Abstract
Viruses alter host–cell gene expression at many biochemical levels, such as transcription, translation, mRNA splicing and mRNA decay in order to create a cellular environment suitable for viral replication. In this review, we discuss mechanisms by which viruses manipulate host–gene expression at the level of mRNA decay in order to enable the virus to evade host antiviral responses to allow viral survival and replication. We discuss different cellular RNA decay pathways, including the deadenylation-dependent mRNA decay pathway, and various strategies that viruses exploit to manipulate these pathways in order to create a virus-friendly cellular environment.
Keywords: cis-acting elements, host mRNA decay, virus
In eukaryotic cells, gene expression is tightly regulated both transcriptionally and post-transcriptionally to assure correct protein production and normal cell function. One important aspect of post-transcriptional regulation is mRNA turnover, in which the cellular mRNA decay machinery works to coordinate the expression of genes by controlling the stability and lifespan of mRNAs through highly regulated mechanisms. The overall importance of mRNA stability in determining gene expression is highlighted by the studies investigating the impact of mRNA decay on gene expression, which estimate that 20–50% of the changes in gene expression in both yeast and mammalian cells upon stimulation are due to altered mRNA decay [1,2].
Viruses have limited genetic material and functional proteins, but manage to overcome this deficit by usurping the host cellular resources to ensure their survival and replication. Following viral infection, cellular gene expression machinery is hijacked by viruses and host–gene expression becomes perturbed as viruses shift the priorities of the cell to viral rather than host–gene expression. To combat this hijacking by viruses, host cells initiate defense responses to try to prevent viral invasion, but viruses have developed mechanisms to turn off or bypass many of these antiviral responses. Virus-induced alteration of host–gene expression plays vital roles in creating a virus-friendly environment to establish and propagate viral infection and this process has important implications for the pathogenesis of virus-causing diseases [3]. In this review, we focus on the interplay between viruses and host mRNA decay machinery, particularly the varieties of strategies viruses have developed to manipulate host mRNA decay in order to facilitate viral infection and inhibit cellular antiviral activities.
Cellular mRNA decay pathways targeted by viruses
Viruses have evolved the ability to control host–gene expression at the level of mRNA turnover in order to evade antiviral responses and exploit the cellular gene expression machineries for viral replication. Virus-encoded factors and enzymes, or even viral genomic material, are able to interact with components of host mRNA decay pathways to modulate the half-life of both viral and cellular mRNAs. In addition, some viruses encode ribonucleases to directly degrade cellular mRNAs. A brief summary of mechanisms by which viruses manipulate host mRNA decay can be found in Table 1. The following sections discuss different cellular mRNA decay pathways that are targeted by viruses and the strategies used by viruses to manipulate these pathways.
Table 1.
Strategies exploited by viruses to manipulate host mRNA decay.
| Strategies | Example viruses | Ref. |
|---|---|---|
| SGs and PBs | ||
| Induce SG formation | Orthoreoviruses and measles virus | [4,5] |
| Block SG formation | IAV and Semliki Forest virus | [6,7] |
| Co-op SG proteins | HCV and HIV | [8,9] |
| Oscillation of SG formation | HCV | [10] |
| Disrupt PBs | Polioviruses | [11] |
| Co-opt PB proteins | HCV | [8] |
| ARE and GRE | ||
| Interact with ARE binding proteins | HSV, adenovirus, alphaviruses, coxsackievirus B3 and EBV | [12–19] |
| Activate signaling pathways to affect ARE or GRE binding proteins | KHSV, HSV1 and HCV | [20–26] |
| Viral protein binds to GRE | HCV | [27] |
| mRNA surveillance pathway | ||
| Interact with NMD pathway components | Human T-lymphotropic virus and HCV | [28–30] |
| RNA interference pathway | ||
| Saturate dicer | Flaviviridae family and adenovirus | [31,32] |
| Mediate target miRNA degradation | Herpesvirus saimiri | [33–36] |
| Interferon-induced RNase L pathway | ||
| Prevent RNase L activation | VACV, influenza virus, mouse hepatitis virus and group C enterovirus | [37–39] |
| Viral ribonucleases | ||
| Viral endonucleases | HSV, KHSV, EBV, IAV | [40–43] |
| Viral protein recruit cellular endonucleases | SARS coronavirus | [44] |
| Cap snatching | IAV, bunyaviruses, orthomyxoviruses and arenaviruses | [45,46] |
| Viral decapping enzymes | VACV and African swine fever virus | [47–49] |
| Viral exonucleases | Lassa fever virus | [50] |
| Viral long noncoding RNA | ||
| Stall and suppress XRN1 | Flaviviridae family | [51,52] |
| RNA methylation | ||
| Alter cellular RNA N6-adenosine methylation | Zika virus | [53] |
ARE: AU-rich element; EBV: Epstein-Barr virus; HCV: Hepatitis C virus; HIV: Human immunodeficiency virus; IAV: Influenza A virus; KHSV: Kaposi’s sarcoma-associated herpesvirus; NMD: Nonsense-mediated decay; PB: Processing body; SG: Stress granule; VACV: Vaccinia virus.
Stress granules & processing bodies
Stress granules (SGs) and processing bodies (PBs) are RNA granules that sequester translationally silenced mRNAs. The cell forms SGs and PBs to regulate mRNA stability, prevent apoptosis and maintain cell homeostasis. SGs are formed upon cellular stresses such as viral infection, heat shock, or other environmental stimuli. They are molecular aggregates composed of translation-initiation factors, small ribosome subunits, RNA binding proteins (RBPs) and mRNAs [54] and are formed transiently to store mRNA at times of stress to help the cell to survive and then quickly recover. Once the stress has ended, SGs disperse rapidly, allowing the previously restrained mRNAs to resume translation [55]. PBs also act to regulate mRNA stability in the cytoplasm but exist constitutively rather than only during responses to stress, and they contain many of the same mRNAs and proteins involved in RNA decay [54]. The molecular mechanism of PB formation is related to aggregation of mRNAs and RBPs [56]. SGs and PBs are related to each other in terms of composition, localization and function [54]. The major difference in terms of composition between PBs and SGs is that PBs do not contain 40S ribosome subunits and most initiation factors like SGs do, which contain stalled 43S preinitiation complexes on mRNAs due to scarcity of stress-induced eIF2-GTP-tRNAMet ternary complex [57].
SGs and PBs are important structures for regulating cellular mRNA stability and translatability, and serve as targets for viral manipulation of host–gene expression. Many viruses induce, inhibit, or modulate SG formation at some point during viral gene expression [56,58–59]. For example, most of the viruses induce SG formation through activating the Protein Kinase R and phosphorylating the eukaryotic translation initiation factor eIF2α, resulting in suppression of host mRNA translation, such as mammalian orthoreovirus and measles virus [4,5]. Other viruses on the other hand, block the formation of SG for viral survival. The influenza A virus (IAV) deploys viral nonstructural protein 1 to inhibit Protein Kinase R activation and eIF2α phosphorylation, blocking SG formation [6]. The Semliki Forest virus nsP3 sequestrates host Ras-GAP SH3-domain-binding protein into the viral replication complex, resulting in suppression of SG formation on viral RNAs and efficient viral mRNA translation [7]. Yet another group of viruses, including hepatitis C virus (HCV) and human immunodeficiency virus, co-opt SG proteins for their own use. HCV hijacks SG components, such as GAP SH3-domain-binding protein and poly(A)-binding protein 1, for viral replication [8]. human immunodeficiency virus protein Gag interacts directly with the host protein Staufen1 for viral encapsidation [9]. Interestingly, HCV also triggers highly dynamic oscillation of SG formation and cellular translation, in order to maintain the survival of infected cell and establish persistent infection [10].
In comparison to SGs, the study on PBs in viral infection has thus far been less comprehensive, leaving the complete interaction between viruses and PBs a mystery for now. Similar to SGs, viruses either disperse, inhibit, or co-opt the PB components to augment infection. For example, polioviruses are found to disrupt PBs, probably by accelerating the cleavage of important mRNA decay elements, such as Xrn1, Dcp1a and Pan3 [11]. Furthermore, HCV redistributes some PB proteins to lipid droplets for viral replication [8].
Some studies in recent years have been questioned the roles of SGs and PBs on regulating mRNA stability since decapping occurs in polysomes. The size and number of the PBs, however, has been correlated with accumulation of mRNA decay proteins and mRNA decay intermediates in PBs, suggesting that decay may also occur in PBs [60]. Even if mRNA decay does not happen in these specialized granules, the sequestration of the decay machinery and mRNAs within these structures might nonetheless regulate the host mRNA pool, providing an opportunity for viruses to affect cellular mRNA and gene expression.
Deadenylation-dependent RNA decay
Deadenylation-dependent RNA decay is the major cellular pathway for cytoplasmic mRNA turnover. Deadenylation is a process of 3′ polyA tail shortening by cellular deadenylase enzyme complexes such as CCR4-NOT, PAN2-PAN3 and PolyA Ribonuclease. The removal of the polyA tail is usually the first and rate limiting step of mRNA decay [61]. The deadenylated transcript is then subject to rapid exonucleolytic degradation, with decay occurring in either the 3′-to-5′ direction by the cytoplasmic exosome and the scavenger decapping enzyme (DcpS), or in the 5′-to-3′ direction by the cellular DcpS DCP2 and exonuclease XRN1 [62–66]. Specific mRNAs are deadenylated at different rates. This differential mRNA deadenylation is not a stochastic process but rather, a precisely controlled process for determining the lifespan of each mRNA. The mechanisms by which the cell selects mRNAs for deadenylation are not fully known [63,67].
During recent decades, advanced techniques such as RNA immunoprecipitation, gene expression microarrays, next generation sequencing, and mass spectrometry have enabled researchers to identify cis-acting elements and trans-acting factors which determine mRNA stability. Cis-acting elements are sequences within mRNA molecules, often in the untranslated regions, that are bound by trans-acting factors, such as RBPs or miRNAs. Their interaction determines the location, translation, and stability of the mRNAs that harbor the cis-acting elements [68]. The study of how these cis-acting elements and trans-acting factors mediate mRNA decay has unraveled a broader understanding of the mechanism of coordinately regulated deadenylation-dependent mRNA decay; mRNA decay rates are determined by the ability of trans-acting factors to recruit or repel components of the mRNA decay machinery. For example, upon binding to cis-acting elements, decay-promoting RBPs can recruit deadenylases such as PolyA Ribonuclease to shorten the polyA tail of a mature mRNA, which then initiates the degradation cascade [69,70].
The interactions between cis-acting elements and trans-acting factors define different post-transcriptional regulatory networks. Different trans-acting factors collectively act on the cis-acting elements within a group of mRNAs that involve the same functional network, thus coordinating the gene expression post-transcriptionally [68,71]. Below, we will review two important mRNA decay regulating cis-acting elements – AU-rich element (ARE) and GU-rich element (GRE), their associated trans-acting factors, and examples of viruses utilizing them to differentially regulate host mRNA stability.
AREs
The best-studied mRNA decay cis-element is the ARE, a sequence found in the 3′UTR of certain transcripts that promotes rapid mRNA decay. The ARE was first identified in 1986 by Caput and colleagues using bioinformatics approaches. They found an evolutionarily-conserved adenine- and uridine-rich consensus sequence that is present in the 3′UTR of both human and mouse TNF mRNAs [72]. Later studies identified more AREs that functioned to mediate mRNA decay, and AREs were organized into three classifications that differ by their sequence composition and decay kinetics [73].
ARE-containing transcripts are found in a variety of cellular processes and account for approximately 5–8% of the whole transcriptome [74]. Numerous cytokine gene (including IFN-γ, IL-2, and TNF-α) and proto-oncogene (v-myc, c-fos) transcripts contain AREs [75]. AREs regulate mRNA stability by interacting with a variety of ARE-binding proteins (AREBPs). Some AREBPs, such as ELAV-like protein 1 or HuR and ELAV-like protein 4 or human antigen D, promote transcript stability when they bind to AREs, while other AREBPs, such as ARE/poly(U) binding/degradation factor 1 (AUF1) and Tristetraprolin (TTP), promote transcript decay [65,76–79]. In addition, microRNAs can directly bind to AREs and regulate the stability of the ARE-containing transcripts [80]. These AREBPs and microRNAs work in concert to determine the biological outcome of the ARE-harboring transcripts [81]. There are over 20 AREBPs from various families and that are spread widely throughout the cell, from nucleus to cytoplasm to subcellular structures such as SGs and PBs. They are also active contributors or effectors in a variety of signaling transduction pathways.
Viruses take advantage of AREs to differentially manipulate the expression of ARE-containing subsets of host transcripts. Viruses produce proteins or noncoding RNAs to inhibit or promote host mRNA degradation, either by affecting the activity of ARE associated trans-acting factors, or by directly binding to the ARE within host mRNAs. For example, herpes simplex virus (HSV) protein UL14, or virion host shutoff (vhs) protein, a viral encoded endonuclease that will be described in detail later, triggers global degradation of host mRNAs [82]. Vhs preferentially cuts mRNAs in the translation initiation region toward the 5′ end, but can also differentially target host ARE-containing transcripts and cleaves them in the 3′UTR [12]. Experiments show that in HSV-infected cells, the ARE-binding protein TTP binds to vhs and guides it to cleave the 3′UTR of ARE-containing stress response mRNAs [13]. In contrast to vhs that promotes the decay of ARE-containing transcripts, adenovirus oncogene product E4orf6 stabilizes ARE-containing mRNAs via an α-helix structure that is also required for the oncogenic activity and ubiquitin E3 ligase assembly of E4orf6. In return, the stabilized ARE-containing mRNAs contribute to the E4orf6 oncogenic activity. E4orf6 lacked oncogenic activity in HuR-knockdown cells that failed to stabilize ARE-containing mRNAs [14], suggesting that the oncogenic activity of E4 or f6 depends on stabilization of ARE-containing transcripts by HuR.
Other examples of virus interacting with AREBPs are alphaviruses, including sindbis virus, ross river virus and chikungunya virus [15,16]. The 3′ UTRs of sindbis virus RNAs bind to HuR with high affinity, which causes the relocation of HuR from the nucleus to the cytoplasm and the sequestration of HuR in the cytoplasm. These changes are associated with the destabilization of cellular mRNAs that are normally targeted by HuR [15].
Interestingly, the coxsackievirus B3 (CVB3) genomic RNA contains AREs and is susceptible to ARE-mediated decay by host factors. To counteract this, CVB3 causes the infected cell to increase expression of the stress-inducible chaperon protein HSP70–1, a host protein involved in the stabilization of ARE-containing mRNAs [17,18]. In turn, the upregulation of HSP70–1 facilitates viral replication by promoting the stabilization of CVB3 genome via the ARE within the 3′UTR of the viral genomic RNA [17]. Additional studies found that the decay promoting AREBP, AUF1, undergoes cytoplasmic redistribution and cleavage upon CVB3 infection [18,19] which may further promote the stabilization of ARE-containing transcripts.
In addition to the AREBPs, viral factors can also compete with AREs to bind to AREBPs, abolishing their mRNA decay regulating capability. For example, cells infected with Epstein-Barr virus (EBV) produce large amount of two noncoding RNAs, EBV-encoded RNA 1 (EBER1) and EBER2. Experiments shown that EBER1 competes with ARE to bind to the p40 isoform of the decay-promoting AREBP, AUF1. Binding by EBER1 to AUF1 prevents AUF1 from binding to its ARE-containing target mRNAs, thus influencing the stability of these ARE-containing transcripts [20].
Numerous signaling pathways, particularly the p38/MK2 mitogen-activated protein kinase (MAPK) pathway, regulate the decay of ARE-containing transcripts by influencing the localization, abundance, and modification of AREBPs [71]. Viral infection is a stimulus for activating the stress-inducible p38/MK2 signaling pathway in the infected cell, and the consequence is usually the stabilization of ARE-containing transcripts that normally decay rapidly. For example, Kaposi’s sarcoma-associated herpesvirus (KSHV) G-protein-coupled receptor (vGPCR) is found to prevent the turnover of host ARE-containing transcripts through activating the p38/MK2 pathway, probably by interacting with MK2. vGPCR also disturbs PB formation during lytic KSHV infection, the net effect of which is the promotion of the secretion of angiogenic factors from the infected cells [21]. Another KSHV protein, kaposin B, binds to MK2 and activates the p38/MK2 pathway, leading to the stabilization of ARE-containing mRNAs that encodes cytokines such as IL-6, TNF-α and IFN-γ [22,23]. The activation of p38/MK2 pathway by Kaposin B also induces accumulation of HuR, an ARE-stabilizing AREBP, leading to the overexpression of ARE-containing trasncripts [24]. Another example of viral manipulation of host signaling is the HSV-1 immediate early protein ICP27 which is responsible for stabilizing the ARE-containing transcripts through activation of the p38/MK2 pathway [25]. In the case of HCV infection, IL-8 (CXCL-8) and other related ARE-containing mRNAs are stabilized via AREs within their 3′UTRs [26]. A later study revealed that the stabilization of CXCL-8 transcript is triggered by HCV activated dsRNA signaling pathways, but the factors responsible for this mechanism are not yet clear [83].
The AREBP, AUF1 is also a target of West Nile Virus. Arginine methylation of AUF1 during West Nile virus infection facilitates viral replication and affects the ability of AUF1 to bind to RNA [84]. Cleavage and cellular relocation of AUF1 plays a role in the infectious cycle of poliovirus or human rhinovirus [85]. However, these studies did not further investigate the impact on host mRNA stability by the viruses, yet we can still speculate that the changes of these AREBPs upon infection would at least affect the decay of some, if not all, of their ARE-containing target transcripts.
GREs
GREs are recently discovered mRNA decay elements that are enriched in the 3′UTR of numerous short-lived transcripts. Inserting a GRE into the 3′UTR of a beta-globin reporter causes the otherwise stable reporter to become highly unstable, indicating that GRE is a functional mediator of mRNA decay [86]. The RNA-binding protein CELF1 has been found to specifically bind to GREs and mediate the subsequent rapid degradation of the GRE-harboring transcripts [87]. Later experiments found that CELF1 also binds to GU-repeat sequences and mediates the decay of their mRNA as well [88]. Thus, the GRE has been defined as of the form UGUU(/G)UGUU(/G)UGU. Transcripts that contain GRE encode important regulators involved in cell growth, proliferation, apoptosis and oncogenesis [89]. The exact mechanism of GRE and CELF1 mediated mRNA decay is not completely understood. It is postulated that in the cytoplasm, the GRE-containing transcript is recognized and bound by CELF1, which recruits deadenylases to initiate the rapid degradation of the transcript [70].
The GRE/CELF1 regulatory networks control a variety of important biological processes, such as cellular growth, differentiation and activation. For example, upon T-cell activation, CELF1 becomes phosphorylated and loses its ability to bind to GREs, the consequence of which is increased stability and expression of GRE-containing mRNAs that are involved in cell activation and proliferation [90]. GRE-containing CELF1 target transcripts also preferentially undergo alternative polyadenylation in activated T cells, resulting in the permanent removal of the 3′UTR regions that hold GREs and thus, the increased stability of these transcripts [91].
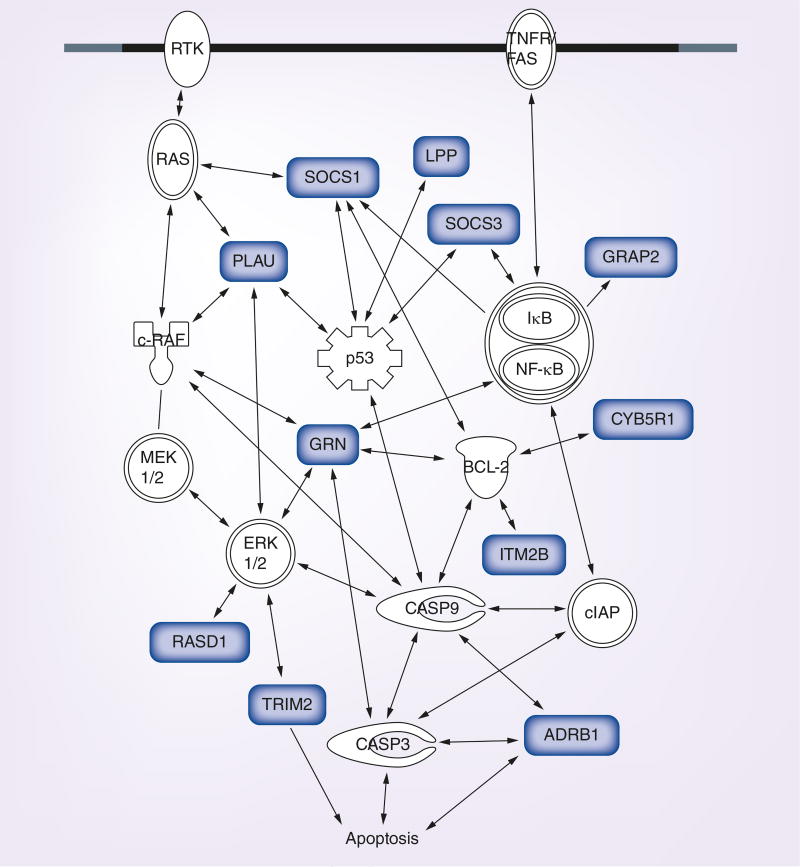
Our laboratory, recently discovered that HCV NS5A binds to GREs and regulates host mRNA decay. NS5A is a multifunctional protein that plays pivotal roles in both viral and cellular processes [92]. Interestingly, NS5A is also an RNA-binding protein that can bind specifically to G- and U-rich sequences within the HCV genomic RNA [93]. Cytoplasmic extracts from human hepatoma cells expressing an HCV subgenomic replicon were immunoprecipitated using an anti-NS5A antibody, and copurified transcripts were identified using RNA-SEQ to identify host transcripts that were bound to NS5A. NS5A target transcripts identified in this manner were found to be enriched for GREs and GRE-like sequences within their 3′UTRs [27]. Pathway analyses of these NS5A target transcripts shows that they participate in many important cellular processes and functions such as regulation of cell growth and apoptosis (Ingenuity Pathway Assistant software). Figure 1 depicts an example of numerous NS5A target transcripts involved in the regulation of apoptosis. Expression of NS5A led to stabilization of GRE-containing reporter transcripts that were otherwise unstable, suggesting that NS5A binding causes stabilization of GRE-containing transcripts. The stabilization and upregulation of host transcripts likely represents an example whereby HCV manipulates host–gene expression to bypass antiviral responses, promote cell growth and prevent cell death in order to establish chronic infection. This could eventually lead to the development of hepatocellular carcinoma since apoptosis is inhibited in chronically infected cells, and additional genetic damage could accumulate in these cells over time. Although, the exact mechanism of the NS5A-mediated mRNA stabilization is not known, NS5A may compete with cellular CELF1 to bind to GREs, thus preventing CELF1-mediated rapid decay.
Figure 1. GU-rich element-containing NS5A target transcripts encode regulators of apoptosis.
Transcripts depicted in grey are NS5A target transcripts that contain GREs. These pathway figures were created using Ingenuity Pathway Assist software (Qiagen Inc).
GRE: GU-rich element.
Specialized endonucleolytic RNA decay
In addition to the major deadenylation-dependent mRNA decay pathway, cytoplasmic mRNAs can also undergo specialized endonucleolytic decay, which is initiated by endonucleolytic cleavage of the mRNA via cellular or viral endonucleases and followed by exonuclease digestion via the cellular exosome and XRN1 [71,77].
mRNA surveillance
Two mRNA surveillance pathways that monitor the quality of cellular mRNAs are nonsense-mediated decay (NMD) and no-go decay, which utilize the endonucleolytic decay mechanism to ensure the fidelity of gene expression. NMD targets mRNAs bearing premature termination codons and mRNAs with abnormally long 3′UTRs. No-go decay eliminates mRNAs with secondary structures that stall ribosomes. Another mRNA surveillance pathway called nonstop decay, on the other hand, targets mRNAs without termination codons for exonucleolytic degradation by the SKI complex and exosome [71].
The primary function of NMD in eukaryotic cells is to degrade endogenous mRNAs that contain premature termination codons, but NMD may also be activated by aberrant structures or by sequences of viral genomic RNA and viral transcripts. Balistreri et al. suggested that NMD serves as a cell-intrinsic barrier to restrict Semliki Forest Virus infection infection because they observed increased infection when NMD components were depleted [28]. In order to protect their RNAs, however, viruses developed mechanisms to circumvent host mRNA surveillance. For example, the HTLV encoded Rex protein interacts with components of NMD effectors, such as UPF1 and EIF3E, suppressing host NMD activity, preventing the degradation of unspliced viral mRNAs by the NMD machinery, and thereby promoting viral replication. The inhibition of NMD also results in the stabilization of host NMD target mRNAs, disturbing host–gene expression [29,30]. HCV infection is also reported to suppress NMD activity as measured by the accumulation of cellular NMD substrates. A combined proteomics/genomics approach reveals that the HCV core protein binds to the host protein within BGCN homolog and prevents its interaction with the key mediators of NMD, partners Y14 and Magoh, thus disrupting host NMD activity [94].
RNA interference pathway
RNAi associated decay pathway facilitated by miRNA and small siRNA also initiates mRNA decay through endonucleolytic cleavage. miRNAs and siRNAs target mRNAs for degradation by partnering other proteins to assemble RNA-induced silencing complex and RNA-induced transcriptional silencing complex. The targeting mRNAs are cleaved by the complexes at sites complementary to the miRNA or siRNA. For mRNAs that do not perfectly base pair with the miRNA, the outcome for miRNA targeting is usually translation inhibition or deadenylation-dependent mRNA decay [95].
A long noncoding RNA of viruses in the flaviviridae family, sfRNA and VA RNA, a noncoding RNA found in adenovirus both influence gene expression by disrupting the RNAi pathway. sfRNA is the 3′ degradation intermediate generated when XRN1 degrades Flavivirus genomic RNA. It possesses a large amount of secondary structures and is highly abundant in the infected cell [96,97]. A recent study has shown that sfRNAs from both Dengue virus and Kunjin virus directly interact with the host RNAi machinery and significantly inhibit the antiviral effect of RNAi in both human cells and mosquitos [31]. In fact, both sfRNA and VA RNA are processed by Dicer, a key effector of the RNAi machinery. sfRNA and VA RNA compete with the cellular Dicer substrates and saturate Dicer, thus blocking the RNAi pathway [32,98].
Herpesvirus saimiri (HVS) encodes seven small nuclear RNAs called HVS U-rich RNAs that also interfere with host–gene expression through RNAi pathways. HVS produces a high abundance of HSURs in infected T lymphocytes [33]. Although, not required for viral replication, the highly conserved HSUR1 and 2 are found to be involved in upregulation of a subset of host mRNAs during latent infection [34]. This is because HSUR1 binds and mediates degradation of host micorRNA-27, leading to increased expression of miRNA-27 targets that are responsible for T-cell activation [34,35]. Interestingly, HSUR1 also contains an ARE and can recruit the host AREBP TTP to degrade HSUR1 itself, but the host ARE-mRNAs levels remain unchanged, suggesting that HVS is using the host ARE-dependent decay pathway to control its HSUR1 expression [36,37].
Interferon-induced RNase L pathway
The interferon-induced RNase L pathway is an innate immunity pathway that is triggered by dsRNAs upon viral infection that lead to apoptosis of the infected cell. The effector in this pathway, RNase L, is an inducible cellular endonuclease that cleaves single-stranded RNA, particularly after UA and UU dinucleotides, to initiate target RNA decay [38]. The dsRNAs (such as viral dsRNA genome and viral replication intermediates) triggers the pathogen recognition receptor 2′,5′-oligoadenylate synthetase enzyme to generate 2′,5′-oligoadenylate (2–5A) from ATP. 2–5A then activates RNase L to cut both viral and cellular mRNAs and mediate endonucleolytic decay pathway [38].
In order to overcome the degradation by cellular RNase L, several viruses such as vaccinia virus and influenza virus produce proteins (E3L protein and NS1 protein, respectively) that bind to the dsRNA, shielding it from being recognized by oligoadenylate synthetase and preventing the activation of the RNase L [38]. Mouse hepatitis virus, on the other hand, expresses nonstructural protein 2 to cleave 2–5A to prevent RNase L activation and thereby protecting the viral RNAs [39]. As another mechanism, group C enterovirus RNAs inhibit the endoribonuclease activity of RNase L by competing with the RNA substrates to bind to its endoribonuclease domain, thereby stabilizing viral as well as cellular RNAs that are RNase L targets [40].
Viral ribonucleases
Instead of interrupting the different host decay pathways to regulate cellular gene expression, some viruses produce ribonucleases to directly cut host mRNAs and induce their degradation. It is a common feature of several alpha- and gamma-herpesviruses to globally destabilize host mRNAs through viral endonucleases. The HSV vhs is an extensively studied viral RNA endonuclease; it not only promotes rapid degradation of cellular mRNAs to shut off host–gene expression, but also eliminates viral early stage mRNA for late stage viral gene expression [41]. The gamma-herpesviruses such as KSHV and EBV encode other endonucleases, shutoff and exonuclease and BGLF5, respectively, that produce host mRNA decay [42,99]. Both vhs and shutoff and exonuclease mediate host mRNA degradation by cleaving host mRNAs into fragments, leaving the cleavage products vulnerable to exonucleolytic degradation in either 5′-3′ or 3′-5′ direction [43,100]. Other viruses produce proteins that also possess endonuclease activity, such as the IAV PA-X protein, which cleaves host mRNA decay using a mechanism that is similar to herpesviruses [44]. In contrast, SARS coronavirus nonstructural protein 1 induces endonucleolytic cleavage within the 5′UTR of host mRNA by activating a cellular endonuclease that is not yet identified. The nonstructural protein 1-induced endonuclease is specific to host mRNA and does not target viral mRNAs for degradation which enables the accumulation of viral mRNAs and proteins for viral replication and infection [45].
In addition, IAV, Bunyaviruses, Orthomyxoviruses and Arenaviruses utilize cap-snatching, a mechanism by which viral enzymes take the 5′ 7-methyl guanosine cap away from cellular mRNA by endonucleolytic cleavage and then integrate the cap to viral transcripts to disguise them as cellular transcripts, leaving the uncapped cellular mRNA susceptible to Xrn1 degradation [46,47]. Instead of cap-snatching, Vaccinia virus (VACV) encodes two DcpS, D9 and D10, to cleave the 5′ 7 mG cap of host mRNA through their Nudix hydrolase domains [48,49]. African swine fever virus also encodes a DcpS that contains a Nudix domain, called g5R [101]. It is likely that the host exonuclease Xrn1 is responsible for the degradation of the decapped RNA substrate, since recent studies have shown the important role of Xrn1 on Vaccina virus growth and replication [50].
Different from the above viruses that encode endonucleases to cleave the cellular mRNAs, Lassa fever virus encodes nucleoprotein, which possesses an exoribonuclease activity and specifically digests dsRNAs in a 3′-5′ direction. Further experiments indicate that this exonuclease activity is essential for nucleoprotein to suppress the activation of innate immunity [51].
Other mechanisms by which viruses manipulate host mRNA stability
In Flaviviridae, two different noncoding viral RNAs are reported to suppress XRN1, the cellular 5′-3′ exoribonuclease. Moon et al. first demonstrated that one short noncoding RNA from the 3′UTR of Dengue or Kunjin viruses, sfRNA, can inhibit XRN1 activity and affect host mRNA stability [52]. The same group also found that transfection of HCV or Bovine viral diarrhea virus genomic RNA 5′UTR alone in the absence of other viral products to living cells is enough to stall and inhibit the enzymatic activity of XRN1, resulting in global increasing of host RNA stability [53]. It is not clear whether the stalling effect is due to direct spatial blockage by the structure of viral 5′UTR, or it is because of the indirect recruitment of other factors by sequences within the viral 5′UTR.
Zika virus affects host–gene expression by altering the N6 - adenosine methylation (m6 A) profile within the cellular RNA, which plays an important role in regulating gene translation, alternative splicing and mRNA stability [102].
Conclusion & future perspective
It is clear from these examples that the mechanism of viral survival in host cells is far more complex than previously understood. Viral manipulation of host mRNA decay has emerged as an important mechanism for viral infection and survival within cells, and there are many opportunities for future research. The battle between viruses and host mRNA decay machinery not only helps us to understand the molecular mechanisms of host–pathogen interaction, but also provides insight and clues for developing therapeutic antiviral drugs. In this review, we have listed several strategies that viruses exert to manipulate host mRNA stability, either by dampening the host RNA decay pathways or by accelerating the decay processes, through either differential regulation or global shutoff. There are also a handful of approaches that we did not discuss here that viruses use to avoid innate host responses to foreign viral RNAs, but these mechanisms have not yet been shown to cause changes in host mRNA stability [103].
It is likely that viruses have developed mechanisms to manipulate host mRNA decay in order to block antiviral responses or to allow viruses to take over the host–gene expression machinery to promote viral replication, but in most cases where viruses have been shown to manipulate host mRNA decay, the outcome of this manipulation on viral infection has not yet been studied. Many questions remain to be answered in terms of the interplay between viral infections and host mRNA turnover pathways. First and foremost, how can these pathways be exploited for therapeutic uses? Also, is it possible that drugs on the market are already working through these pathways? For example, drugs like ledipasvir that target NS5A for HCV infection but whose mechanism of action remain unclear might function by perturbing the ability of NS5A to bind GREs and modulate host–gene expression. Furthermore, how do viral ribonucleases mark some subsets of host mRNAs for decay but not others? And how can viral ribonucleases avoid cutting their own viral RNAs? How do viral RNAs, which contain sequences that resemble cellular mRNA stability determinants, escape the host decay processes that specifically target their cellular counterparts? How many additional aspects of mRNA decay pathways can be regulated by viruses? Given that this is a relatively new avenue of research, one can assume that there are many more viral mechanisms yet to be discovered. Current and future studies focusing on the host-virus interaction, and altered host mRNA decay induced by viral infection, will ultimately lead to new insights.
Executive summary.
Viruses affecting stress granules (SGs) & processing bodies (PBs)
Stress granules and processing bodies are RNA-containing granules that regulate mRNA translation and stability.
Viruses induce or inhibit SG and PB formation, or co-opt SG and PB proteins for promoting viral infection and affecting host mRNA stability.
Deadenylation-dependent RNA decay pathways that are targeted by viruses
Deadenylation-dependent decay is the major pathway for cytoplasmic mRNA turnover.
Cis-acting elements and trans-acting factors determine mRNA stability.
AU-rich element and GU-rich element-mediated mRNA decay are manipulated by viruses.
Specialized endonucleolytic RNA decay pathways that are targeted by viruses
Nonsense-mediated decay, an mRNA surveillance pathway, is suppressed by viruses leading to stabilization of both viral and cellular Nonsense-mediated decay RNA substrates.
RNA interference pathway is inhibited by viral factors to control cellular mRNA stability.
Viruses inhibit activation of the interferon-induced RNase L pathway to stabilize viral as well as cellular RNAs that are RNase L targets.
Viruses encode ribodonucleases to directly tailor host mRNA stability.
Other mechanisms that viruses develop to affect host mRNA stability
Viral long noncoding RNA from flaviviridae family viruses stall and suppress cellular exonuclease XRN1, resulting in a global increase of host RNA stability.
Viruses alter the methylation profile (such as the N6 - adenosine methylation) within the cellular RNA to change their stability, splicing and translatability.
Conclusion
Several examples of viruses that manipulate cellular mRNA decay are discussed.
Understanding the interaction between viruses and the host mRNA decay machinery provides insight and clues for developing therapeutic antiviral drugs.
Acknowledgments
PR Bohjanen was supported by NIH grants AI057484 and AI072068. L Guo was funded through NIH grant T32 AI83196. I Vlasova St Louis was funded through a fellowship from the Lymphoma Research Foundation and supported by start-up funds from the Department of Medicine at the University of Minnesota.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell. 2011;22(15):2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwanhausser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Su C, Zhang J, Zheng C. Herpes simplex virus 1 UL41 protein abrogates the antiviral activity of hZAP by degrading its mRNA. Virol. J. 2015;12:203. doi: 10.1186/s12985-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin Q, Hastings C, Miller CL. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 2009;83(21):11090–11101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okonski KM, Samuel CE. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 2013;87(2):756–766. doi: 10.1128/JVI.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, Mccormick C. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog. 2014;10(7):e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panas MD, Varjak M, Lulla A, et al. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell. 2012;23(24):4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariumi Y, Kuroki M, Kushima Y, et al. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 2011;85(14):6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrahamyan LG, Chatel-Chaix L, Ajamian L, et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell. Sci. 2010;123(Pt 3):369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 10.Ruggieri A, Dazert E, Metz P, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12(1):71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougherty JD, White JP, Lloyd RE. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 2011;85(1):64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esclatine A, Taddeo B, Evans L, Roizman B. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl Acad. Sci. USA. 2004;101(10):3603–3608. doi: 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu M, Taddeo B, Roizman B. Tristetraprolin recruits the herpes simplex virion host shutoff RNase to AU-rich elements in stress response mRNAs to enable their cleavage. J. Virol. 2015;89(10):5643–5650. doi: 10.1128/JVI.00091-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroshima T, Aoyagi M, Yasuda M, et al. Viral-mediated stabilization of AU-rich element containing mRNA contributes to cell transformation. Oncogene. 2011;30(26):2912–2920. doi: 10.1038/onc.2011.14. [DOI] [PubMed] [Google Scholar]
- 15.Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 2013;5(4):909–917. doi: 10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson AM, Anderson JR, Barnhart MD, et al. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J. Biol. Chem. 2012;287(43):36229–36238. doi: 10.1074/jbc.M112.371203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu Y, Ye X, Hanson PJ, et al. Hsp70–1: upregulation via selective phosphorylation of heat shock factor 1 during coxsackieviral infection and promotion of viral replication via the AU-rich element. Cell Mol Life Sci. 2016;73(5):1067–1084. doi: 10.1007/s00018-015-2036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284(5413):499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 19.Wong J, Si X, Angeles A, et al. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J. 2013;27(7):2777–2787. doi: 10.1096/fj.12-226498. [DOI] [PubMed] [Google Scholar]
- 20.Lee N, Pimienta G, Steitz JA. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein–Barr virus. RNA. 2012;18(11):2073–2082. doi: 10.1261/rna.034900.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corcoran JA, Khaperskyy DA, Johnston BP, et al. Kaposi’s sarcoma-associated herpesvirus G-protein-coupled receptor prevents AU-rich-element-mediated mRNA decay. J. Virol. 2012;86(16):8859–8871. doi: 10.1128/JVI.00597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang HW, Boshoff C. Linking Kaposi virus to cancer-associated cytokines. Trends Mol. Med. 2005;11(7):309–312. doi: 10.1016/j.molmed.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Mccormick C, Ganem D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science. 2005;307(5710):739–741. doi: 10.1126/science.1105779. [DOI] [PubMed] [Google Scholar]
- 24.Yoo J, Kang J, Lee HN, et al. Kaposin-B enhances the PROX1 mRNA stability during lymphatic reprogramming of vascular endothelial cells by Kaposi’s sarcoma herpes virus. PLoS Pathog. 2010;6(8):e1001046. doi: 10.1371/journal.ppat.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcoran JA, Hsu WL, Smiley JR. Herpes simplex virus ICP27 is required for virus-induced stabilization of the ARE-containing IEX-1 mRNA encoded by the human IER3 gene. J. Virol. 2006;80(19):9720–9729. doi: 10.1128/JVI.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green J, Khabar KS, Koo BC, Williams BR, Polyak SJ. Stability of CXCL-8 and related AU-rich mRNAs in the context of hepatitis C virus replication in vitro. J. Infect. Dis. 2006;193(6):802–811. doi: 10.1086/500510. [DOI] [PubMed] [Google Scholar]
- 27.Guo L, Sharma SD, Debes J, et al. The hepatitis C viral nonstructural protein 5A stabilizes growth-regulatory human transcripts. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky061. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balistreri G, Horvath P, Schweingruber C, et al. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe. 2014;16(3):403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Mocquet V, Neusiedler J, Rende F, et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J. Virol. 2012;86(14):7530–7543. doi: 10.1128/JVI.07021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano K, Ando T, Yamagishi M, et al. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect. 2013;15(6–7):491–505. doi: 10.1016/j.micinf.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Moon SL, Dodd BJ, Brackney DE, Wilusz CJ, Ebel GD, Wilusz J. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology. 2015;485:322–329. doi: 10.1016/j.virol.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnettler E, Sterken MG, Leung JY, et al. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 2012;86(24):13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54(5):599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 34.Cook HL, Lytle JR, Mischo HE, et al. Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr. Biol. 2005;15(10):974–979. doi: 10.1016/j.cub.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 35.Cazalla D, Steitz JA. Down-regulation of a host microRNA by a viral noncoding RNA. 2010;75:321–324. doi: 10.1101/sqb.2010.75.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan XC, Myer VE, Steitz JA. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11(19):2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol. Cell. Biol. 2004;24(10):4522–4533. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman RH. Viral encounters with 2´,5´-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, Jha BK, Wu A, et al. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11(6):607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA. 2008;14(6):1026–1036. doi: 10.1261/rna.958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elgadi MM, Hayes CE, Smiley JR. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 1999;73(9):7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe M, Glaunsinger B, Van Leeuwen D, et al. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl Acad. Sci. USA. 2007;104(9):3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012;86(17):9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khaperskyy DA, Schmaling S, Larkins-Ford J, Mccormick C, Gaglia MM. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog. 2016;12(2):e1005427. doi: 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7(12):e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimura T, Esteban R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl Acad. Sci. USA. 2011;108(43):17667–17671. doi: 10.1073/pnas.1111900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang J, Naitow H, Gardner NA, et al. The structural basis of recognition and removal of cellular mRNA 7-methyl G ‘caps’ by a viral capsid protein: a unique viral response to host defense. J. Mol. Recognit. 2005;18(2):158–168. doi: 10.1002/jmr.724. [DOI] [PubMed] [Google Scholar]
- 48.Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc. Natl Acad. Sci. USA. 2007;104(7):2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parrish S, Moss B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J. Virol. 2007;81(23):12973–12978. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess HM, Mohr I. Cellular 5´-3´ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe. 2015;17(3):332–344. doi: 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hastie KM, Kimberlin CR, Zandonatti MA, Macrae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3´ to 5´ exonuclease activity essential for immune suppression. Proc. Natl Acad. Sci. USA. 2011;108(6):2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moon SL, Anderson JR, Kumagai Y, et al. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18(11):2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moon SL, Blackinton JG, Anderson JR, et al. XRN1 stalling in the 5´ UTR of hepatitis C virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog. 2015;11(3):e1004708. doi: 10.1371/journal.ppat.1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169(6):871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisinger-Mathason TSK, Andrade J, Groehler AL, et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Molecular Cell. 2008;31(5):722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reineke LC, Lloyd RE. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436(2):255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30(Pt 6):963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 58.Malinowska M, Niedzwiedzka-Rystwej P, Tokarz-Deptula B, Deptula W. Stress granules (SG) and processing bodies (PB) in viral infections. Acta Biochim Pol. 2016;63(2):183–188. doi: 10.18388/abp.2015_1060. [DOI] [PubMed] [Google Scholar]
- 59.Onomoto K, Yoneyama M, Fung G, Kato H, Fujita T. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014;35(9):420–428. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coller J, Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 61.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 62.Song MG, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol. Cell. 2010;40(3):423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7(8):1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 64.Hsu CL, Stevens A. Yeast cells lacking 5´–>3´ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5’ cap structure. Mol. Cell. Biol. 1993;13(8):4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen CY, Gherzi R, Ong SE, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107(4):451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 66.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21(17):4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011;2(2):167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8(7):533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 69.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003;23(11):3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12(6):1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500(1):10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3’-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl Acad. Sci. USA. 1986;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20(11):465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 74.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29(1):246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gillis P, Malter JS. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J. Biol. Chem. 1991;266(5):3172–3177. [PubMed] [Google Scholar]
- 76.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19(6):4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13(4):246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58(2):266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deschenes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays. 2006;28(8):822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- 80.Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120(5):623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 81.Raghavan A, Robison RL, Mcnabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J. Biol. Chem. 2001;276(51):47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 82.Rivas HG, Schmaling SK, Gaglia MM. Shutoff of host–gene expression in influenza A virus and herpesviruses: similar mechanisms and common themes. Viruses. 2016;8(4):102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagoner J, Austin M, Green J, et al. Regulation of CXCL-8 (interleukin-8) induction by double-stranded RNA signaling pathways during hepatitis C virus infection. J. Virol. 2007;81(1):309–318. doi: 10.1128/JVI.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Friedrich S, Schmidt T, Schierhorn A, et al. Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45. RNA. 2016;22(10):1574–1591. doi: 10.1261/rna.055269.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rozovics JM, Chase AJ, Cathcart AL, et al. Picornavirus modification of a host mRNA decay protein. MBio. 2012;3(6):e00431–00412. doi: 10.1128/mBio.00431-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vlasova IA, Tahoe NM, Fan D, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29(2):263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vlasova-St Louis I, Dickson AM, Bohjanen PR, Wilusz CJ. CELFish ways to modulate mRNA decay. Biochim Biophys Acta. 2013;1829(6–7):695–707. doi: 10.1016/j.bbagrm.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rattenbacher B, Beisang D, Wiesner DL, et al. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol. Cell. Biol. 2010;30(16):3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Halees AS, Hitti E, Al-Saif M, et al. Global assessment of GU-rich regulatory content and function in the human transcriptome. RNA Biol. 2011;8(4):681–691. doi: 10.4161/rna.8.4.16283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beisang D, Rattenbacher B, Vlasova-St Louis IA, Bohjanen PR. Regulation of CUG-binding protein 1 (CUGBP1) binding to target transcripts upon T cell activation. J. Biol. Chem. 2012;287(2):950–960. doi: 10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beisang D, Reilly C, Bohjanen PR. Alternative polyadenylation regulates CELF1/CUGBP1 target transcripts following T cell activation. Gene. 2014;550(1):93–100. doi: 10.1016/j.gene.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Chassey B, Navratil V, Tafforeau L, et al. Hepatitis C virus infection protein network. Mol. Syst. Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang L, Hwang J, Sharma SD, et al. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 2005;280(43):36417–36428. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 94.Ramage HR, Kumar GR, Verschueren E, et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol. Cell. 2015;57(2):329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 96.Charley PA, Wilusz J. Standing your ground to exoribonucleases: function of Flavivirus long non-coding RNAs. Virus Res. 2016;212:70–77. doi: 10.1016/j.virusres.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pijlman GP, Funk A, Kondratieva N, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4(6):579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 98.Schnettler E, Tykalova H, Watson M, et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 2014;42(14):9436–9446. doi: 10.1093/nar/gku657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host–gene expression by accelerating global mRNA turnover. Mol. Cell. 2004;13(5):713–723. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 100.Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog. 2011;7(10):e1002339. doi: 10.1371/journal.ppat.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parrish S, Hurchalla M, Liu SW, Moss B. The African swine fever virus g5R protein possesses mRNA decapping activity. Virology. 2009;393(1):177–182. doi: 10.1016/j.virol.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lichinchi G, Zhao BS, Wu Y, et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20(5):666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Narayanan K, Makino S. Interplay between viruses and host mRNA degradation. Biochim Biophys Acta. 2013;1829(6–7):732–741. doi: 10.1016/j.bbagrm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]