Evolution in blockade antibody epitopes in human norovirus GII.4 2015 strains resulted in virus with antigenicity and ligand-binding properties that are significantly distinct from those for GII.4 2012 strains, potentially accounting for the endemic spread of this virus.
Keywords: Norovirus, blockade antibody, antibody neutralization, viral evolution, antigenic drift
Abstract
Background
Human noroviruses are the leading cause of acute gastroenteritis. Strains of the GII.4 genotype cause pandemic waves associated with viral evolution and subsequent antigenic drift and ligand-binding modulation. In November 2015, a novel GII.4 Sydney recombinant variant (GII.P16-GII.4 Sydney) emerged and replaced GII.Pe-GII.4 Sydney as the predominant cause of acute gastroenteritis in the 2016–2017 season in the United States.
Methods
Virus-like particles of GII.4 2012 and GII.4 2015 were compared for ligand binding and antibody reactivity, using a surrogate neutralization assay.
Results
Residue changes in the capsid between GII.4 2012 and GII.4 2015 decreased the potency of human polyclonal sera and monoclonal antibodies. A change in epitope A resulted in the complete loss of reactivity of a class of blockade antibodies and reduced levels of a second antibody class. Epitope D changes modulated monoclonal antibody potency and ligand-binding patterns.
Conclusions
Substitutions in blockade antibody epitopes between GII.4 2012 and GII.4 2015 influenced antigenicity and ligand-binding properties. Although the impact of polymerases on fitness remains uncertain, antigenic variation resulting in decreased potency of antibodies to epitope A, coupled with altered ligand binding, likely contributed significantly to the spread of GII.4 2015 and its replacement of GII.4 2012 as the predominant norovirus outbreak strain.
Human noroviruses are the primary cause of acute gastroenteritis [1–3], which has significant financial and societal costs [4]. Although >30 genotypes are known to infect humans, strains of the GII.4 genotype cause 70%–80% of norovirus outbreaks [5, 6]. About every 2–5 years, new GII.4 variant strains emerge with altered antigenicity and ligand-binding patterns [7–11]. These new viral characteristics alter susceptible populations and drive escape from herd immunity, resulting in cyclical norovirus pandemics about every 3 years over the past decade [12]. The most recent pandemic strain, GII.Pe-GII.4 Sydney (GII.4 2012), emerged in 2012 and became the dominant variant globally [13, 14], accounting for 53% of norovirus outbreaks reported in the United States during September–December 2012 [13].
Recombinant norovirus strains are frequently detected, particularly between pandemic peaks [15–17]. These recombinant strains typically consist of the polymerase of one norovirus strain coupled with the capsid gene of another norovirus strain. In several countries in East Asia, GII.4 2012 was replaced by GII.17 Kawasaki in the 2014–2015 season [18–20]. The success of this new GII.17 strain was likely driven by changes in blockade antibody epitopes [21] and, possibly, other viral features, including acquisition of a different/mutated polymerase type [20, 22]. During winter 2016–2017, a recombinant GII.2 strain with a GII.P16 polymerase emerged in Asia and Germany [23–25]. Although GII.P16 polymerases have also been found with GII.13 [17, 26] and GII.3 capsids [27], the GII.P16-GII.2 polymerase forms a different subclade [27]. Emergence of strains with capsids of rare genotypes with the GII.P16 polymerase have led some groups to suggest that the pathogenicity of these newly emergent recombinants is largely driven by fitness conferred by the GII.P16 polymerase [15, 27, 28]. Currently, there are no means of testing the effect of human norovirus polymerase activity in viral fitness or pathogenicity.
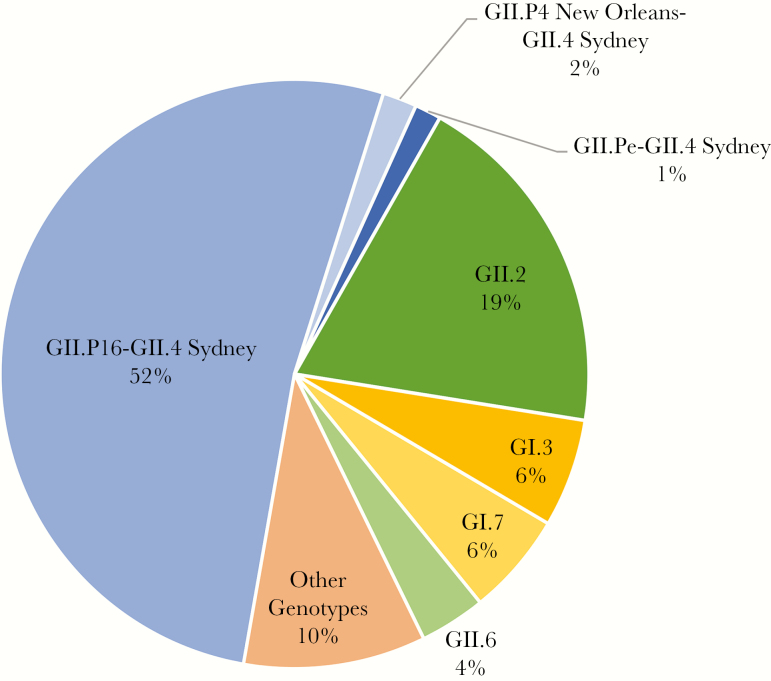
In 2014, viruses with a GII.P16 polymerase and a GII.4 2012 capsid gene were reported as a new recombinant strain, GII.P16-GII.4 Sydney (GII.4 2015) [15, 27, 29, 30]. In the United States, GII.P16-GII.4 Sydney (GII.4 2015) has been the predominant strain, accounting for >50% of norovirus outbreaks between September 2016 and October 2017 (Figure 1) [31]. Limited sequence change between GII.4 2012 and GII.4 2015 capsids and high population immunity to GII.4 2012 have been suggested to support a hypothesis of polymerase-driven fitness (eg, increased transmission and timing of viral clearance [32, 33]) as key for emergence of these strains [15, 27].
Figure 1.
Genotype distribution of 693 norovirus outbreaks in the United States, 1 September 2016–31 August 2017. Data are from [31].
Reported GII.4 2015 viruses differ from GII.4 2012 in the amino acid sequences that encode GII.4 evolving blockade antibody epitopes A and D [15, 27]. Direct testing of the effect of these sequence changes on viral antigenicity has not been reported. Epitope A is an immunodominant antigenic site. Historically, changes in epitope A at positions 294, 368, 372, and 373 correlate with loss of blockade antibody binding and emergence of new epidemiologically significant GII.4 strains [7, 8, 34, 35]. Evolving blockade antibody epitope D (residues 391 and 393–395) includes the histo-blood group antigen (HBGA) carbohydrate-binding site 2 [11, 12]. These amino acids form weak interactions with saccharide side chains distal to the L-fucose bound in the conserved carbohydrate binding site 1 [36]. Residue substitutions within epitope D have a dual effect of modulating blockade antibody potency and ligand binding through stabilizing interactions. Historically, change at residue 393 in GII.4 virus-like particles (VLPs) resulted in loss of binding of a human monoclonal antibody (mAb) [7, 34] and modulation of Lewis and B antigen binding [9, 37]. Further, substitution at residue 395 also affected binding to H type 3, Lewis, A, and B antigens [9, 37, 38]. These data illustrate that single amino acid substitutions at key residues in functional domains can have significant phenotypic influences on GII.4 strains. In this study, we compared the antigenic and ligand-binding characteristics of GII.4 2012 and GII.4 2015 and found that evolution in blockade antibody epitopes in GII.4 2015 resulted in virus with antigenicity and ligand-binding properties that were significantly distinct from those for GII.4 2012, potentially accounting for the endemic spread of GII.4 2015.
MATERIALS AND METHODS
VLPs
Norovirus ORF2 sequences were synthesized by Bio Basic (Amherst, NY), and VLPs were expressed in baby hamster kidney cells, using Venezuelan equine encephalitis virus replicons expressing norovirus ORF2, as described previously [7]. Particle integrity was confirmed by visualization of particles approximately 40 nm in diameter, using electron microscopy.
mAb Development
mAbs were developed by Genscript, using the MonoExpress protocol and GII.4.2012 VLP as immunogen. Seven mAbs were double subcloned and purified, each from a different single fused cell. mAbs were isotyped using the Pierce Rapid Isotyping Kit–Mouse (Thermo Scientific), following the manufacturer’s directions.
Enzyme Immunoassay (EIA) and Blockade of VLP-Ligand Binding Assays
EIA and blockade antibody assays were performed at 37◦C. VLPs were used at a concentration of 0.25 µg/mL [39, 40]. Mean half maximal effective concentrations (EC50 titers) and 95% confidence intervals (CIs) were determined from dose-response sigmoidal curve fits, using GraphPad 7.02 [41, 42]. An OD450 >3 times the background value, after background subtraction, was scored as a positive EIA result. Samples with values below this limit or that did not block at least 50% of VLP binding to pig gastric mucin type III (PGM; Sigma Aldrich, St. Louis) at the lowest serum dilution tested were assigned a titer of >8 (EIA) or 0.5 times the limit of detection (blockade), for statistical analysis.
VLP-Ligand Binding Assays
VLP binding to PGM was detected by rabbit polyclonal antiserum to GII.4 2012 (Cocalico Biologicals, Stevens, PA), as described elsewhere [40]. Biotinylated HBGAs (10 µg/mL; Glycotech, Gaithersberg, MD) were bound to NeutrAvidin-coated plates (ThermoFisher) for 1 hour before the addition of VLP (2 µg/mL) for 1 hour [7, 9]. Biotinylated HBGA–bound VLPs were detected as described for antibody blockade of ligand binding. Incubations were done at 37°C.
Ethics Statement
Thirty-five archived human sera samples estimated to have been collected during 2014–2016 and 14 sera collected from healthy human adult volunteers in 2016 who provided samples according to established institutional review board guidelines after informed consent were used in this study. Additional data on donor demographic characteristics are unavailable. Before use, sera were heat inactivated for 30 minutes at 56◦C. Experimentation guidelines of the Department of Health and Human Services were followed.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7.02 [7, 41]. EC50 values were log transformed for analysis. Antibody titer and PGM binding measurements were compared by means of an unpaired t test with the Welch correction, the Wilcoxon test, or the Mann-Whitney U test. A difference was considered significant if P < .05.
RESULTS
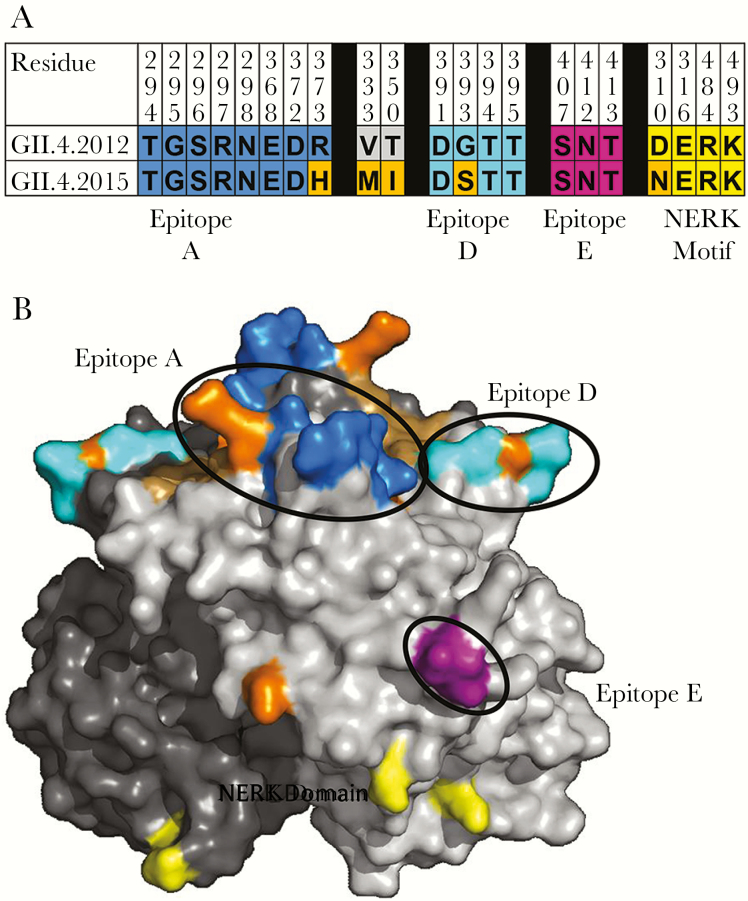
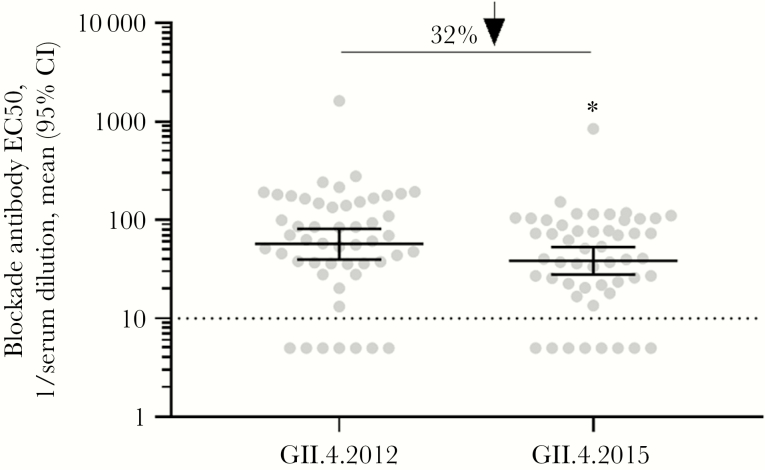
Changes in blockade antibody epitopes are associated with escape from antibody-mediated immunity. Compared with GII.4 2012, the new GII.4 2015 Sydney strain varies at 5 residues in the P2 domain (Figure 2A); residue 373 in epitope A, residue 333 in predicted epitope B, residue 350, and residue 393 in epitope D, and residue 310 in the NERK motif, a domain that regulates particle conformation and, subsequently, antibody access to occluded epitopes (Figure 2B). VP1 changes outside the P2 domain include valine to isoleucine at residues 119 and 145. Changes in residues 368 and 393 between GII.4 2012 VP1 and the preceding GII.4 variant strains GII.4 Den Haag (GII.4 2006b) and GII.4 New Orleans (GII.4 2009) were key to loss of antibody immunity [34]. To evaluate whether the changes in blockade antibody epitopes between GII.4 2012 and GII.4 2015 are significant enough to cause phenotypic changes in blockade antibody potency, we compared the blockade antibody titers of 49 human serum samples collected from 2014 through 2016 for GII.4 2012 and GII.4 2015. At the polyclonal sera level, GII.4 2015 titers were significantly less than GII.4 2012 titers (P < .001, by the Wilcoxon test; Figure 3). Geometric mean titers decreased 32%, from 56.9 (95% CI, 53.14–81.24) for GII.4 2012 to 38.54 (95% CI, 27.96–39.85) for GII.4 2015.
Figure 2.
Capsid residues defining antigenic and ligand-binding sites differ between GII.4 2012 and GII.4 2015. A, GII.4 2012 and GII.4 2015 differ at 5 residues in the P2 subdomain of the major capsid protein (orange). B, Characterized blockade antibody epitopes and residue changes between GII.4 2012 (JX459908) and GII.4 2015 (KX907727) were highlighted onto the P domain dimer of GII.4 2012 [34] (residue 333 is not visible in this view). Residues within epitopes A (blue) and D (cyan) and the NERK motif (yellow) of the major capsid protein differ by single amino acid changes between GII.4 2012 and GII.4 2015 (orange). The carbohydrate-binding domain remains conserved (brown).
Figure 3.
Population preexisting antibody responses are less robust for GII.4 2015 as compared to GII.4 2012. Archived sera collected between 2014 and 2016 from healthy blood donors were evaluated for blockade of GII.4 2012 and GII.4 2015 virus-like particle binding to ligand. The mean percentage of control binding as compared to no antibody was fit with sigmoidal dose-response curve analysis, and half-maximal effective concentrations (EC50 values) were calculated. CI, confidence interval. *P < .05, by the Wilcoxon test, compared with GII.4 2012.
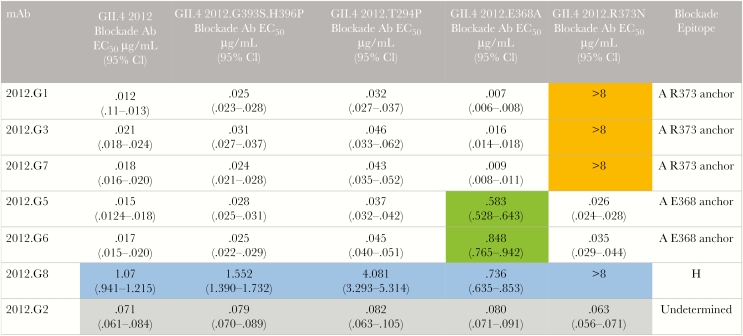
Blockade antibody epitope A accounts for approximately 40% of the total serum blockade antibody activity [35]. To determine whether the change at residue 373 of epitope A may account for the 32% decrease in the blockade antibody geometric mean titer, we developed a panel of mouse mAbs to GII.4 2012 and characterized these mAbs by EIA, measurement of blockade potency, and epitope mapping. Five mAbs were specific for GII.4 2012. One mAb cross-reacted with GII.4 2009 and another with 2009, 1997, and 1987 GII.4 viruses (Supplementary Table 1). All 7 mAbs blocked binding of GII.4 2012 to carbohydrate ligand (Figure 4). The mAbs were evaluated for blockade potency, using a panel of GII.4 2012 VLPs with mutations in epitopes A and D [34]. Five GII.4 2012–specific mAbs lost blockade potency when epitope A residues were modified. Blockade function of mAbs 2012.G1, G3, and G7 was lost in GII.4 2012 R373N. Further, mAb blockade potency decreased by 39% for 2012.G5 and by 50% for 2012.G6 for GII.4 2012.E368A. These data identified 5 epitope A–specific mAbs with 2 different contact footprints, one anchoring at residue 373 and another at residue 368 (Figure 4). 2012.G8 has a unique binding and blockade pattern, indicating that it recognizes an undefined epitope H. Although 2012.G8 is able to bind several GII.4 strain VLPs, it only blocks GII.4 2012. The blockade potency of 2012.G8 was ablated by the R373N change, identifying epitope H to be influenced by epitope A, as has been shown for GII.4 2006.G6 and GII.4 1987.G1 [8, 35]. 2012.G8 binding did not compete with binding of epitope A human mAb NVB43.9 [7] to GII.4 2009 (data not shown), supporting its new designation as epitope H. Targeted changes in epitope A or D did not affect blockade potency of 2012.G2, and this epitope is yet undefined.
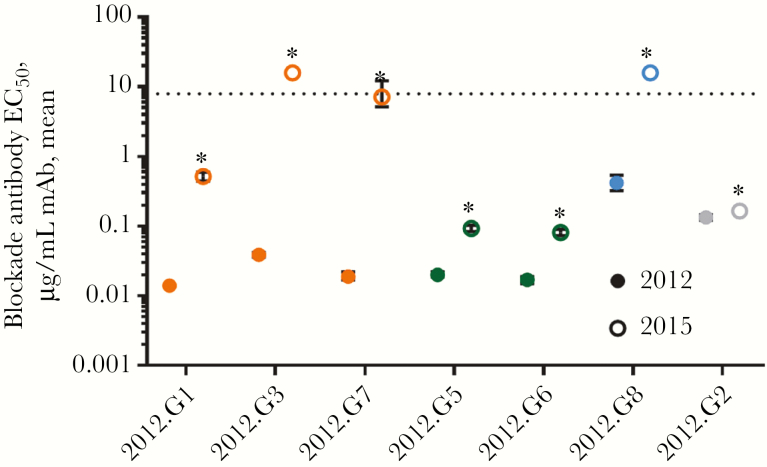
Figure 4.
Epitope mapping of GII.4 2012 monoclonal antibodies (mAbs). mAbs to GII.4 2012 were evaluated for reactivity to a panel of virus-like particles with mutations in epitopes A and D in GII.4 2012, to map binding residues. Five mAbs recognize epitope A. Three anchor at residue 373 (orange) and 2 at 368 (green). 2012.G8 recognizes a novel cross-reactive undefined epitope H (blue). The epitope for 2012.G2 is undetermined (gray). Ab, antibody; CI, confidence interval; EC50, half-maximal effective concentration.
Characterization of 7 antibodies yielded 4 binding patterns to GII.4 2012 blockade antibody epitopes. An unpaired t test with the Welch correction revealed that antibody blockade of GII.4 2015 VLP-ligand binding was significantly reduced for each mAb tested, compared with blockade of GII.4 2012 (Figure 5). Blockade by epitope A mAbs anchoring on residue 373 were most affected. For these mAbs, blockade Ab titers decreased 36.9-fold (for 2012.G1), 410.3-fold (for 2012.G3), and 377.4-fold (for 2012.G7). mAbs to epitope A anchoring at residue 368 were less affected, with reduction in the blockade antibody titer by 4.7-fold (for 2012.G5) and 4.8-fold (for 2012.G6). The 2012.G8 blockade titer was reduced 38-fold. The 2012.G2 titer changed only 1.2-fold, demonstrating that some blockade epitopes are conserved between GII.4 2012 and GII.4 2015 and that the sharp loss of blockade potency for high-avidity epitope A mAbs is not the result of particle malformation, but instead the product of viral evolution in evolving blockade antibody epitopes.
Figure 5.
GII.4 2015 is antigenically distinguishable from GII.4 2012 at multiple blockade antibody epitopes. Monoclonal antibodies (mAbs) to GII.4 2012 were evaluated for the blockade potency of GII.4 2012 (closed circles) and GII.4 2015 (open circles) binding to ligand. The mean percentage of control binding as compared to no antibody was fit with sigmoidal dose-response curve analysis, and half-maximal effective concentrations (EC50 values) were calculated. Blockade potency was severely diminished for mAbs that anchor on residue 373 of epitope A (orange) and epitope H (blue) and were reduced for mAbs that anchor on residue 368 (green). The epitope for 2012.G2 is undetermined (gray). Error bars represent the 95% confidence intervals. *P < .05, by an unpaired t test with the Welch correction, compared with GII.4 2012.
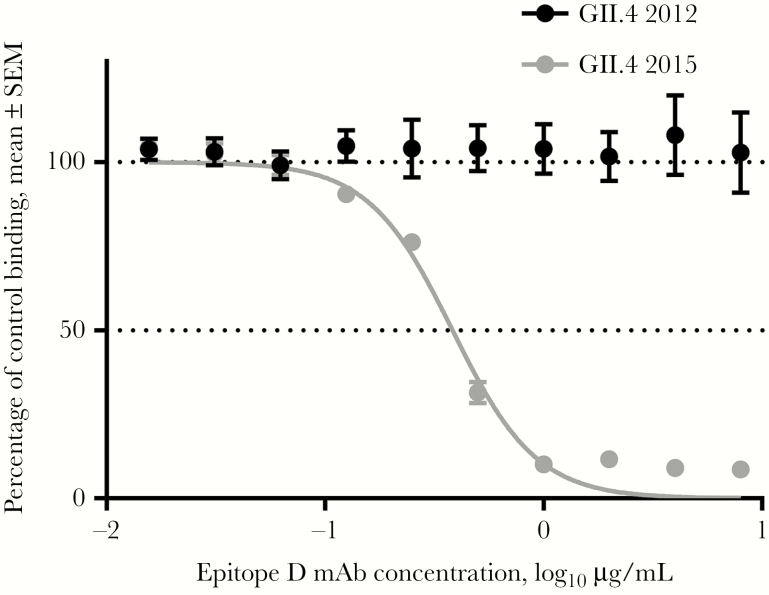
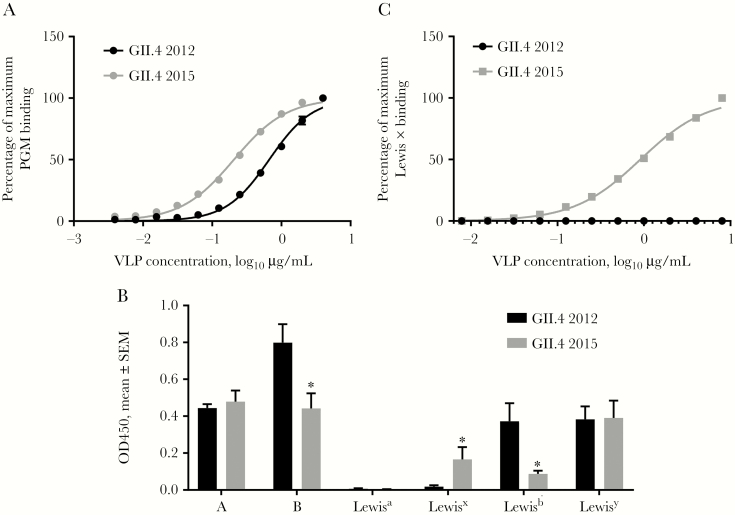
In addition to epitope A changes, a key change at residue 393 between GII.4 2012 and the preceding pandemic strain GII.4 2009 resulted in loss of binding of human mAb NVB 97 [7] to epitope D [34]. In the GII.4 2015 VP1, G393 reverted back to S393, and blockade of NVB 97 was restored (Figure 6), confirming residue 393 as an anchor for this human mAb. Residue 393 also stabilizes the interaction between branched carbohydrate moieties extending out of the conserved carbohydrate pocket, mediating affinity for different ligands and, potentially, susceptible populations [9, 37, 38]. GII.4 Sydney 2015 bound 3 times more tightly to PGM (EC50, 0.21; 95% CI, .19–.22), compared with GII.4 2012 (EC50, 0.67; 95% CI, .62–.71; P < .0001, by an unpaired t test with the Welch correction; Figure 7A). The fucose-binding domain of GII.4 2015 is unchanged as compared to other GII.4 VLPs, maintaining the H antigen binding site. Both GII.4 2012 and GII.4 2015 VLPs bind to a broad range of HBGA moieties, including A, B, and Lewis antigens (Figure 7B), as previously reported for GII.4 strains [9]. GII.4 2015 uniquely bound to Lewis x. Lewis x binding was confirmed to be dose dependent (Figure 7C). These data support residue 393 as a key modulator of carbohydrate binding and demonstrate the effect of a single residue substitution on ligand-binding preferences and potentially susceptible host populations.
Figure 6.
Changes in epitope D of GII.4 2015 alter blockade antibody binding. GII.4 2012 and 2015 virus-like particles were evaluated for blockade of ligand binding by human monoclonal antibody (mAb) to epitope D, NVB 97. The mean percentage of control binding as compared to no antibody was fit with sigmoidal dose-response curve analysis. SEM, standard error of the mean.
Figure 7. .
Changes in epitope D of GII.4 2015 alter ligand binding. GII.4 2012 and 2015 virus-like particles (VLPs) were evaluated for binding to carbohydrate moieties. A, VLP binding to PGM. B, VLP binding to synthetic biotinylated histo-blood group antigens. SEM, standard error of the mean. *P < .05, by the Mann-Whitney U test, compared with GII.4 2012. C, VLP binding to biotinylated Lewis x.
DISCUSSION
Mechanisms of the emergence of pandemic GII.4 human norovirus include antigenic drift (ie, evolution in specific epitopes) [7, 43], recombination between polymerase and capsid sequences from different strains [14], expanded host range (ie, broad binding of HBGAs types) [9, 44], and increased symptoms/extended duration of shedding relative to other genotypes [45, 46]. By comparing the capsid features of GII.4 2012 and GII.4 2015, we demonstrated that antigenic drift is a major driving force for new GII.4 strains even during periods of endemic (subpandemic) levels of disease. The question of how much change is needed between strains to generate a new dominant variant is just beginning to be explored. The threshold is dependent on the position of the changed residue [7, 47]. How the number of residue changes and the order of these changes affect viral fitness are unknown. Yet, this information is vital to our ability to develop effective surveillance approaches that could influence public health outcomes through behavior changes that could mitigate disease spread or through vaccination programs in which strain reformulation may be needed.
Substitutions in epitope A are associated with pandemic strain emergence [7, 47]. Eight known residues of epitope A have been identified, with 4 footprints for antibody binding: linear 294–298 anchor, 368 anchor, 373 anchor, and a rare group of antibodies that have some cross-reactivity with other GII.4 strains but are still dependent on epitope A residues for binding [8, 34, 35]. Here, the GII.4 2015 R373H substitution results in loss of blockade for the 3 mAbs that anchor at 373. Of note, E368 is conserved between GII.4 2012 and GII.4 2015, yet the 2 mAbs that anchor at 368 were reduced in potency by 5-fold. 368 and 373 are on opposite sides of epitope A, making it unlikely that the R373H change in epitope A is accounting for the decrease in 368 anchor mAbs. Modeling of the polar interactions identified 2 bonds between E368 and either T350, found in GII.4 2012, or I350, found in GII.4 2015. The exchange of the larger hydrophilic threonine for the smaller, more hydrophobic isoleucine may be affecting the conformation of E368 presentation for antibody binding, thus reducing antibody affinity for epitope A mAbs that anchor at E368. If true, then 350 is another example of a residue outside an epitope that can regulate antibody blockade potency, likely by regulating local epitope conformation [40]. Awareness of how and which residues outside antibody epitopes may influence antibody neutralization will be an important component in developing sophisticated surveillance systems able to detect prepandemic strain changes.
GII.4 2015 viruses restored epitope D (393–395) to STT and subsequently restored blockade potency of the human mAb to epitope D. Affinity for different HBGAs is mediated by residues of epitope D [9, 38]. Here, G393S translated to improved binding to the biologically relevant carbohydrates in PGM and synthetic Lewis x. It is not known whether altered carbohydrate preference coupled with the loss of 373 mAbs is enough to expand the virus into new populations in the context of high herd immunity. At the population level, these changes decreased sera blockade potency by 32%. In comparison, blockade antibody titer in convalescent serum from people infected with GII.4 2009 lost approximately 70% of the blockade titer for GII.4 2012 [34].
The primary limitation of this study is the lack of ability to test the effects of specific residue changes on virus binding, entry, and neutralization. It is also important to note that the sequences studied here are representative strains. In a pandemic environment, a strain exists as a quasi-species with some variation in sequence [27]. For example, GII.4 2012 strains with G393 and S393 cocirculate. It is possible that cocirculation of strains with varied ligand-binding preferences may serve to expand the susceptible host pool at the genetic level. Furthermore, viruses within the quasi-species will escape antibody-mediated protection and spread, be attenuated by low-titer cross-reactive antibodies, or be neutralized and become extinct. The strains in the middle between escape and extinction may extend herd immunity by boosting cross-reactive antibodies to slightly altered epitopes, possibly extending the life-span of either natural or vaccine-induced immunity.
Like other RNA viruses, including influenza A virus and human immunodeficiency virus [48, 49], recombinant noroviruses contribute to global disease burden [15]. The role of human norovirus RNA polymerases in strain dominance is not clear, and replication models are needed to determine the effect of polymerase activity on viral fitness. Early studies suggested that GII.4 polymerases had higher mutation rates, potentially explaining high evolution rates [50]. At this time, it is uncertain whether any of the other human norovirus RNA polymerases have altered fidelity rates as compared to the early GII.4 strains. Nor is it clear whether human norovirus polymerases encode any virulence determinants or are the target for strong T-cell responses. Further investigation of the role of polymerases are warranted, but this study clearly demonstrates that GII.4 Sydney strains are undergoing significant antigenic and ligand-binding change over time, providing direct evidence for the dominance of the GII.4 2015 strains as compared to GII.4 2012. These data support the fundamental role of viral evolution of the capsid gene in norovirus emergence and persistence. Time will reveal whether the GII.P16/GII.2012 capsid strain is a final variant of the GII.4 Sydney viruses as herd immunity drives them toward extinction or whether continued evolution at residue 368 could play an important role and eliminate a second class of epitope A blockade antibodies, potentially negating herd immunity and initiating a new norovirus pandemic.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Victoria Madden of the Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, University of North Carolina–Chapel Hill, for expert technical support.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the National Institute of Health, Allergy, and Infectious Diseases, National Institutes of Health (NIH; grants R56 A15-0756 and U19 AI109761 CETR), the Wellcome Trust (grant A17-0915-001), and the NIH (grant P30 AI50410 to the University of North Carolina at Chapel Hill Center for AIDS Research).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Riddle MS, Walker RI. Status of vaccine research and development for norovirus. Vaccine 2016; 34:2895–9. [DOI] [PubMed] [Google Scholar]
- 2. Pringle K, Lopman B, Vega E, Vinje J, Parashar UD, Hall AJ. Noroviruses: epidemiology, immunity and prospects for prevention. Future Microbiol 2015; 10:53–67. [DOI] [PubMed] [Google Scholar]
- 3. Hall AJ, Glass RI, Parashar UD. New insights into the global burden of noroviruses and opportunities for prevention. Expert Rev Vaccines 2016; 15:949–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One 2016; 11:e0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoa Tran TN, Trainor E, Nakagomi T, Cunliffe NA, Nakagomi O. Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants. J Clin Virol 2013; 56:185–93. [DOI] [PubMed] [Google Scholar]
- 6. Zhou HL, Zhen SS, Wang JX et al. Burden of acute gastroenteritis caused by norovirus in China: A systematic review. J Infect 2017; 75:216–24. [DOI] [PubMed] [Google Scholar]
- 7. Lindesmith LC, Beltramello M, Donaldson EF et al. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog 2012; 8:e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindesmith LC, Donaldson EF, Baric RS. Norovirus GII.4 strain antigenic variation. J Virol 2011; 85:231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindesmith LC, Donaldson EF, Lobue AD et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med 2008; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Rougemont A, Ambert-Balay K, Belliot G, Pothier P. [Norovirus infections: an overview]. Med Sci (Paris) 2010; 26:73–8. [DOI] [PubMed] [Google Scholar]
- 11. Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J Virol 2011; 85:8635–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol 2010; 8:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emergence of new norovirus strain GII.4 Sydney--United States, 2012. MMWR Morb Mortal Wkly Rep 2013; 62:55. [PMC free article] [PubMed] [Google Scholar]
- 14. Eden JS, Hewitt J, Lim KL et al. The emergence and evolution of the novel epidemic norovirus GII.4 variant Sydney 2012. Virology 2014; 450–451:106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cannon JL, Barclay L, Collins NR et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol 2017; 55:2208–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Supadej K, Khamrin P, Kumthip K et al. Wide variety of recombinant strains of norovirus GII in pediatric patients hospitalized with acute gastroenteritis in Thailand during 2005 to 2015. Infect Genet Evol 2017; 52:44–51. [DOI] [PubMed] [Google Scholar]
- 17. Lim KL, Hewitt J, Sitabkhan A et al. A multi-site study of norovirus molecular epidemiology in Australia and New Zealand, 2013–2014. PLoS One 2016; 11:e0145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parra GI, Green KY. Genome of emerging norovirus GII.17, United States, 2014. Emerg Infect Dis 2015; 21:1477–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu J, Fang L, Zheng H et al. The evolution and transmission of epidemic GII.17 noroviruses. J Infect Dis 2016; 214:556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Graaf M, van Beek J, Vennema H et al. Emergence of a novel GII.17 norovirus - End of the GII.4 era?Euro Surveill 2015; 20 pii: 21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindesmith LC, Kocher JF, Donaldson EF et al. Emergence of novel human norovirus GII.17 strains correlates with changes in blockade antibody epitopes. J Infect Dis 2017; 216:1227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen H, Qian F, Xu J et al. A novel norovirus GII.17 lineage contributed to adult gastroenteritis in Shanghai, China, during the winter of 2014–2015. Emerg Microbes Infect 2015; 4:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizukoshi F, Nagasawa K, Doan YH et al. Molecular evolution of the RNA-dependent RNA polymerase and capsid genes of human norovirus genotype GII.2 in Japan during 2004–2015. Frontiers Microbiol 2017; 8:705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu LT, Kuo TY, Wu CY, Liao WT, Hall AJ, Wu FT. Recombinant GII.P16-GII.2 Norovirus, Taiwan, 2016. Emerg Infect Dis 2017; 23:1180–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niendorf S, Jacobsen S, Faber M et al. Steep rise in norovirus cases and emergence of a new recombinant strain GII.P16-GII.2, Germany, winter 2016. Euro Surveill 2017; 22 pii: 30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khamrin P, Kumthip K, Supadej K et al. Noroviruses and sapoviruses associated with acute gastroenteritis in pediatric patients in Thailand: increased detection of recombinant norovirus GII.P16/GII.13 strains. Arch Virol 2017; 162:3371–80. [DOI] [PubMed] [Google Scholar]
- 27. Ruis C, Roy S, Brown JR, Allen DJ, Goldstein RA, Breuer J. The emerging GII.P16-GII.4 Sydney 2012 norovirus lineage is circulating worldwide, arose by late-2014 and contains polymerase changes that may increase virus transmission. PLoS One 2017; 12:e0179572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tohma K, Lepore CJ, Ford-Siltz LA, Parra GI. Phylogenetic analyses suggest that factors other than the capsid protein play a role in the epidemic potential of GII.2 norovirus. mSphere 2017; 2 pii: e00187-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bidalot M, Thery L, Kaplon J, De Rougemont A, Ambert-Balay K. Emergence of new recombinant noroviruses GII.p16-GII.4 and GII.p16-GII.2, France, winter 2016 to 2017. Euro Surveill 2017; 22 pii: 30508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi YS, Koo ES, Kim MS, Choi JD, Shin Y, Jeong YS. Re-emergence of a GII.4 norovirus Sydney 2012 variant equipped with GII.P16 RdRp and Its predominance over novel variants of GII.17 in South Korea in 2016. Food Environ Virol 2017; 9:168–78. [DOI] [PubMed] [Google Scholar]
- 31. Centers for Disease Control and Prevention. Number of confirmed norovirus outbreaks submitted to CaliciNet by state September 1, 2017–October 31, 2017 https://www.cdc.gov/norovirus/reporting/calicinet/data.html. Accessed 2 December 2017.
- 32. Arias A, Thorne L, Ghurburrun E, Bailey D, Goodfellow I. Norovirus polymerase fidelity contributes to viral transmission in vivo. MSphere 2016; 1:e00279–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arias A, Thorne L, Goodfellow I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife 2014; 3:e03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Debbink K, Lindesmith LC, Donaldson EF et al. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J Infect Dis 2013; 208:1877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindesmith LC, Costantini V, Swanstrom J et al. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J Virol 2013; 87:2803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao S, Lou Z, Tan M et al. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 2007; 81:5949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J Virol 2011; 85:8635–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Rougemont A, Ruvoen-Clouet N, Simon B et al. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J Virol 2011; 85:4057–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindesmith LC, Beltramello M, Swanstrom J et al. Serum immunoglobulin A cross-strain blockade of human noroviruses. Open Forum Infect Diseas 2015; 2:ofv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lindesmith LC, Donaldson EF, Beltramello M et al. Particle conformation regulates antibody access to a conserved GII.4 norovirus blockade epitope. J Virol 2014; 88:8826–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lindesmith LC, Ferris MT, Mullan CW et al. Broad blockade antibody responses in human volunteers after immunization with a multivalent norovirus VLP candidate vaccine: immunological analyses from a phase I clinical trial. PLoS Med 2015; 12:e1001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindesmith LC, Mallory ML, Jones TA et al. Impact of pre-exposure history and host genetics on antibody avidity following norovirus vaccination. J Infect Dis 2017; 215:984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Debbink K, Lindesmith LC, Donaldson EF, Baric RS. Norovirus immunity and the great escape. PLoS Pathog 2012; 8:e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan M, Jiang X. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog 2010; 6:e1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Desai R, Hembree CD, Handel A et al. Severe outcomes are associated with genogroup 2 genotype 4 norovirus outbreaks: a systematic literature review. Clin Infect Dis 2012; 55:189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zelner JL, Lopman BA, Hall AJ, Ballesteros S, Grenfell BT. Linking time-varying symptomatology and intensity of infectiousness to patterns of norovirus transmission. PLoS One 2013; 8:e68413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Allen DJ, Gray JJ, Gallimore CI, Xerry J, Iturriza-Gómara M. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 2008; 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McDonald SM, Nelson MI, Turner PE, Patton JT. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat Rev Microbiol 2016; 14:448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev 2009; 73:451–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog 2010; 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.