Abstract
Due to its simplicity, versatility, and high efficiency, the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technology has emerged as one of the most promising approaches for treatment of a variety of genetic diseases, including human cancers. However, further translation of CRISPR/Cas9 for cancer gene therapy requires development of safe approaches for efficient, highly specific delivery of both Cas9 and single guide RNA to tumors. Here, novel core–shell nanostructure, liposome-templated hydrogel nanoparticles (LHNPs) that are optimized for efficient codelivery of Cas9 protein and nucleic acids is reported. It is demonstrated that, when coupled with the minicircle DNA technology, LHNPs deliver CRISPR/Cas9 with efficiency greater than commercial agent Lipofectamine 2000 in cell culture and can be engineered for targeted inhibition of genes in tumors, including tumors the brain. When CRISPR/Cas9 targeting a model therapeutic gene, polo-like kinase 1 (PLK1), is delivered, LHNPs effectively inhibit tumor growth and improve tumor-bearing mouse survival. The results suggest LHNPs as versatile CRISPR/Cas9-delivery tool that can be adapted for experimentally studying the biology of cancer as well as for clinically translating cancer gene therapy.
Keywords: brain cancer, CRISPR/Cas9, gene therapy, liposomes, nanogels
1. Introduction
The clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein 9 (CRISPR/Cas9) nuclease system has recently evolved as the most promising genome editing technology. The system uses an engineered single guide RNA (sgRNA) to direct the Cas9 nuclease to complementary regions, where Cas9 cleaves the recognized DNA and generates double-stranded breaks (DSBs), leading to insertions or deletions at specific target genomic loci.[1] The CRISPR/Cas9 technology, which has been explored for a variety of biological applications, such as transcription regulation,[2] and genome editing,[1b] has great potential for cancer gene therapy. Progression of many cancers is driven by activated oncogenes. For these cancers, effective treatment can be potentially achieved through suppression of the activated oncogenes, which has been traditionally accomplished using RNA interference (RNAi) technology. However, the RNAi approach suffers from a major limitation in that it targets mRNAs and thus, produces a transient genetic regulation but leaves the original copy of the oncogene intact. Unlike the RNAi approach, CRISPR technology induces genetic knockout at the genomic DNA level for permanent elimination of targeted genes. Consequently, delivery of Cas9/sgRNA-based gene therapy makes it possible to produce a persistent therapeutic benefit.
Despite its potential, translation of the Cas9/sgRNA technology for cancer gene therapy is contingent upon the development of a delivery system that can efficiently and safely target tumor cells.[3] For delivery of Cas9/sgRNA to tumors in the brain, further engineering is required for facilitating penetration of the blood-brain barrier (BBB).[4] Currently, CRISPR/Cas9 is often delivered using viral vectors, such as adenoassociated virus.[5] The viral approaches have a major advantage because of its high transduction efficiency. Unfortunately, clinical translation of the viral approaches has been challenging due to safety concerns.[6] To overcome these limitations, delivery of CRISPR/Cas9 using nonviral vectors is a promising alternative.[7] Recently, several nonviral approaches have been explored for delivery of CRISPR/Cas9, including hydrodynamic injection,[8] cell penetrating peptides,[9] and synthetic nanoparticles.[10] Among them, some demonstrated excellent efficiency in delivery of CRISPR/Cas9 in vivo; however, none were able to penetrate the BBB.
In this study, we designed and synthesized liposome-templated hydrogel nanoparticles (LHNPs) for efficient codelivery of protein and nucleic acids. By using polo-like kinase 1 (PLK1) as a model gene, we demonstrated that LHNPs were capable of efficient delivery of CRISPR/Cas9 for effective cancer therapy in a mouse flank tumor model. We further showed that the LHNPs can be engineered through an autocatalytic brain tumor-targeting (ABTT) mechanism, which we recently developed for drug delivery to tumors in the brain,[11] for targeted delivery of CRISPR/Cas9 to tumors in the brain. Our results suggest that LHNPs are capable of targeted delivery of CRISPR/Cas9 for cancer gene therapy.
2. Results and Discussion
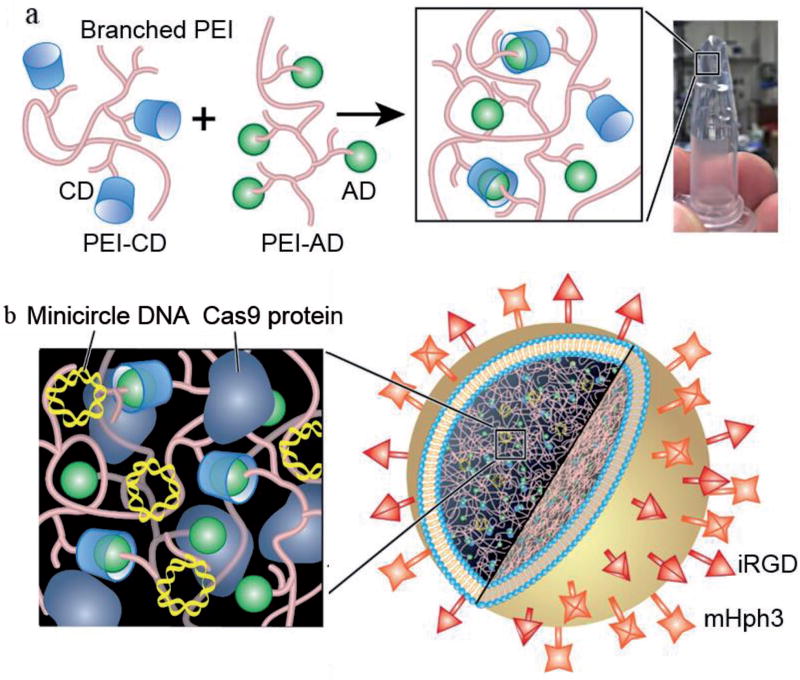
CRISPR/Cas9 is a two-component system, which includes Cas9 and gRNA functioning at the protein level and RNA level, respectively. Although Cas9 can be delivered in the form of DNA, mRNA, or protein, previous studies suggest that Cas9 delivered in the form of protein maximizes the efficiency and reduces the off-target effects.[12] Delivery of Cas9 as protein is also advantageous because constitutive expression of Cas9, which may induce undesired side effects,[13] is avoided. To establish a system for simultaneous delivery of protein and nucleic acids, we designed and synthesized LHNPs (Figure 1). The core of LHNPs was formed by polyethylenimine (PEI) hydrogel that was designed for encapsulation of Cas9 protein. The shell consisted of cationic 1,2-dioleoyl-3-trimethylammonium-propane chloride salt (DOTAP) lipids that were selected for efficient delivery of genetic materials (Figure 1a). Molecular structures of major chemicals used for LHNP synthesis were showed in Figure S1 in the Supporting Information. Schematic diagram of LHNP synthesis was included in Figure S2 in the Supporting Information. The resulting LHNPs were capable of efficient encapsulation of both Cas9 protein and sgRNA with high efficiency (Figure 1b).
Figure 1.
Fabrication of LHNPs. a) Formation of PEI hydrogel through CD-AD mediated host–guest interaction. b) Schematics of LHNPs. Core and shell of LHNPs are formed by DOTAP liposomes and PEI hydrogel, respectively.
2.1. Rational Design of LHNPs for Codelivery of Cas9 Protein and Nucleic Acids
DOTAP liposome is a classical gene delivery nanocarrier which has demonstrated high safety favorable for clinical use.[14] However, it is limited by its low encapsulation of Cas9 protein, with an encapsulation efficiency of 6.3% (Figure 2a). To improve its encapsulation efficiency, we used DOTAP liposomes as a template. Hydrogel was then formed by crosslinking cyclodextrin (CD)-engrafted PEI (25 kDa) (PEI-CD) with adamantine (AD)-engrafted PEI (25 kDa) (PEI-AD) through CD-AD mediated host–guest interaction (Figure 1a). Conjugation of CD and AD to PEI was confirmed by 1H NMR (Figure S3, Supporting Information). PEI-based hydrogel was selected for two reasons. First, PEI-CD and PEI-AD were able to noncovalently crosslink without further addition of initiators or/and ultraviolet irradiation, a condition which is often required for hydrogel synthesis but harmful for protein activity. Second, PEI is a well-characterized gene delivery agent and can enhance the encapsulation and delivery of nucleic acids. Rheology test revealed that the resulting hydrogel has a mechanically soft structure (Figure S4, Supporting Information), which is favorable for maintenance of protein activity.[15] We found that with the inclusion of hydrogel, the encapsulation efficiency of Cas9 increased to 62.8% (Figure 2a).
Figure 2.
Synthesis of LHNPs for codelivery of protein and DNA. a) Cas9 encapsulation efficiency of DOTAP liposomes with and without hydrogel-core. b) Gene delivery efficiency of pGL4.13-loaded, PEI hydrogel-core DOTAP liposomes and Lipofectamine 2000 (Lip2k) on U87 cells. Experiments were carried out in triplicate and the standard deviation is denoted using error bars. Luciferase signal was detected at 72 h after transfection and normalized to the amount of total protein for comparison. * and ** represent p < 0.05 and 0.005, respectively. c) A representative TEM image of LHNPs. Scale bar: 500 nm. d) Controlled release of Cas9 and sgRNA DNA from LHNPs. e) Cytotoxicity of LHNPs and Lip2k on U87 cells when the same amount of DNA was delivered. Cytotoxicity studies were carried out in U87 cells plated in a 96-well plate. For each well, 60 ng DNA was used.
We evaluated the hydrogel-core DOTAP liposomes for gene delivery in human brain cancer U87 cells. By using the expression of luciferase gene as a reporter, we found that transfection of U87 cells was 4.3 times lower compared to lipofectamine 2000 (Lip2k) (Figure 2b). To enhance their gene delivery ability, we tested two ligands, mHph1 and mHph3, by conjugating them to the surface of liposomes. mHph1 is a cell penetration peptide that we recently developed for enhancing the gene delivery ability of nanocarriers.[11,16] mHph3 is a modified form of mHph1 consisting of mHph1 fused with a CM18 fragment.[17] We found that the resulting mHph3-conjugated nanoparticles exhibited the greatest efficiency and delivered luciferase gene 1.3 times more efficiently than Lip2k (p < 0.05) (Figure 2b). To simplify the nomenclature, we designated the mHph3-conjugated, DOTAP liposome-templated hydrogel nanoparticles as LHNPs.
Transmission electron microscopy (TEM) revealed that LHNPs are spherical and have a diameter of 95 nm (Figure 2c). LHNPs released 91.5% of DNA and 85.2% of protein over 3 d in a controlled manner (Figure 2d). The release of DNA could be potentially further extended through encapsulation of DNA within LHNPs (Figure S5, Supporting Information). Flow cytometry analysis showed that LNHPs could be uptaked by U87 cells with efficiency up to 100% (Figure S6, Supporting Information). Confocal microscopy confirmed that LHNPs efficiently delivered Cas9 protein to cells and transfected cells with high efficiency after encapsulation of green fluorescent protein (GFP) (Figure S7, Supporting Information). Additionally, LHNPs have a favorable toxicity profile. When the same amount of DNA was delivered, LHNPs exhibited significantly lower toxicity than Lip2K (Figure 2e). We found that LHNPs could be synthesized with high reproducibility (Table S1, Supporting Information).
2.2. Design and Selection of gRNAs for Efficient Inhibition of PLK1
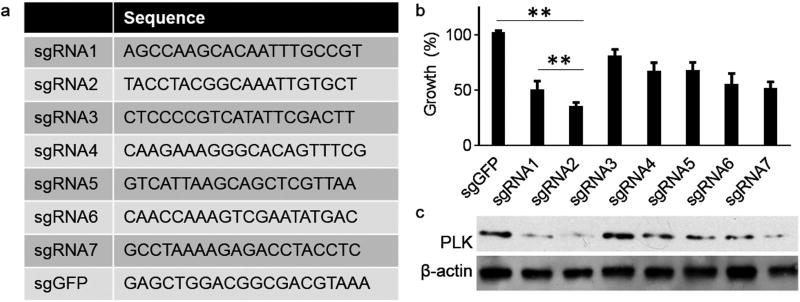
In a recent report, we demonstrated that targeted delivery of siRNA targeting PLK1 using octa-functional poly(lactic-co-glycolic acid) nanoparticles significantly inhibited cancer progression.[16] To assess if the CRISPR/Cas9-based gene therapy can produce a comparable efficacy, we selected PLK1 as a model gene. In order to identify sgRNAs for efficient inhibition of PLK1, we analyzed the genomic sequence of PLK1 and identified seven sgRNAs, sgPLK1-1–7, with high scores (Figure 3a). Candidate sgRNAs were synthesized and cloned into lentiGuide-Puro, a vector for sgRNA expression.[18] Next, we evaluated the impact of the candidate sgRNAs on U87 cell growth and use a sgRNA targeting GFP (sgGFP) as a control. sgRNAs were delivered through transfection by Lip2k to Cas9-expressing U87 cells, which were generated through transduction with lentiCas9-Blast lentivirus.[18] After 4 d, cell growth was determined by the standard 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Results in Figure 3b showed that several sgRNAs, including sgRNA1-1, sgRNA1-2, and sgRNA1-7, significantly inhibited U87 cell proliferation. Among them, sgRNA1-2 demonstrated the greatest efficiency and inhibited cell growth by 64.0%. Consistent with the growth results, Western Blot analysis revealed that sgPLK1-2 inhibited the expression of PLK1 with the highest efficiency (Figure 3c). Surveyor assay confirmed that Cas9/sgPLK1-2 inhibited PLK1 through genome editing (Figure S8, Supporting Information). Therefore, sgPLK1-2 was selected for the remainder of the study.
Figure 3.
Design and selection of gRNAs for efficient inhibition of PLK1. a) Sequences of candidate PLK1-targeting sgRNAs and control GFP-targeting sgRNA. b,c) Evaluation of candidate sgRNAs on U87 cell proliferation (b, MTT assay) and PLK1 expression (c, Western Blot).
2.3. Optimization of the Delivery of CRISPR/Cas9 via LHNPs
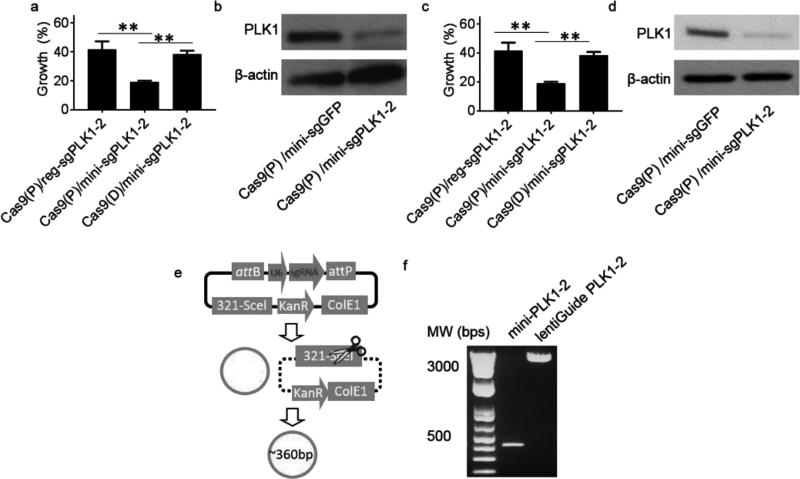
We synthesized LHNPs that encapsulated Cas9 protein and sgPLK1-2 in the regular lentiGuide vector, designated as Cas9(P)/reg-sgPLK1-2, and evaluated them in U87 cells and GS5 cells, a human brain cancer stem cell line.[19] Treatment with Cas9(P)/reg-sgPLK1-2- encapsulated LHNPs decreased cell proliferation by 52.8% and 58.3% for U87 cells and GS5 cells, respectively (Figure 4a,c). Compared to 64.0% for U87 cells in the study described in Figure 3b, the efficiency of Cas9(P)/reg-sgPLK1-2-encapsulated LHNPs is relatively low. To enhance the efficiency, we utilized minicircle DNA technology[20] and replaced the sgPLK1-2 in lentiGuide with minicircle sgPLK1-2. The sgPLK1-2 expression cassette in the lentiGuide vector was subcloned into pMC.BESPX, a minicircle DNA expression system,[20] though which we produced 364 bp minicircle-sgPLK1-2, which contains a U6 promoter (249 bp), targeted sequence (20 bp), and chimeric guide RNA scaffold (75 bp) (Figure 4e). The quality of minicircle-sgPLK1-2 was confirmed by gel analysis (Figure 4f). We synthesized and evaluated LHNPs with encapsulation of Cas9 protein and minicircle-sgPLK1-2, designated as Cas9(P)/mini-sgPLK1-2, and found that the resulting Cas9(P)/mini-sgPLK1-2- encapsulated LHNPs inhibited cell growth by 79.3% and 80.2% for U87 cells and GS5 cells, respectively (Figure 4a,c). We confirmed that the observed growth inhibition effects resulted from inhibition of PLK1 gene, as treatment with Cas9(P)/mini-sgPLK1-2- encapsulated LHNPs markedly inhibited the expression of PLK1 in U87 cells (Figure 4b,d). Inclusion of Cas9 in the form of protein is beneficial, as LHNPs encapsulated with Cas9 in the form of DNA in lentiCas9-Blast only inhibited the proliferation of U87 cells and GS5 cells by 61.7% and 61.6%, respectively (Figure 4a,c). Encapsulation of different forms of Cas9 or sgRNA did not significantly alter the morphology or the size of LHNPs (Figure S9, Supporting Information).
Figure 4.
Optimization of the delivery of CRISPR/Cas9 via LHNPs. a–d) Effects of LHNPs in the indicated formulation on cell proliferation (a: U87; c: GS5) and PLK1 expression (b: U87; d: GS5). Cas9(P): Cas9 in form of protein. Cas9(D): Cas9 in form of DNA in lentiCas9-Blast. e) Schematic diagram of minicircle sgRNA production. Detailed information about the construct and miniDNA production can be found in the reference by Kay et al.[20] f) Quality of the minicircle sgPLK1-2 determined by gel analysis.
2.4. Targeted Delivery of CRISPR/Cas9 for Cancer Gene Therapy
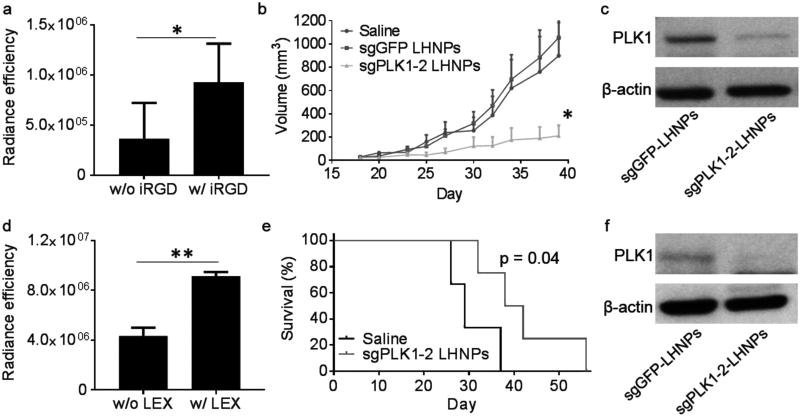
Delivery of nanoparticles to tumors can be improved through conjugation of cancer-targeting ligands, such as internalizing RGD (iRGD), a modified form of RGD (Arg-Gly-Asp) peptide with high affinity for αvβ3/αvβ5 integrins and neuropilin-1.[16,21] To assess if iRGD enhanced delivery of LHNPs to tumors, we synthesized LHNPs with and without conjugation of iRGD and evaluated them in mice bearing U87 flank tumors. LHNPs encapsulated IR780, a near-infrared fluorescent dye that allows for noninvasive detection using in vivo imaging system (IVIS). We found that conjugation of iRGD significantly enhanced the accumulation of LHNPs in tumors. Based on fluorescence intensity, the concentration of nanoparticles in tumors in mice treated with iRGD-conjguated LHNPs was 2.6 times greater than that of mice treated with LHNPs without modification (Figure 5a). The IVIS images were included in Figure S10a in the Supporting Information. Conjugation of iRGD did not change the gene delivery ability of LHNPs (Figure S10b, Supporting Information). We determined the in vivo uptake of LHNPs. Tumor-bearing mice were established through subcutaneous injection of GFP-labeled U87 cells. After tumor volumes reached ≈100 mm3, LHNPs encapsulated with Rodamine-B-labeled cas9 protein were administrated to mice daily for three consecutively days through tail vein injection. One day after the last injection, mice were euthanized. Tumors were isolated and digested. Flow cytometry analysis revealed that 33.1% of tumor cells uptaked LHNPs (Figure S6, Supporting Information). Due to the great tumor-targeting effect, we employed the iRGD-conjguated LHNPs for the following in vivo studies.
Figure 5.
Targeted delivery of CRISPR/Cas9-encapsulated LHNPs for cancer gene therapy. a) Semi-quantification of LHNPs with and without conjugation of iRGD in flank tumors based on fluorescence intensity. “Radiance efficiency” (p sec−1 cm−2 sr−1 uw−1 cm−2) on the y-axis was determined using Living Image 3.0 by dividing the fluorescence signal intensity with area of region of interest (ROI). All experiments were carried out in triplicate and the standard deviation is denoted using error bars. b) Changes of tumor volume versus time in mice that received intravenous administration of the indicated treatments (n =6). c) Western Blot analysis of the expression of PLK1 in residual tumors (n =3). d) Semi-quantification of iRGD-conjugated LHNPs with and without encapsulation of LEX in intracranial tumors based on fluorescence intensity. e) Kaplan–Meier survival curves for intracranial U87 tumor-bearing mice with indicated treatments. f) Western Blot analysis of the expression of PLK1 in residual tumors (n =3).
We evaluated the therapeutic benefit of targeted delivery of Cas9/minicircle-sgPLK1-2-loaded LHNPs in tumor-bearing mice. After the average tumor volume reached ≈50 mm3, mice were randomly assigned into three groups, which received treatment of Cas9/minicircle-sgPLK1-2-loaded LHNPs (designated as sgPLK1-2 LHNPs), Cas9/minicircle-sgGFP-loaded LHNPs (designated as sgGFP LHNPs), and phosphate-buffered saline (PBS), respectively. Mice were monitored for tumor size and body weight. Results in Figure 5b showed that intravenous administration of sgPLK1-2 LHNPs significantly inhibited tumor growth (p < 0.05). By the end of the study, the average tumor volume in the group receiving treatment of sgPLK1-2 LHNPs was 23.5% of that in the control groups receiving saline treatment. The tumor growth inhibtion was due to the inhibition of PLK1. Western Blot analysis of the residual tumors showed that the expression of PLK1 in the treatment group was 36.3% of those in the control groups (Figure 5c). No significant difference in weight was found among all three groups, suggesting that intravenous administration of LHNPs has limited systemic toxicity (Figure S10d, Supporting Information). Histochemical analysis by TUNEL staining identified a significant increase in the number of apoptotic cells after treatment with Cas9/sgPLK1. Additionally, H&E staining showed that tumors from animals in the treatment groups had significantly decreased cellular mass with a lower nuclear-cytoplasmic ratio than those from control treatments (Figure S10e, Supporting Information).
We next assessed if the LHNPs could be further engineered for targeted delivery of gene therapy to intracranial tumors through the ABTT mechanism that we recently developed.[11] LHNPs were modified through encapsulation of Lexiscan (LEX), a small molecule known to transiently enhance BBB permeability.[22] The resulting LHNPs, which included iRGD and LEX for tumor targeting and autocatalysis, respectively, were evaluated in mice bearing intracranial U87 tumors. We found that inclusion of LEX increased the accumulation of nanoparticles 2.1-fold, confirming the autocatalysis effect (Figure 5d). The IVIS images were included in Figure S10a in the Supporting Information. Encapsulation of LEX did not alter the gene delivery ability of LHNPs (Figure S10b, Supporting Information). Next, we evaluated the engineered LHNPs for delivery of CRISPR/Cas9 to intracranial tumors for gene therapy. Mice bearing intracranial U87 gliomas received intravenous administration of Cas9/mini-sgPLK1-2-encapsulated LHNPs three times a week for three consecutive weeks. Results in Figure 5e showed that LHNP treatment effectively enhanced tumor-bearing survival. The median survival time for mice receiving LHNP treatment was 40 d, which was significantly longer than that for mice receiving treatment with saline (29 d) (p < 0.05). Consistent with the survival data, we found that compared to the control group, the expression of PLK1 in the nanoparticle treatment group was inhibited by 60.4% (Figure 5f), suggesting that the observed survival benefit resulted from the inhibition of PLK1 in tumors.
2.5. Discussion
In this study, we developed LHNPs for targeted delivery CRISPR/Cas9 for cancer gene therapy. LHNPs consist of a core–shell structure, in which PEI-based hydrogel was formed within cationic liposomes. This unique structure allowed for simultaneous encapsulation of Cas9 and plasmid DNA with high efficiency. We found that LHNPs were capable of delivery of CRISPR/Cas9 and that the delivery efficiency was maximized when sgRNA, in form of minicircle DNA, was used. Minicircle sgRNA is superior to sgRNA in the regular vector due to two reasons. First, elimination of prokaryote cassettes in the plasmid backbone, which contains CpG islands, may reduce inflammation and enhance plasmid propagation in mammalian cells.[20,23] Second, the DNA loading capacity of LHNPs depends on the amount of charge on their surface. In the same condition, LHNPs encapsulated sgRNA in vector lentiGuide-Puro and minicircle sgRNA with comparable weights (Table S1, Supporting Information). However, compared to sgRNA in vector lentiGuide-Puro, minicircle sgRNA is over 20 times smaller in size (364 bps vs 8318 bps). Therefore, the use of minicircle sgRNA allows LHNPs to deliver over 20 times greater copies of sgRNA. We found that LHNPs encapsulated with Cas9 in form of DNA exhibited an efficiency significantly lower than LHNPs encapsulated with Cas9 in form of protein. This finding is consistent with previous findings that concluded codelivery of both Cas9 and sgRNA in DNA form is not optimal.[12] Finally, we demonstrated that the resulting LHNPs with coencapsulation of Cas9 protein and minicircle gRNA were capable of efficiently inhibiting target genes both in vitro and in vivo and could be employed for delivery of effective cancer gene therapy.
Compared to other nanoparticle approaches previously documented,[10a–g] LHNPs have two major advantages. First, LHNPs allow delivery of Cas9 in the form of protein, making it possible to maximize the efficiency and specificity of CRISPR/Cas9[12] and minimize the undesired side effects associated with constitutive activation of Cas9.[13] Delivery of Cas9 in the form of protein has been previously attempted by other groups using lipid nanoparticles,[10h] gold nanoparticles,[10f] and DNA nanoclews.[10g] Although all of them demonstrated the ability to knock out targeted genes in vitro, none of them were tested in animals, and, thus, it is unknown whether they can mediate efficient gene editing in vivo. Second, LHNPs can be engineered for efficient delivery of CRISPR/Cas9 to tumors in the brain, a major group of tumors for which novel therapies are in great demand.[4b,24] By contrast, none of the reported nanocarriers were shown to have comparable ability.
We are aware that, despite the demonstration of significant tumor-inhibition efficacy, the CRISPR/Cas9-mediated, PLK1-targeted gene therapy failed to eliminate tumors (Figure 5). This result highlights a limitation of CRISPR/Cas9-gene editing for cancer gene therapy, as it is unlikely that CRISPR/Cas9 efficiently edits targeted genes in all cells within bulk tumors. In this case, the remained, unkilled cells may lead to tumor relapse. The clinical benefit of LHNP- mediated CRISPR/Cas9 cancer therapy could be potentially maximized by employing it as a tumor relapse-prevention therapy following the standard clinical regimens. In this case, the standard regimens eliminate majority of tumor cells. After this, CRISPR/Cas9 is delivered to residual tumor cells in a targeted manner by LHNPs, leading to elimination of residual tumors and, hence, prevention of tumor relapse. We are also aware of the potential offtarget effects of CRISPR/Cas9, which may induce side effects. Recently, tremendous efforts have been made to enhance the targeting specificity of CRISPR/Cas9. These efforts have led to the development of a variety of improved CRISPR/Cas9 strategies, such as double nicking[25] and Fok1-dCas9.[26] We expect that the combination of the improved CRISPR/Cas9 strategies with targeted drug delivery by LHNPs will minimize the offtarget effects of CRISPR/Cas9 for cancer treatment.
3. Conclusion
In summary, we have synthesized novel LHNPs for efficient delivery of CRISPR/Cas9. Due to their high delivery efficiency, their capacity for targeted delivery to tumors within the brain, and their construction from biomaterials with known minimal toxicity, LHNPs have the potential to serve as a versatile tool to advance CRISPR/Cas9-based experimental cancer research and clinical translation of cancer gene therapy.
4. Experimental Section
Materials and Cell Culture
The human glioma cell line U87 was obtained from American Type Culture Collection and cultured in dulbecco’s modified eagle medium (DMEM) medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 units mL−1 penicillin, and 100 µg mL−1 streptomycin (Invitrogen) in a 37 °C incubator containing 5% CO2. Human brain cancer stem cell line GS5, which was previously characterized,[19] was maintained in Neurobasal A medium (Invitrogen) supplemented with B27 (Invitrogen), fibroblast growth factor-2 (20 ng mL−1, Peprotech), and epidermal growth factor (20 ng mL−1, Peprotech). Peptides were synthesized by AnaSpec. Cas9 protein was expressed and purified according to published procedures.[27] sgRNA oligos were synthesized at the Yale Keck facility. All chemicals were purchased from Sigma-Aldrich unless otherwise noted.
Synthesis of PEI-AD and PEI-CD
For synthesis of PEI-CD, branched polyethyleneimine (78 mg, M.W. 25 000, 0.0031 mmol, Sigma-Aldrich) was dissolved in ddH2O. Next, randomized mono-Tos-cyclodextrin (77 mg, M.W. 1230, 0.063 mmol, Trappsol) was added into the solution. The mixture was heated to 60 °C and stirred for 8 h. By the end of the reaction, the reactant was dialyzed with ddH2O to remove unreacted cyclodextrin. The resulting PEI-CD was collected after lyophilization. For synthesis of PEI-AD, branched polyethyleneimine (70 mg, M.W. 25 000, 0.0028 mmol, Sigma-Aldrich) was dissolved in chloroform, followed by adding 1-adamantyl isocyanate (5 mg, M.W. 177.24, 0.028 mmol). The mixture was heated to 70 °C and stirred for 6 h. After cooling down to room temperature, chloroform was removed by rotor evaporation. Raw product was redissolved in ddH2O. Unreacted admantyl isocyanate was removed by centrifugation at 4000 rpm for 10 min. The resulting PEI-AD was collected after lyophilization.
Fabrication of LHNPs
DOTAP (Avanti Polar Lipids), cholesterol (Avanti Polar Lipids), and 1,2-distearroyl-sn-glycero-3-phosphoethanolamine (DSPE)-polyethylene glycol (PEG)2000-maleimide (MAL) (Avanti Polar Lipids) were dissolved in chloroform/methanol mixed solution in a 2:1 ratio at a concentration of 14.3, 26.0, and 6.8 µmol mL−1, respectively. For a typical synthesis, 700 µL DOTAP solution, 400 µL cholesterol solution, and 40 µL DSPE-PEG2000-MAL were mixed in 7 mL glass vial by vortex. Solvents were removed by constant air flow. Next, the mixed lipids were resuspended in 1 mL aqueous solution containing 490 µL PEI-AD (10 mg mL−1 in ddH2O), 490 µL PEI-CD (10 mg mL−1 in ddH2O), and 20 µL Cas9 protein solution (2 mg mL−1 in PBS). Cycles of 30 s vortexing followed by 5 s idle sitting at room temperature were repeated five times. The resulting multilamellar liposomes were extruded five times through a 200 nm polycarbonate membrane (Whatman) and seven times through a 100 nm membrane using a Avanti Mini Extruder (Avanti Polar Lipids). Immediately after extrusion, the mixture was diluted into 35 mL ddH2O and subjected to centrifugation at 15 000 rpm for 30 min. Liposomes were then collected, washed with ddH2O twice, and resuspended in ddH2O at 6 mg mL−1. To load DNA, 1 mL Lipo-gel solution was incubated with 1.2 µg DNA and incubated on ice for 1 h. For conjugation of mHph3, 30 µg peptide was added into LHNPs suspension and reacted for 60 min. For conjugation of both mHph3 and iRGD, 30 µg mHph3 peptide was added into LHNPs suspension. After reaction for 30 min, 30 µg iRGD was added and reacted for additional 30 min. Unreacted peptides were removed by centrifugation at 15 000 rpm for 30 min. For encapsulation of LEX, the same synthesis procedures were used except that 0.1 mg LEX was added to the lipid mixture prior to solvent evaporation.
Characterization of Encapsulation Efficiency and Drug Release
For characterization of encapsulation efficiency, 1 mg LHNPs were lysed in 10 µL dimethyl sulfoxide (DMSO) and diluted in 90 µL PBS. The amounts of Cas9 protein and DNA were determined based on the standard bicinchoninic acid (BCA) assay and Pico-Green assay, respectively, using commercial kits (ThermoFisher Scientific). LHNPs without encapsulants were used as background. For characterization of drug release, LHNPs (3–5 mg) were suspended in 1 mL PBS (pH 7.4), and incubated at 37 °C with gentle shaking. At each sampling time, the LHNP suspension was centrifuged for 15 min at 15 000 rpm. The supernatant was removed for quantification of Cas9 protein or DNA and replaced with an equivalent volume of PBS for continued monitoring of release. Detection of Cas9 protein or DNA was conducted using the same BCA and Pico-Green assays described above.
TEM
After synthesis, LHNPs were applied to holey carbon-coated copper grids (Electron Microscopy Sciences) and dried by air flow. Images were captured using a TEM microscope (FEI Tecnai TF20 TEM).
In Vitro Gene Transfection
U87 cells were plated at a density of 4000 cells in 100 µL per well in 96-well plates. The plasmid encoding luciferase pGL4.13 (Promega) was used as the model gene to be encapsulated into LHNPs in different formulations for evaluating in vitro gene transfection. Control transfection using Lipofectamine 2000 (Lip2k, Invitrogen) was performed per the standard protocol described in the manufacturer’s manual. Briefly, Lip2k was mixed with DNA at a v/m ratio of 2.5. After incubation at room temperature for 20 min, the complexes were added to cells. For evaluation of LHNPs, free LNHPs were added to each well at 1 mg mL−1. 24 h later, medium containing Lip2k complexes or LHNPs was replaced with fresh cell culture medium. Luciferase assays were performed 72 h post-treatment when the cells were lysed with 100-µL-reporter lysis buffer (Promega). After a freeze–thaw cycle, the cell lysate was collected. After centrifugation at 15 000 rpm for 5 min, 20 µL of the supernatant was collected for luciferase assay using the Luciferase Assay Reagent (Promega) according to the standard protocol described in manufacturer’s manual. An additional 20 µL was collected and used to quantify protein concentration using a BCA protein assay kit (ThermoFisher Scientific). The luciferase signal was divided by the amount of total protein for normalization.
In Vitro Cytotoxicity Evaluation
Cells were plated at a density of 4000 cells in 100 µL per well in 96-well plates. After overnight incubation, cells were treated with LHNPs at 1 mg mL−1. After 72 h, cell proliferation was quantified using the standard dimethyl thiazolyl diphenyl tetrazolium salt (MTT) assay. Briefly, 10 mg mL−1 MTT in PBS was added to the cells resulting in a final MTT concentration of 1 mg mL−1. The cells were incubated for an additional 4 h at 37 °C. Afterward, the media were removed and 150 µL DMSO was added to each well to dissolve the formazen crystals. The absorption was measured at 570 nm using a BioTek Instrument ELx800 microplate reader.
Design and Selection of sgPLKs
sgRNAs targeting PLK1 was designed using a CRISPR Design tool developed by the Feng Zhang lab at massachusetts institute of technology (MIT) (http://crispr.mit.edu/). Evaluation of candidate sgRNAs was performed using U87 cells with stable overexpression of Cas9 protein, which were obtained through lentiviral transduction according to published procedures.[28] sgRNAs were delivered to cells by Lip2k. 4 d after transfection, the impact of sgRNAs on cell proliferation and PLK1 expression was determined by the standard MTT assay and Western Blot assay, respectively. The MTT assay was carried out as described above. For the Western Blot assay, cells were collected, washed with PBS, and lysed in radio immunoprecipitation assay (RIPA) buffer (Thermo Scientific) supplemented with protease Inhibitor Tablet (Roche). About 10 µg of each protein were run on an sodium dodecyl sulfate (SDS)-polyacrylamide gel, and after electrophoresis, the proteins were transferred onto nitrocellulose membranes (Bio-Rad). Membranes were probed with primary antibodies against PLK1 (F-8, mouse monoclonal IgG28, Santa Cruz Biotechnology) and then secondary antibody polyclonal goat antimouse immunoglobulins/horseradish peroxidase (HRP) (Dako). The color reaction was developed with detection reagent (Thermo Scientific).
Production of Minicircle DNA
The sgRNA expression cassette within sgPLK1-2 lentiGuide-Puro was amplified by polymerase chain reaction (PCR) and cloned into the BglII and Sal1 restriction sites of vector pMC.BESPX.[20] The forward and reverse primers used for PCR amplification are GTCGGTAGATCTGAGGGCCTATTTCCCATGAT and GTCATTGTCGACAAAAAAGCACCGACTCGGTG, respectively. The production of minicircle-sgPLK1-2 was carried out according to previously published procedures.[20] Briefly, sgPLK1-2 pMC.BESPX construct was transformed into E. coli strain ZYCY10P3S2T. A single clone of transformed E. coli was selected and cultured at 37 °C in 400 mL Terrific Broth supplemented with kanamycin (Thermo Scientific). In the following day, minicircle induction media comprising 400 mL of lysogeny broth, 16 mL 1 n NaOH and 400 µL 20% l-arabinose (Sigma-Aldrich) was added to the culture. After 6 h incubation at 32 °C, the E. coli culture was collected through centrifugation. Minicircle-sgPLK1-2 DNA was purified with EndfoFree Plasmid kits (Qiagen).
Bio-Distribution of LHNPs in Tumors
Female athymic (NCr-nu/nu) nude mice were maintained in a sterile environment. This project was approved by the Yale University Institutional Animal Care and Use Committee (IACUC). To establish the flank tumor model, mice received subcutaneous flank injections of 1 × 106 U87 cells. Tumor size was measured three times a week using traceable digital vernier calipers (Fisher). Tumor volume was determined by measuring the length (l) and width (w), and then calculating the volume (V) using the following formula: V =l*w2*0.5. Tumor homing study began when the volume reached ≈200 mm3 (day 1). Mice were randomly divided into two groups (n =5) and received intravenous treatment of IR780-loaded LHNPs with and without conjugation of iRGD. In order to ensure each mouse received the same amount of IR780 dye, the actual dosage was slightly adjusted and normalized according to the fluorescence intensity. 24 h later, mice were euthanized and the tumors were harvested for imaging using IVIS. To establish the intracranial U87 model, mice were anesthetized via intraperitoneal injection of ketamine and xylazine. 20 000 U87 cells in 5 µL of PBS were injected into the right striatum 2 mm lateral and 0.5 mm posterior to the bregma and 4 mm below the dura using a stereotactic apparatus with a UltraMicroPump (UMP3) (World Precision Instruments, FL). Evaluation was performed 3 w after tumor inoculation according the same procedures described above, except that LHNPs with and without encapsulation of LEX were used.
Therapeutic Evaluation of LHNPs
For evaluation in flank tumors, tumor bearing mice were established according to the procedures described above. Treatments started when the volume reached ≈200 mm3. For evaluation in intracranial tumors, nude mice were anesthetized via intraperitoneal injection of ketamine and xylazine. Twenty thousand luciferase-expressing U87 cells in 2 µL of PBS were injected into the right striatum 2 mm lateral and 0.5 mm posterior to the bregma and 3 mm below the dura using a stereotactic apparatus with a UltraMicroPump (UMP3) (World Precision Instruments, FL). One week after tumor inoculation, mice with comparable luciferase expression intensity in the brain were randomly divided into treatment groups and subjected to treatments. For both studies, treatments were performed through intravenous injection of nanoparticles at a dose of 1 mg in 100 µL PBS 3 d a week for three weeks. The animals’ weight, grooming, and general health were monitored on a daily basis. Animals were euthanized after either a 15% loss in body weight or when it was humanely necessary due to clinical symptoms. The Kaplan–Meier survival curves were plotted.
Statistical Analysis
All data were collected in triplicate and reported as mean and standard deviation. Comparison of two conditions was evaluated by the unpaired t-test. p < 0.05 (*), 0.01 (**), and 0.0001 (****) were considered significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants NS095817, NS095147, American Brain Tumor Association, and the State of Connecticut. This research was partially supported by scholarships from the Chinese Scholarship Council to Y.C., X.W., and J.L.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Zeming Chen, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin 300071, P. R. China.
Dr. Fuyao Liu, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Yanke Chen, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA.
Dr. Jun Liu, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Dr. Xiaoying Wang, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Ann T. Chen, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Dr. Gang Deng, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Dr. Hongyi Zhang, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Dr. Jie Liu, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA
Prof. Zhangyong Hong, State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin 300071, P. R. China
Prof. Jiangbing Zhou, Department of Neurosurgery, Yale University, New Haven, CT 06510, USA Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA.
References
- 1.a) Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Science. 2012;337:816. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Science. 2013;339:819. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Cheng AW, Wang HY, Yang H, Shi LY, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Cell Res. 2013;23:1163. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gilbert LA, Larson MH, Morsut L, Liu ZR, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. Cell. 2013;154:442. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Cell. 2013;152:1173. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Swiech L, Heidenreich M, Banerjee A, Habib N, Li YQ, Trombetta J, Sur M, Zhang F. Nat. Biotechnol. 2015;33:102. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson CE, Gersbach CA. Annu. Rev. Chem. Biomol. Eng. 2016;7:637. doi: 10.1146/annurev-chembioeng-080615-034711. [DOI] [PubMed] [Google Scholar]
- 4.a) Deeken JF, Loscher W. Clin. Cancer Res. 2007;13:1663. doi: 10.1158/1078-0432.CCR-06-2854. [DOI] [PubMed] [Google Scholar]; b) Zhou J, Atsina KB, Himes BT, Strohbehn GW, Saltzman WM. Cancer J. 2012;18:89. doi: 10.1097/PPO.0b013e318244d8ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senis E, Fatouros C, Grosse S, Wiedtke E, Niopek D, Mueller AK, Borner K, Grimm D. Biotechnol. J. 2014;9:1402. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 6.Li SD, Huang L. J. Controlled Release. 2007;123:181. doi: 10.1016/j.jconrel.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Zhou J, Liu J, Cheng CJ, Patel TR, Weller CE, Piepmeier JM, Jiang Z, Saltzman WM. Nat. Mater. 2012;11:82. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. Nature. 2014;514:380. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramakrishna S, Dad AK, Beloor J, Gopalappa R, Lee SK, Kim H. Genome Res. 2014;24:1020. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Nat. Biotechnol. 2015;33:73. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J. Nat. Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jiang C, Mei M, Li B, Zhu X, Zu W, Tian Y, Wang Q, Guo Y, Dong Y, Tan X. Cell Res. 2017;27:440. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Li L, Song L, Liu X, Yang X, Li X, He T, Wang N, Yang S, Yu C, Yin T, Wen Y, He Z, Wei X, Su W, Wu Q, Yao S, Gong C, Wei Y. ACS Nano. 2017;11:95. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]; e) Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ. Angew. Chem., Int. Ed. Engl. 2017;56:1059. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, Rotello VM. ACS Nano. 2017;11:2452. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z. Angew. Chem., Int. Ed. Engl. 2015;54:12029. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Wang M, Zuris JA, Meng FT, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu QB. Proc. Natl. Acad. Sci. USA. 2016;113:2868. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han L, Kong DK, Zheng MQ, Murikinati S, Ma C, Yuan P, Li L, Tian D, Cai Q, Ye C, Holden D, Park JH, Gao X, Thomas JL, Grutzendler J, Carson RE, Huang Y, Piepmeier JM, Zhou J. ACS Nano. 2016;10:4209. doi: 10.1021/acsnano.5b07573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Kouranova E, Forbes K, Zhao G, Warren J, Bartels A, Wu Y, Cui X. Hum. Gene Ther. 2016;27:464. doi: 10.1089/hum.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD. J. Biotechnol. 2015;208:44. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 13.a) Kelton WJ, Pesch T, Matile S, Reddy ST. Chimia. 2016;70:439. doi: 10.2533/chimia.2016.439. [DOI] [PubMed] [Google Scholar]; b) Gwiazda KS, Grier AE, Sahni J, Burleigh SM, Martin U, Yang JG, Popp NA, Krutein MC, Khan IF, Jacoby K, Jensen MC, Rawlings DJ, Scharenberg AM. Mol. Ther. 2016;24:1570. doi: 10.1038/mt.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Lu C, Stewart DJ, Lee JJ, Ji L, Ramesh R, Jayachandran G, Nunez MI, Wistuba II, Erasmus JJ, Hicks ME, Grimm EA, Reuben JM, Baladandayuthapani V, Templeton NS, McMannis JD, Roth JA. PloS One. 2012;7:e34833. doi: 10.1371/journal.pone.0034833. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Porteous DJ, Dorin JR, McLachlan G, Davidson-Smith H, Davidson H, Stevenson BJ, Carothers AD, Wallace WA, Moralee S, Hoenes C, Kallmeyer G, Michaelis U, Naujoks K, Ho LP, Samways JM, Imrie M, Greening AP, Innes JA. Gene Ther. 1997;4:210. doi: 10.1038/sj.gt.3300390. [DOI] [PubMed] [Google Scholar]
- 15.Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. Cell. 2015;163:829. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Patel TR, Fu M, Bertram JP, Saltzman WM. Biomaterials. 2012;33:583. doi: 10.1016/j.biomaterials.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomone F, Cardarelli F, Di Luca M, Boccardi C, Nifosi R, Bardi G, Di Bari L, Serresi M, Beltram F. J. Controlled Release. 2012;163:293. doi: 10.1016/j.jconrel.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 18.Sanjana NE, Shalem O, Zhang F. Nat. Methods. 2014;11:783. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Patel TR, Sirianni RW, Strohbehn G, Zheng M-Q, Duong N, Schafbauer T, Huttner AJ, Huang Y, Carson RE, Zhang Y, Sullivan DJ, Jr, Piepmeier JM, Saltzman WM. Proc. Natl. Acad. Sci. USA. 2013;110:11751. doi: 10.1073/pnas.1304504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kay MA, He CY, Chen ZY. Nat. Biotechnol. 2010;28:1287. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Cancer Cell. 2009;16:510. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carman AJ, Mills JH, Krenz A, Kim DG, Bynoe MS. J. Neurosci. 2011;31:13272. doi: 10.1523/JNEUROSCI.3337-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney M, Ong SG, Padilla A, Yao Z, Goodman S, Wu JC, Yang F. ACS Nano. 2013;7:7241. doi: 10.1021/nn402657d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Tu Y. Cancer Transl. Med. 2015;1:176. [Google Scholar]
- 25.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Cell. 2013;154:1380. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Nat. Biotechnol. 2014;32:569. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Science. 2012;337:816. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana NE, Shalem O, Zhang F. Nat. Methods. 2014;11:783. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 The ORCID identification number(s) for the author(s) of this article can be found under
The ORCID identification number(s) for the author(s) of this article can be found under