of primary human hepatocytes in vitro. J Biomol Screen. 2016;21:897. doi: 10.1177/1087057116660277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna M. Microscopic anatomy of the liver. Clin Liver Dis. 2013;2:S4. doi: 10.1002/cld.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappaport AM, Borowy ZJ, Lougheed WM, Lotto WN. Subdivision of hexagonal liver lobules into a structural and functional unit. Role in hepatic physiology and pathology. Anat Rec. 1954;119:11. doi: 10.1002/ar.1091190103. [DOI] [PubMed] [Google Scholar]
- 36.Wisse E, Braet F, Dianzhong Luo D, De Zanger R, Jans D, Crabbe E, Vermoesen A. Structure and function of sinusoidal lining cells in the liver. Toxicol Pathol. 1996;24:100. doi: 10.1177/019262339602400114. [DOI] [PubMed] [Google Scholar]
- 37.Wisse E, Braet F, Luo D, Vermijlen D, Eddouks M, Konstandoulaki M, Empsen C, de Zanger RB. Endothelial cells of the hepatic sinusoids: a review. In: Tanikawa K, Ueno T, editors. Liver Diseases and Hepatic Sinusoidal Cells. Springer; Tokyo, Japan: 1999. pp. 17–53. [Google Scholar]
- 38.Bilzer M, Roggel F, Gerbes AL. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006;26:1175. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 39.Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 40.Malarkey DE, Johnson K, Ryan L, Boorman G, Maronpot RR. New insights into functional aspects of liver morphology. Toxicol Pathol. 2005;33:27. doi: 10.1080/01926230590881826. [DOI] [PubMed] [Google Scholar]
- 41.Buckberg G, Hoffman JIE, Mahajan A, Saleh S, Coghlan C. Cardiac mechanics revisited: the relationship of cardiac architecture to ventricular function. Circulation. 2008;118:2571. doi: 10.1161/CIRCULATIONAHA.107.754424. [DOI] [PubMed] [Google Scholar]
- 42.Young AA, Cowan BR. Evaluation of left ventricular torsion by cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2012;14:49. doi: 10.1186/1532-429X-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poveda F, Gil D, Martí E, Andaluz A, Ballester M, Carreras F. Helical structure of the cardiac ventricular anatomy assessed by diffusion tensor magnetic resonance imaging with multiresolution tractography. Rev Española Cardiol Engl Ed. 2013;66:782. doi: 10.1016/j.rec.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 44.Kocica MJ, Corno AF, Carreras-Costa F, Ballester-Rodes M, Moghbel MC, Cueva CNC, Lackovic V, Kanjuh VI, Torrent-Guasp F. The helical ventricular myocardial band: global, three-dimensional, functional architecture of the ventricular myocardium. Eur J Cardio-Thorac Surg. 2006;29:S21. doi: 10.1016/j.ejcts.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 45.Korecky B, Hai CM, Rakusan K. Functional capillary density in normal and transplanted rat hearts. Can J Physiol Pharmacol. 1982;60:23. doi: 10.1139/y82-003. [DOI] [PubMed] [Google Scholar]
- 46.Cheung DY, Duan B, Butcher JT. Current progress in tissue engineering of heart valves: multiscale problems, multiscale solutions. Expert Opin Biol Ther. 2015;15:1155. doi: 10.1517/14712598.2015.1051527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoen FJ, Levy RJ. Tissue heart valves: current challenges and future research perspectives. J Biomed Mater Res. 1999;47:439. doi: 10.1002/(sici)1097-4636(19991215)47:4<439::aid-jbm1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 48.McNulty W, Usmani OS. Techniques of assessing small airways dysfunction. Eur Clin Respir J. 2014;1:25898. doi: 10.3402/ecrj.v1.25898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsia CCW, Hyde DM, Weibel ER. Lung structure and the intrinsic challenges of gas exchange. Compr Physiol. 2016;6:827. doi: 10.1002/cphy.c150028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mercer RR, Russell ML, Crapo JD. Alveolar septal structure in different species. J Appl Physiol. 1994;77:1060. doi: 10.1152/jappl.1994.77.3.1060. [DOI] [PubMed] [Google Scholar]
- 51.Itoh H, Nishino M, Hatabu H. Architecture of the lung: morphology and function. J Thorac Imaging. 2004;19:221. doi: 10.1097/01.rti.0000142835.06988.b0. [DOI] [PubMed] [Google Scholar]
- 52.Weibel ER, Knight BW. A morphometric study on the thickness of the pulmonary air-blood barrier. J Cell Biol. 1964;21:367. doi: 10.1083/jcb.21.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West JB. Comparative physiology of the pulmonary blood-gas barrier: the unique avian solution. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1625. doi: 10.1152/ajpregu.00459.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts MA, Tran D, Coulombe KLK, Razumova M, Regnier M, Murry CE, Zheng Y. Stromal cells in dense collagen promote cardiomyocyte and microvascular patterning in engineered human heart tissue. Tissue Eng A. 2016;22:633. doi: 10.1089/ten.tea.2015.0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scarritt ME, Pashos NC, Bunnell BA. A review of cellularization strategies for tissue engineering of whole organs. Front Bioeng Biotechnol. 2015;3:43. doi: 10.3389/fbioe.2015.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 59.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 60.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, Herzog E, Niklason LE. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song JJ, Guyette JP, Gilpin SE, Gonzalez G, Vacanti JP, Ott HC. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19:646. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang YS, Dokmeci MR, Khademhosseini A. Microfluidic techniques for development of 3D vascularized tissue. Biomaterials. 2014;35:7308. doi: 10.1016/j.biomaterials.2014.04.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 64.Keane TJ, Swinehart IT, Badylak SF. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods. 2015;84:25. doi: 10.1016/j.ymeth.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 65.Nakayama KH, Batchelder CA, Lee CI, Tarantal AF. Decellularized rhesus monkey kidney as a Three-Dimensional Scaffold for Renal Tissue Engineering. Tissue Eng A. 2010;16:2207. doi: 10.1089/ten.tea.2009.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Q, Bao J, Zhou Y, Wang Y, Du Z, Shi Y, Li L, Bu H. Optimizing perfusion-decellularization methods of porcine livers for clinical-scale whole-organ bioengineering. Biomed Res Int. 2015;2015:785474. doi: 10.1155/2015/785474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods Mol Biol. 2013;997:23. doi: 10.1007/978-1-62703-348-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2016;16:115. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren X, Moser PT, Gilpin SE, Okamoto T, Wu T, Tapias LF, Mercier FE, Xiong L, Ghawi R, Scadden DT, Mathisen DJ, Ott HC. Engineering pulmonary vasculature in decellularized rat and human lungs. Nat Biotechnol. 2015;33:1097. doi: 10.1038/nbt.3354. [DOI] [PubMed] [Google Scholar]
- 71.Huang SXL, Islam MN, O’Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, Snoeck HW. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. 2013;32:84. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu TY, Lin B, Kim J, Sullivan M, Tobita K, Salama G, Yang L. Repopulation of decellularized mouse heart with human induced pluripotent stem cell-derived cardiovascular progenitor cells. Nat Commun. 2013;4:2307. doi: 10.1038/ncomms3307. [DOI] [PubMed] [Google Scholar]
- 73.Sutherland ML, Fabre KM, Tagle DA. The National Institutes of Health Microphysiological Systems Program focuses on a critical challenge in the drug discovery pipeline. Stem Cell Res Ther. 2013;4(Suppl 1):I1. doi: 10.1186/scrt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stokes CL, Cirit M, Lauffenburger DA. Physiome-on-a-chip: the challenge of ‘scaling’ in design, operation, and translation of microphysiological systems. CPT Pharmacometrics Syst Pharmacol. 2015;4:559. doi: 10.1002/psp4.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marx U, Andersson TB, Bahinski A, Beilmann M, Beken S, Cassee FR, Cirit M, Daneshian M, Fitzpatrick S, Frey O, Gaertner C, Giese C, Griffith L, Hartung T, Heringa MB, Hoeng J, de Jong WH, Kojima H, Kuehnl J, Leist M, Luch A, Maschmeyer I, Sakharov D, Sips AJAM, Steger-Hartmann T, Tagle DA, Tonevitsky A, Tralau T, Tsyb S, van de Stolpe A, Vandebriel R, Vulto P, Wang J, Wiest J, Rodenburg M, Roth A. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. ALTEX. 2016;33:272. doi: 10.14573/altex.1603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep. 2017;7:42296. doi: 10.1038/srep42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol Bioeng. 2016;113:2213. doi: 10.1002/bit.25989. [DOI] [PubMed] [Google Scholar]
- 78.Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat Protoc. 2010;5:491. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 79.Sia SK, Whitesides GM. Microfluidic devices fabricated in Poly (dimethylsiloxane) for biological studies. Electrophoresis. 2003;24:3563. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 80.Berthier E, Young EWK, Beebe D. Engineers are from PDMS-land, Biologists are from Polystyrenia. Lab Chip. 2012;12:1224. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- 81.Xia Y, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153. [Google Scholar]
- 82.Song JW, Gu W, Futai N, Warner KA, Nor JE, Takayama S. Computer-controlled microcirculatory support system for endothelial cell culture and shearing. Anal Chem. 2005;77:3993. doi: 10.1021/ac050131o. [DOI] [PubMed] [Google Scholar]
- 83.Zheng C, Zhang X, Li C, Pang Y, Huang Y. Microfluidic device for studying controllable hydrodynamic flow induced cellular responses. Anal Chem. 2017;89:3710. doi: 10.1021/acs.analchem.7b00013. [DOI] [PubMed] [Google Scholar]
- 84.Smith Q, Gerecht S. Going with the flow: microfluidic platforms in vascular tissue engineering. Curr Opin Chem Eng. 2014;3:42. doi: 10.1016/j.coche.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee SA, Chung SE, Park W, Lee SH, Kwon S. Three-dimensional fabrication of heterogeneous microstructures using soft membrane deformation and optofluidic maskless lithography. Lab Chip. 2009;9:1670. doi: 10.1039/b819999j. [DOI] [PubMed] [Google Scholar]
- 86.Chueh B, Huh D, Kyrtsos CR, Houssin T, Futai N, Takayama S. Leakage-free bonding of porous membranes into layered microfluidic array systems. Anal Chem. 2007;79:3504. doi: 10.1021/ac062118p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kane R. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 89.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 90.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol. 2013;5:1119. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 92.Kim S, LesherPerez SC, Kim BC, Yamanishi C, Labuz JM, Leung B, Takayama S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication. 2016;8:15021. doi: 10.1088/1758-5090/8/1/015021. [DOI] [PubMed] [Google Scholar]
- 93.Gori M, Simonelli MC, Giannitelli SM, Businaro L, Trombetta M, Rainer A. Investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. PLoS ONE. 2016;11:e0159729. doi: 10.1371/journal.pone.0159729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, Alves SE, Salmon M, Ferrante TC, Weaver JC, Bahinski A, Hamilton GA, Ingber DE. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2015;13:151. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 95.Lee PJ, Hung PJ, Lee LP. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol Bioeng. 2007;97:1340. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 96.Legendre A, Baudoin R, Alberto G, Paullier P, Naudot M, Bricks T, Brocheton J, Jacques S, Cotton J, Leclerc E. Metabolic characterization of primary rat hepatocytes cultivated in parallel microfluidic bio-chips. J Pharm Sci. 2013;102:3264. doi: 10.1002/jps.23466. [DOI] [PubMed] [Google Scholar]
- 97.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A. 2010;107:565. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tanaka Y, Sato K, Shimizu T, Yamato M, Okano T, Kitamori T, Healy KE, Folch A, Okano T. A micro-spherical heart pump powered by cultured cardiomyocytes. Lab Chip. 2007;7:207. doi: 10.1039/b612082b. [DOI] [PubMed] [Google Scholar]
- 99.Carson D, Hnilova M, Yang X, Nemeth CL, Tsui JH, Smith AST, Jiao A, Regnier M, Murry CE, Tamerler C, Kim DH. Nanotopography-induced structural anisotropy and sarcomere development in human cardiomyocytes derived from induced pluripotent stem cells. ACS Appl Mater Interfaces. 2016;8:21923. doi: 10.1021/acsami.5b11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Macadangdang J, Guan X, Smith AST, Lucero R, Czerniecki S, Childers MK, Mack DL, Kim DH. Nanopatterned human iPSC-based model of a dystrophin-null cardiomyopathic phenotype. Cell Mol Bioeng. 2015;8:320. doi: 10.1007/s12195-015-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Hydrogels in regenerative medicine. Adv Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 103.O’Brien FJ. Biomaterials & scaffolds for tissue engineering. Mater Today. 2011;14:88. [Google Scholar]
- 104.Tien J. Microfluidic approaches for engineering vasculature. Curr Opin Chem Eng. 2014;3:36. [Google Scholar]
- 105.Chrobak KM, Potter DR, Tien J. Formation of perfused, functional microvascular tubes in vitro. Microvasc Res. 2006;71:185. doi: 10.1016/j.mvr.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 106.Wong KHK, Truslow JG, Khankhel AH, Chan KLS, Tien J. Artificial lymphatic drainage systems for vascularized microfluidic scaffolds. J Biomed Mater Res A. 2013;101A:2181. doi: 10.1002/jbm.a.34524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Golden AP, Tien J. Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab Chip. 2007;7:720. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- 108.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huling J, Ko IK, Atala A, Yoo JJ. Fabrication of biomimetic vascular scaffolds for 3D tissue constructs using vascular corrosion casts. Acta Biomater. 2016;32:190. doi: 10.1016/j.actbio.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 110.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci U S A. 2012;109:9342. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang B, Montgomery M, Chamberlain MD, Ogawa S, Korolj A, Pahnke A, Wells LA, Massé S, Kim J, Reis L, Momen A, Nunes SS, Wheeler AR, Nanthakumar K, Keller G, Sefton MV, Radisic M. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat Mater. 2016;15:669. doi: 10.1038/nmat4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee EJ, Kim DE, Azeloglu EU, Costa KD. Engineered cardiac organoid chambers: toward a functional biological model ventricle. Tissue Eng A. 2008;14:215. doi: 10.1089/tea.2007.0351. [DOI] [PubMed] [Google Scholar]
- 113.Bajaj P, Schweller RM, Khademhosseini A, West JL, Bashir R. 3D biofabrication strategies for tissue engineering and regenerative medicine. Annu Rev Biomed Eng. 2014;16:247. doi: 10.1146/annurev-bioeng-071813-105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Ying Shun T, Gough A, Taylor DL. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med. 2016;241:101. doi: 10.1177/1535370215592121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ligresti G, Nagao RJ, Xue J, Choi YJ, Xu J, Ren S, Aburatani T, Anderson SK, MacDonald JW, Bammler TK, Schwartz SM, Muczynski KA, Duffield JS, Himmelfarb J, Zheng Y. A Novel three-dimensional human peritubular microvascular system. J Am Soc Nephrol. 2016;27:2370. doi: 10.1681/ASN.2015070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chaturvedi RR, Stevens KR, Solorzano RD, Schwartz RE, Eyckmans J, Baranski JD, Stapleton SC, Bhatia SN, Chen CS. Patterning vascular networks in vivo for tissue engineering applications. Tissue Eng C Methods. 2015;21:509. doi: 10.1089/ten.tec.2014.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mihic A, Li J, Miyagi Y, Gagliardi M, Li SH, Zu J, Weisel RD, Keller G, Li RK. The effect of cyclic stretch on maturation and 3D tissue formation of human embryonic stem cell-derived cardiomyocytes. Biomaterials. 2014;35:2798. doi: 10.1016/j.biomaterials.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 118.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Murphy SV, Skardal A, Atala A. Evaluation of hydrogels for bio-printing applications. J Biomed Mater Res A. 2013;101A:272. doi: 10.1002/jbm.a.34326. [DOI] [PubMed] [Google Scholar]
- 120.Skardal A, Atala A. Biomaterials for Integration with 3-D bioprinting. Ann Biomed Eng. 2014;43:730. doi: 10.1007/s10439-014-1207-1. [DOI] [PubMed] [Google Scholar]
- 121.Pati F, Jang J, Ha D-H, Won Kim S, Rhie J-W, Shim J-H, Kim D-H, Cho D-W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat Commun. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.He Y, Yang F, Zhao H, Gao Q, Xia B, Fu J. Research on the printability of hydrogels in 3D bioprinting. Sci Rep. 2016;6:29977. doi: 10.1038/srep29977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahmed EM. Hydrogel: preparation, characterization, and applications: a review. J Adv Res. 2015;6:105. doi: 10.1016/j.jare.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rutz AL, Hyland KE, Jakus AE, Burghardt WR, Shah RN. A multimaterial bioink method for 3D printing tunable, cell-compatible hydrogels. Adv Mater. 2015;27:1607. doi: 10.1002/adma.201405076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Skardal A, Devarasetty M, Kang HW, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo J, Soker S, Atala A. A hydrogel bioink tool-kit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater. 2015;25:24. doi: 10.1016/j.actbio.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 126.Mandrycky C, Wang Z, Kim K, Kim DH. 3D bioprinting for engineering complex tissues. Biotechnol Adv. 2016;34:422. doi: 10.1016/j.biotechadv.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ozbolat IT, Hospodiuk M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials. 2016;76:321. doi: 10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 128.Raman R, Bashir R. Chapter 6—stereolithographic 3D bioprinting for biomedical applications. In: Atala A, Yoo JJ, editors. Essentials of 3D Biofabrication and Translation. Academic Press; 2015. pp. 89–121. [Google Scholar]
- 129.Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci Rep. 2016;6:34845. doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng GS, Sheikh F, Chien S, Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A. 2016;113:2206. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Horváth L, Umehara Y, Jud C, Blank F, Petri-Fink A, Rothen-Rutishauser B. Engineering an in vitro air-blood barrier by 3D bioprinting. Sci Rep. 2015;5:7974. doi: 10.1038/srep07974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Faulkner-Jones A, Fyfe C, Cornelissen DJ, Gardner J, King J, Courtney A, Shu W. Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D. Biofabrication. 2015;7:44102. doi: 10.1088/1758-5090/7/4/044102. [DOI] [PubMed] [Google Scholar]
- 133.Nguyen DG, Funk J, Robbins JB, Crogan-Grundy C, Presnell SC, Singer T, Roth AB. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS ONE. 2016;11:e0158674. doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Shrike Zhang Y, Shin SR, Calzone G, Annabi N, Shupe TD, Bishop CE, Atala A, Dokmeci MR, Khademhosseini A. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8:14101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 135.Gaetani R, Doevendans PA, Metz CHG, Alblas J, Messina E, Giacomello A, Sluijter JPG. Cardiac tissue engineering using tissue printing technology and human cardiac progenitor cells. Biomaterials. 2012;33:1782. doi: 10.1016/j.biomaterials.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 136.Gaetani R, Feyen DAM, Verhage V, Slaats R, Messina E, Christman KL, Giacomello A, Doevendans PAFM, Sluijter JPG. Epicardial application of cardiac progenitor cells in a 3D-printed gelatin/hyaluronic acid patch preserves cardiac function after myocardial infarction. Biomaterials. 2015;61:339. doi: 10.1016/j.biomaterials.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 137.Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016;110:45. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 139.Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol. 2015;33:1193. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Vander Werff R, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RRG, Grompe M, Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by WNT-driven regeneration. Nature. 2013;494:247. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 143.Khademhosseini A, Eng G, Yeh J, Kucharczyk PA, Langer R, Vunjak-Novakovic G, Radisic M. Microfluidic patterning for fabrication of contractile cardiac organoids. Biomed Microdevices. 2007;9:149. doi: 10.1007/s10544-006-9013-7. [DOI] [PubMed] [Google Scholar]
- 144.Iyer RK, Chui J, Radisic M. Spatiotemporal tracking of cells in tissue-engineered cardiac organoids. J Tissue Eng Regen Med. 2009;3:196. doi: 10.1002/term.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Voges HK, Mills RJ, Elliott DA, Parton RG, Porrello ER, Hudson JE. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development. 2017;144:1118. doi: 10.1242/dev.143966. [DOI] [PubMed] [Google Scholar]
- 146.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR, Shroyer N, Wells J, Helmrath M, Kotton D, Elefanty A, Stanley E, Chen Q, Prabhakar S, Weissman I, Lim B. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4:327. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dye BR, Dedhia PH, Miller AJ, Nagy MS, White ES, Shea LD, Spence JR, Ellis J, Rossant J, Sun Y, Grabowski G, Finkbeiner S, Spence J, Shroyer N, Wells J, Helmrath M, Mense M, Rowe S, Engelhardt J, Hsu Y, Rajagopal J. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. Elife. 2016;5:876. doi: 10.7554/eLife.19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Takahashi H, Nakayama M, Shimizu T, Yamato M, Okano T. Anisotropic cell sheets for constructing three-dimensional tissue with well-organized cell orientation. Biomaterials. 2011;32:8830. doi: 10.1016/j.biomaterials.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 149.Jiao A, Trosper NE, Yang HS, Kim J, Tsui JH, Frankel SD, Murry CE, Kim DH. Thermoresponsive nanofabricated substratum for the engineering of three-dimensional tissues with layer-by-layer architectural control. ACS Nano. 2014;8:4430. doi: 10.1021/nn4063962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gupta R, Van Rooijen N, Sefton MV. Fate of endothelialized modular constructs implanted in an omental pouch in nude rats. Tissue Eng A. 2009;15:2875. doi: 10.1089/ten.tea.2008.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat Rev Genet. 2014;15:82. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 154.Briquez PS, Clegg LE, Martino MM, Mac Gabhann F, Hubbell JA. Design principles for therapeutic angiogenic materials. Nat Rev Mater. 2016;1:15006. [Google Scholar]
- 155.Asti A, Gioglio L. Natural and synthetic biodegradable polymers: different scaffolds for cell expansion and tissue formation. Int J Artif Organs. 2014;37:187. doi: 10.530/ijao.5000307. [DOI] [PubMed] [Google Scholar]
- 156.Gilpin A, Yang Y. Decellularization strategies for regenerative medicine: from processing techniques to applications. Biomed Res Int. 2017;2017:1. doi: 10.1155/2017/9831534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Saldin LT, Cramer MC, Velankar SS, White LJ, Badylak SF. Extracellular matrix hydrogels from decellularized tissues: structure and function. Acta Biomater. 2017;49:1. doi: 10.1016/j.actbio.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]