Abstract
Background: HPV16 is a common sexually transmitted infection although few infections lead to cervical precancer/cancer; we cannot distinguish nor mechanistically explain why only certain infections progress. HPV16 can be classified into four main evolutionary-derived variant lineages (A, B, C, D) that have been previously suggested to have varying disease risks.
Methods: We used a high-throughput HPV16 whole-genome sequencing assay to investigate variant lineage risk among 3215 HPV16-infected women. Using sublineages A1/A2 as the reference, we assessed all variant lineage associations with infection outcome over three or more years of follow-up: 1107 control subjects (<CIN2), 906 CIN2, 1008 CIN3, 69 squamous cell carcinomas (SCC), 85 adenocarcinomas in situ (AIS), and 40 adenocarcinomas. Logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs). All statistical tests were two-sided.
Results: A4 sublineage was associated with an increased risk of cancer, specifically adenocarcinoma (OR = 9.81, 95% CI = 2.02 to 47.69, P = 4.7x10 −03 ). Lineage B had a lower risk of CIN3 (OR = 0.51, 95% CI = 0. 28 to 0.91, P = .02) while lineage C showed increased risk (OR = 2.06, 95% CI = 1.09 to 3.89, P = .03). D2/D3 sublineages were strongly associated with an increased risk of CIN3 and cancer, particularly D2 (OR for cancer = 28.48, 95% CI = 9.27 to 87.55, P = 5.0x10 −09 ). D2 had the strongest increased risk of glandular lesions, AIS (OR = 29.22, 95% CI = 8.94 to 95.51, P = 2.3x10 −08 ), and adenocarcinomas (OR = 137.34, 95% CI = 37.21 to 506.88, P = 1.5x10 −13 ). Moreover, the risk of precancer and cancer for specific variant lineages varied by a women’s race/ethnicity; those women whose race/ethnicity matched that of the infecting HPV16 variant had an increased risk of CIN3 + ( P < .001).
Conclusions: Specific HPV16 variant sublineages strongly influence risk of histologic types of precancer and cancer, and viral genetic variation may help explain its unique carcinogenic properties.
Most of the approximately 200 types of human papillomavirus (HPV) infections cause no evident pathology or only benign papillomas (warts). Nevertheless, 13 types from one clade of the alpha genus ( 1 ) of HPV (including HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 [ 2 ]) cause virtually all cases of cervical cancer and a large proportion of other anogenital and oropharyngeal cancers. More than half a million people each year are afflicted with an HPV-related cancer worldwide ( 3 ). These carcinogenic (or high risk [HR]) HPV infections are extremely common and easily transmitted to susceptible epithelia by direct physical contact ( 4 ). Only a small proportion of HR infections persist and progress to precancer that in turn can ultimately lead to cancer ( 5 , 6 ). We do not know why these common and typically benign infections sometimes cause cancer. Previous studies suggest that viral genetic variation is associated with cancer risk (for review, see [ 7 ]).
HPV types differ from each other by at least 10% in the highly conserved L1 (major capsid protein) gene sequence ( 4 ). Of the HR types, HPV16 is uniquely prevalent in virtually all regions of the world (suggesting greater “fitness”) and, when persistent, is uniquely likely to cause precancer/cancer. As a result, HPV16 causes half of cervical cancers worldwide and is one of the most important human carcinogens ( 8 , 9 ).
While recognizing the eventual long-term promise of cancer prevention by prophylactic vaccination against HPV16, we continue to study the genotypic determinants of HPV16-induced cervical precancer and cancer to understand viral carcinogenesis and possibly to improve preventive strategies further. Our newly developed, high-throughput, next-generation HPV16 whole-genome sequencing method ( 10 ) permits much larger studies and more detailed genotype-phenotype examinations of the HPV genome than previously possible.
HPV16 can be divided into four main variant lineages and nine sublineages, differing in L1 sequence by less than 10% for main variants and as little as 0.5% for sublineages ( 11 ). This evolutionary genetic variation within HPV16 has already been linked by studies using partial sequencing to substantial differences in cervical carcinogenicity ( 12–22 ). Some authors ( 21 , 23–25 ), but not others ( 26 , 27 ), suggest that different HPV16 variants might be associated with glandular vs squamous histology. More controversially, there have been a few reports suggesting that co-evolution of HPV16 and humans influences cancer risk ( 14 , 28 ) while other studies have not confirmed this relationship ( 29 , 30 ).
Using a novel next-generation sequencing method, we conducted a very large case-control study of HPV16 variant lineages and risk of precancer/cancer, taking advantage of the routine use of HPV testing in cervical screening and linkage to electronic medical records at Kaiser Permanente Northern California (KPNC). This study extends what is known about HPV16 genetic variation and its impact on cervical carcinogenicity.
Methods
Study Population
Beginning in 2003, women age 30 years and older at KPNC have been routinely screened every three years for cervical cancer by “co-testing,” ie, by testing for a pool of 13 carcinogenic HPV types using the Hybrid Capture 2 assay (HC2; Qiagen) along with cervical cytology. In addition, women age 21 to 30 years with an equivocal cytologic result were triaged using the HC2 test. The Persistence and Progression (PaP) cohort is a repository of more than 110 000 residual HC2 specimens from exfoliated cervical cells from approximately 55 000 women, obtained during routine cervical cancer testing at KPNC between December 2006 and January 2011. Approximately 45 000 HPV-positive enrollment specimens were collected in specimen transport medium (STM; Qiagen Inc., Gaithersburg, MD) ( 31 ). This represents the majority of HC2-positive women during that time period. A random sample of HPV-negative specimens from the same time period was stored as well. We contacted women whose residual specimens were stored and asked them to opt out if they wished not to participate in testing for HPV-related biomarkers, including HPV genotypes; approximately 8% did not participate, and their specimens were destroyed. De-identified data including age, race and ethnicity, and follow-up cytologic and pathologic results were obtained from electronic health records. The study protocol was reviewed and approved by the National Cancer Institute Institutional Review Board.
This present study of HPV16 variant lineages included specimens from 3579 HC2-positive women, previously found to contain HPV16 DNA ( 31 ): The study population included 1032 women diagnosed with cervical intraepithelial neoplasia grade 2 (CIN2), 1079 CIN grade 3 (CIN3), and 71 squamous cell carcinoma (SCC) cases, 91 adenocarcinoma in situ (AIS) and 41 adenocarcinoma, and 1265 women with less than CIN2 (including 902 with <CIN2 histology and 363 with no histology during our study, biopsy not indicated in these women). The case patients were diagnosed at enrollment or during the study follow-up period after the baseline specimens were collected. The control subjects were defined as women having enrollment specimens with HPV16 DNA and no histologic evidence of equivocal precancer or worse (CIN2+) during the follow-up study period. Women included in our study were followed for a median of 4.3 years (interquartile range = 3.36, range = 7.85). Women are followed as long as possible and only censored if they received treatment for a CIN2+ lesion or if they were lost to follow-up. Self-reported race and ethnicity for women in the PaP cohort population were obtained from the electronic health records. Women were categorized as white, Asian or Pacific Islander, African American, Hispanic, multiracial, or other/unknown.
HPV16 Detection and DNA Isolation
DNA was extracted from the banked STM specimens as previously described ( 35 ). Typing methods varied for different subsets of the cohort, and some were tested with multiple methods. The Burk laboratory (The Bronx, NY) used MY09/M11 L1 degenerate primer PCR (MY09/11 PCR) and type-specific dot-blot hybridization methods to type HC2-positive STM specimens (n = 1013) ( 35 , 36 ). Another large group of enrollment specimens was tested using Linear Array and Cobas (n = 2035) by Roche Molecular Systems (Pleasanton, CA). A third group of specimens was typed by BD using Onclarity (n = 911; Sparks, MD).
Ion Torrent Library Preparation and Sequencing
We used a custom Thermo Fisher Ion Torrent AmpliSeq HPV16 panel approach to amplify the entire 7906 bp HPV16 genome as previously described ( 10 ). In brief, our next-generation sequencing (NGS) assay used the Thermo Fisher Life Sciences’ Ion Torrent Proton and a custom HPV16 Ion Ampliseq panel of 48 multiplexed primers. After amplification, an Ion Torrent adapter-ligated library was generated following the manufacturer’s Ion AmpliSeq Library Preparation kit 2.0-96LV protocol (Life Technologies, Part #4480441) and sequenced on the Proton. Raw sequencing reads generated by the Ion Torrent Proton were quality and adaptor trimmed by the Ion Torrent Suite and aligned to the HPV16 reference sequence (7906bp [ 37 ]). All BAM files were processed through a custom quality control and in-house analysis pipeline. The HPV16 sequences were genotyped by the GATK UnifiedGenotyper. SNP calls were made by the GATK UnifiedGenotyper (SNP discovery mode). Low-quality SNP calls and SNP clusters were filtered and masked. A custom in-house HPV16 annotation database was used to annotate identified nucleotide variants to HPV gene/region.
HPV16 Variant Lineage Classification
HPV16 variant lineage assignment was based on the maximum likelihood (ML) tree topology constructed using RAxML MPI v7.2.8.27 ( 38 ), including 16 HPV16 A, B, C, and D variant lineage reference sequences ( 11 ), and lineage assignments were confirmed with SNP patterns. In case of multiple variants present in a specimen, a predominant variant was assigned based on presence in at least 60% of the sequence reads. A total of 364 specimens (10.2%) were removed because of poor read depth (as previously described [10]), poor or spotty coverage across the genome, or poor resolution in the phylogenetic tree (ie, a definitive lineage could not be assigned); we used a stringent quality control threshold to minimize genotyping or lineage errors in our analysis.
HPV16 Phylogenetic Nomenclature
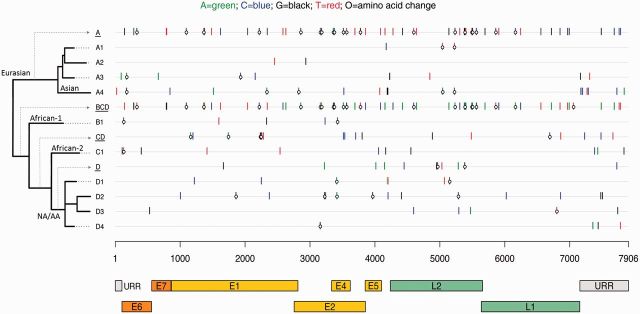
HPV16 variant lineages have co-evolved with specific human populations ( 32 , 33 ) and were named originally based on epidemiologic observations that variants were most prevalent in a particular geographic region. The HPV16 major evolutionary branchings are now known as A, B, C, and D ( 11 ). “European” variants (now sublineages A1, A2, and A3) have been found commonly in European populations, “African” variants (lineages B [African-1] and C [African-2]) in Africa, “North-American” variant (sublineage D1) in North American populations, “Asian-American” variants (sublineages D2 and D3) in Asia, American Indians, Central and South America, and Spain, and the “Asian” variant (sublineage A4) in Southeast Asia ( 33 , 34 ). Current phylogenic approaches abandon geographic assignments because of their increasing uncertainty. HPV16 variant lineages/sublineages and sublineage-specific nucleotide and amino acid differences are illustrated in Figure 1 .
Figure 1.
Phylogenetic tree and sublineage-specific nucleotide and amino acid changes in HPV16 sublineages. The x-axis shows the HPV16 genome positions and gene regions, aligned according to sublineage in the phylogenetic tree on the y-axis . Sublineage-specific single-nucleotide polymorphisms (SNPs) were determined from an alignment of 63 complete-genome nucleotide sequences by selecting the nucleotides that occurred only in members of a given sublineage. The nucleotide of each SNP is color-coded as shown at the top figure legend , and the open circles represent nonsynonymous changes. Amino acid changes in the E2/E4 gene region are changes observed in E2. SNPs for a given lineage are cumulative as the tree is traversed from deepest node out to finer sublineage branches. For example, A ( underlined, first row ) changes are found in all A sublineage genomes plus the changes on each specific A sublineage row; and the D4 sublineage ( last row ) contains all changes shown on the BCD ( underlined, row 6 ), CD ( underlined, row 8 ), D ( underlined, row 10 ), and D4 ( row 14 ) lines. E1 = early gene 1; E2 = early gene 2; E4 = early gene 4; E5 = early gene 5; E6 = early gene 6; E7 = early gene 7; L1 = late gene 1; L2 = late gene 2; NA/AA = North American/Asian-American; URR = upstream regulatory region.
Statistical Analyses
A logistic regression model was used to obtain the odds ratios (ORs) and 95% confidence intervals (CIs) for precancer and cancer risk using the control subjects (ie, women with HPV16 and <CIN2) as the referent group. Risk associations were also performed, stratified by the women’s self-reported race/ethnicity. For the variant lineage risk associations stratified by race/ethnicity, the D2 and D3 lineages were combined because of small numbers and similarity in their risk associations. Statistical analyses were performed with SPSS version 21.0 and R version 3.1.2; all statistical tests were two-sided. A P value of less than .05 was considered statistically significant.
Results
Characteristics of the Study Population
The analysis set consisted of 1107 control subjects (<CIN2), 906 CIN2, 1008 CIN3, 69 SCC, 85 AIS, and 40 adenocarcinoma. The study population and characteristics of the women by HPV16 variant lineage are shown in Table 1 . Most women in our study (2805, 87.2%) had an HPV16 A variant lineage infection, including: 2300 A1 (71.5%), 423 A2 (13.2%), 19 A3 (0.6%), and 63 A4 (2.0%). There were 410 women (12.8%) with an HPV16 non-A variant lineage infection: 79 B (2.5%), 75 C (2.3%), and 256 D (8.0%).
Table 1.
Counts and characteristics of each HPV16 variant sublineage detected in 3215 women in the NCI-KPNC PaP cohort
| Characteristics | A1 | A2 | A3 | A4 | B1 | C1 | D1 | D2 | D3 | D4 | Total | P * |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||
| Age at enrollment, y | ||||||||||||
| 21-29 | 674 (71.5) | 143 (15.2) | 2 (0.2) | 3 (0.3) | 17 (1.8) | 35 (3.7) | 3 (0.3) | 7 (0.7) | 51 (5.4) | 7 (0.7) | 942 (100) | |
| 30-39 | 944 (71.2) | 176 (13.3) | 6 (0.5) | 24 (1.8) | 27 (2.0) | 31 (2.3) | 7 (0.5) | 18 (1.4) | 86 (6.5) | 7 (0.5) | 1326 (100) | |
| 40-49 | 383 (70.4) | 74 (13.6) | 5 (0.9) | 15 (2.8) | 18 (3.3) | 5 (0.9) | 2 (0.4) | 11 (2.0) | 29 (5.3) | 2 (0.4) | 544 (100) | |
| 50-59 | 189 (70.5) | 17 (6.3) | 6 (2.2) | 18 (6.7) | 15 (5.6) | 3 (1.1) | 2 (0.7) | 6 (2.2) | 12 (4.5) | 0 (0.0) | 268 (100) | |
| 60+ | 110 (81.5) | 13 (9.6) | 0 (0.0) | 3 (2.2) | 2 (1.5) | 1 (0.7) | 1 (0.7) | 1 (0.7) | 3 (2.2) | 1 (0.7) | 135 (100) | <.001 |
| Self-reported race | ||||||||||||
| White | 1255 (73.1) | 261 (15.2) | 4 (0.2) | 13 (0.8) | 29 (1.7) | 29 (1.7) | 7 (0.4) | 14 (0.8) | 93 (5.4) | 11 (0.6) | 1716 (100) | |
| Hispanic | 451 (69.2) | 76 (11.7) | 3 (0.5) | 5 (0.8) | 20 (3.1) | 16 (2.5) | 3 (0.5) | 20 (3.1) | 56 (8.6) | 2 (0.3) | 652 (100) | |
| African American | 134 (66.7) | 19 (9.5) | 0 (0.0) | 0 (0.0) | 20 (10.0) | 18 (9.0) | 1 (0.5) | 1 (0.5) | 7 (3.5) | 1 (0.5) | 201 (100) | |
| Asian | 296 (69.2) | 40 (9.3) | 11 (2.6) | 43 (10.0) | 8 (1.9) | 7 (1.6) | 3 (0.7) | 5 (1.2) | 15 (3.5) | 0 (0.0) | 428 (100) | |
| Multiracial | 24 (80.0) | 2 (6.7) | 0 (0.0) | 0 (0.0) | 1 (3.3) | 2 (6.7) | 0 (0.0) | 0 (0.0) | 1 (3.3) | 0 (0.0) | 30 (100) | |
| Unknown | 136 (74.7) | 24 (13.2) | 1 (0.5) | 1 (0.5) | 1 (0.5) | 3 (1.6) | 1 (0.5) | 3 (1.6) | 9 (4.9) | 3 (1.6) | 182 (100) | <.001 |
| Histology outcome | ||||||||||||
| Control | 839 (75.8) | 142 (12.8) | 7 (0.6) | 20 (1.8) | 37 (3.3) | 15 (1.4) | 6 (0.5) | 5 (0.5) | 33 (3.0) | 3 (0.3) | 1107 (100) | |
| CIN2 | 667 (73.6) | 125 (13.8) | 5 (0.6) | 13 (1.4) | 22 (2.4) | 30 (3.3) | 4 (0.4) | 1 (0.1) | 34 (3.8) | 5 (0.6) | 906 (100) | |
| CIN3 | 706 (70.0) | 135 (13.4) | 6 (0.6) | 23 (2.3) | 17 (1.7) | 27 (2.7) | 4 (0.4) | 21 (2.1) | 60 (6.0) | 9 (0.9) | 1008 (100) | |
| SCC | 41 (59.4) | 11 (15.9) | 0 (0.0) | 2 (2.9) | 2 (2.9) | 2 (2.9) | 0 (0.0) | 2 (2.9) | 9 (13.0) | 0 (0.0) | 69 (100) | |
| AIS | 38 (44.7) | 9 (10.6) | 1 (1.2) | 3 (3.5) | 0 (0.0) | 1 (1.2) | 1 (1.2) | 7 (8.2) | 25 (29.4) | 0 (0.0) | 85 (100) | |
| Adeno | 9 (22.5) | 1 (2.5) | 0 (0.0) | 2 (5.0) | 1 (2.5) | 0 (0.0) | 0 (0.0) | 7 (17.5) | 20 (50.0) | 0 (0.0) | 40 (100) | <.001 |
| Total | 2300 (71.5) | 423 (13.2) | 19 (0.6) | 63 (2.0) | 79 (2.5) | 75 (2.3) | 15 (0.5) | 43 (1.3) | 181 (5.6) | 17 (0.5) | 3215 (100) | – |
* P value based on a two-sided Fisher’s exact test. Adeno = adenocarcinoma; AIS = adenocarcinoma in situ; CIN2 = cervical intraepithelial neoplasia grade 2; CIN3 = cervical intraepithelial neoplasia grade 3; SCC = squamous cell carcinoma.
Age at enrollment differed by variant lineages ( P < .001); the percentage with A1 was especially high among women age 60 years and older ( Table 1 ; Supplementary Figure 1 , available online). Women from self-reported race/ethnicity groups tended to have higher prevalences of HPV variants sharing the same geographic origins, ie, A1 and A2 with whites (European), A4 with Asians, B and C with African Americans, and D2 and D3 with Hispanics ( P < .001) ( Table 1 ). Having multiple HPV genotypes (in other words, having another type in addition to HPV16) was common (36.3%) but not associated with HPV16 variant (data not shown). Histology outcome was different among the variant lineages ( P < .001) ( Table 1 ).
HPV16 Variant Associations With Disease Risks
We assessed each HPV16 variant lineage association with cervical precancer (CIN2 and CIN3 separately) and cancer compared with the most common sublineage, A1 ( Table 2 ). Within the A lineage infections, sublineages A2 and A3 were not associated with outcome ( P > .1) compared with A1. Because A2 had sufficient sample size to conclude that disease risk was similar to A1, these two sublineages were combined and used as the referent. The A4 sublineage was associated with an increased risk of cancer (odds ratio [OR] = 3.16, 95% confidence intervals [CI] = 1.05 to 9.54, P = .04). Women with an HPV16 non-A variant lineage (B/C/D) infection had an increased risk of CIN3 and cancer compared with women with an A1/A2 lineage infection (OR for cancer = 6.87, 95% CI = 4.42 to 10.68, P = 9.7x10 −18 ). However, risk varied qualitatively by non-A lineage: The B lineage was associated with a lower risk of CIN3 (OR = 0.51, 95% CI = 0.28 to 0.91, P = .02) whereas the C lineage was associated with an increased risk of both CIN2 (OR = 2.48, 95% CI = 1.32 to 4.64, P = .005) and CIN3 (OR = 2.06, 95% CI = 1.09 to 3.89, P = .03). The D1 and D4 sublineages were not associated with CIN3 (there were no cancers in these women) although the positive association between D4 and CIN3 risk approached statistical significance (OR = 3.31, 95% CI = 0.89 to 12.28, P = .07). Women with D2 and D3 variant lineage infections had the highest risks of CIN3 and cancer compared with A1/A2: For example, D2 had an odds ratio for cancer of 28.48 (95% CI = 9.27 to 87.55, P = 5.0x10 −09 ), and D3 had an odds ratio for cancer of 13.90 (95% CI = 7.93 to 24.37, P = 3.7x10 −20 ). Risks of CIN2 were not associated with D2 or D3 variants. If we adjust for multiple comparisons among our 10 sublineages, a P value threshold of .005, some of these borderline P values are not considered statistically significant; however, all other P values are still statistically significant.
Table 2.
HPV16 variant lineage associations with precancer and cancer
| Tested variant | No. (%) | Reference variant | No. (%) | Status | P * | OR (95% CI) | P trend † |
|---|---|---|---|---|---|---|---|
| A2 | 142 (14.5) | A1 | 839 (85.5) | Control | – | – | |
| A2 | 125 (15.8) | 667 (84.2) | CIN2 | .44 | 1.11 (0.85 to 1.44) | ||
| A2 | 144 (16.2) | 744 (83.8) | CIN3 | .30 | 1.14 (0.89 to 1.47) | ||
| A2 | 12 (19.4) | 50 (80.6) | Cancer | .30 | 1.42 (0.74 to 2.73) | .20 | |
| A2 | 156 (16.4) | 794 (83.6) | CIN3+ | .24 | 1.16 (0.91 to 1.49) | ||
| A3 | 7 (0.7) | A1/A2 | 981 (99.3) | Control | – | – | |
| A3 | 7 (0.8) | 888 (99.2) | CIN3 | .85 | 1.10 (0.39 to 3.16) | ||
| A4 | 20 (2.0) | A1/A2 | 981 (98.0) | Control | – | – | |
| A4 | 13 (1.6) | 792 (98.4) | CIN2 | .55 | 0.81 (0.40 to 1.63) | ||
| A4 | 26 (2.8) | 888 (97.2) | CIN3 | .23 | 1.44 (0.80 to 2.59) | ||
| A4 | 4 (6.1) | 62 (93.9) | Cancer | .04 | 3.16 (1.05 to 9.54) | .06 | |
| A4 | 30 (3.1) | 950 (96.9) | CIN3+ | .13 | 1.55 (0.87 to 2.75) | ||
| B/C/D | 99 (9.2) | A1/A2 | 981 (90.8) | Control | – | – | |
| B/C/D | 96 (10.8) | 792 (89.2) | CIN2 | .22 | 1.20 (0.89 to1.61) | ||
| B/C/D | 172 (16.2) | 888 (83.8) | CIN3 | 1.2E-06 | 1.92 (1.47 to 2.50) | ||
| B/C/D | 43 (41.0) | 62 (59.0) | Cancer | 9.7E-18 | 6.87 (4.42 to 10.68) | 9.2E-15 | |
| B/C/D | 215 (18.5) | 950 (81.5) | CIN3+ | 4.8E-10 | 2.24 (1.74 to 2.89) | ||
| B | 37 (3.6) | A1/A2 | 981 (96.4) | Control | – | – | |
| B | 22 (2.7) | 792 (97.3) | CIN2 | .26 | 0.74 (0.43 to 1.26) | ||
| B | 17 (1.9) | 888 (98.1) | CIN3 | .02 | 0.51 (0.28 to 0.91) | ||
| B | 3 (4.6) | 62 (95.4) | Cancer | .69 | 1.28 (0.38 to 4.28) | .06 | |
| B | 20 (2.1) | 950 (97.9) | CIN3+ | .04 | 0.56 (0.32 to 0.97) | ||
| C | 15 (1.5) | A1/A2 | 981 (98.5) | Control | – | – | |
| C | 30 (3.6) | 792 (96.4) | CIN2 | .005 | 2.48 (1.32 to 4.64) | ||
| C | 28 (3.1) | 888 (96.9) | CIN3 | .03 | 2.06 (1.09 to 3.89) | ||
| C | 2 (3.1) | 62 (96.9) | Cancer | .33 | 2.11 (0.47 to 9.43) | .04 | |
| C | 30 (3.1) | 950 (96.9) | CIN3+ | .02 | 2.07 (1.10 to 3.86) | ||
| D1 | 6 (0.6) | A1/A2 | 981 (99.4) | Control | – | – | |
| D1 | 5 (0.6) | 888 (99.4) | CIN3 | .89 | 0.92 (0.28 to 3.03) | ||
| D2 | 5 (0.5) | A1/A2 | 981 (99.5) | Control | – | – | |
| D2 | 1 (0.1) | 792 (99.9) | CIN2 | .20 | 0.25 (0.03 to 2.12) | ||
| D2 | 28 (3.1) | 888 (96.9) | CIN3 | 1.9E-04 | 6.19 (2.38 to 16.09) | ||
| D2 | 9 (12.7) | 62 (87.3) | Cancer | 5.0E-09 | 28.48 (9.27 to 87.55) | 4.2E-10 | |
| D2 | 37 (3.7) | 950 (96.3) | CIN3+ | 2.2E-05 | 7.64 (2.99 to 19.53) | ||
| D3 | 33 (3.3) | A1/A2 | 981 (96.7) | Control | – | – | |
| D3 | 34 (4.1) | 792 (95.9) | CIN2 | .33 | 1.28 (0.78 to 2.08) | ||
| D3 | 85 (8.7) | 888 (91.3) | CIN3 | 6.6E-07 | 2.85 (1.88 to 4.30) | ||
| D3 | 29 (31.9) | 62 (68.1) | Cancer | 3.7E-20 | 13.90 (7.93 to 24.37) | 1.3E-16 | |
| D3 | 114 (10.7) | 950 (89.3) | CIN3+ | 3.6E-10 | 3.57 (2.40 to 5.31) | ||
| D4 | 3 (0.3) | A1/A2 | 981 (99.7) | Control | – | – | |
| D4 | 9 (1.0) | 888 (99.0) | CIN3 | .07 | 3.31 (0.89 to 12.28) |
* P values were calculated using a two-sided logistic regression model. CI = confidence interval; CIN2 = cervical intraepithelial neoplasia grade 2; CIN3 = cervical intraepithelial neoplasia grade 3, including AIS; CIN3 + = includes CIN3, AIS, and all cancers; OR = odds ratio.
† Ptrend values using a two-sided regression model of the CIN2, CIN3, and cancer associations.
HPV16 Variant Associations With Histologic Subtypes
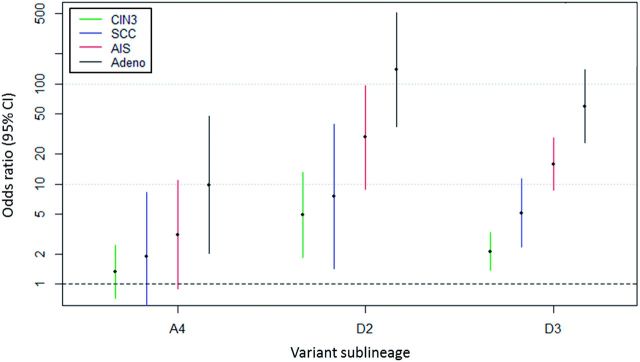
We further evaluated risk associations by histologic subtype ( Figure 2 , Table 3 ) and observed that variant lineage was associated with precancer/cancer histology ( P < .001). The D2/D3 variants were most common in glandular lesions, identified in 67.5% of the adenocarcinomas, whereas the A1/A2 variants were identified in 75.4% of SCCs. Compared with A1/A2, D2 had the strongest increased risk of AIS (OR = 29.22, 95% CI = 8.94 to 95.51, P = 2.3x10 −08 ) and adenocarcinoma (OR = 137.34, 95% CI = 37.21 to 506.88, P = 1.5x10 −13 ) in contrast to squamous precancer (CIN3: OR = 4.90, 95% CI = 1.84 to 13.05, P = .001) and SCC (OR = 7.55, 95% CI = 1.43 to 39.82, P = .02). D3 showed a similar increased risk of glandular lesions (adenocarcinoma: OR = 59.45, 95% CI = 25.81 to 136.98, P = 8.5x10 −22 ) compared with squamous lesions (SCC: OR = 5.15, 95% CI = 2.34 to 11.31, P = 4.6x10 −5 ). The A4 sublineage also showed an increased risk of AIS (OR = 3.13, 95% CI = 0.90 to 10.91, P = .07) and adenocarcinoma (OR = 9.81, 95% CI = 2.02 to 47.69, P = .005) in contrast to CIN3 (OR = 1.34, 95% CI = 0.73 to 2.46, P = .34) and SCC (OR = 1.89, 95% CI = 0.43 to 8.29, P = .40). Unlike the other lineages, B and C were not associated with glandular (AIS/adenocarcinoma) vs squamous (CIN3/SCC) lesions.
Figure 2.
HPV16 variant lineage associations with squamous and glandular precancer/cancer subtypes. Squamous lesions include CIN3 ( green ) and SCC ( blue ); glandular lesions include AIS ( red ) and Adeno ( black ). Y-axis shows the odds ratios and 95% confidence intervals ( colored bars ) on a log scale. Adeno = adenocarcinoma; AIS = adenocarcinoma in situ; CI = confidence interval; CIN3 = cervical intraepithelial neoplasia grade 3; SCC = squamous cell carcinoma.
Table 3.
HPV16 variant lineage associations with precancer and cancer subtypes
| Tested variant | No. (%) | Reference variant | No. (%) | Status | P * | OR (95% CI) |
|---|---|---|---|---|---|---|
| A4 | 20 (2.0) | A1/2 | 981 (98.0) | Control | – | – |
| A4 | 23 (2.7) | 841 (97.3) | CIN3 | .34 | 1.34 (0.73 to 2.46) | |
| A4 | 2 (3.7) | 52 (96.3) | SCC | .40 | 1.89 (0.43 to 8.29) | |
| A4 | 3 (6.0) | 47 (94.0) | AIS | .07 | 3.13 (0.90 to 10.91) | |
| A4 | 2 (16.7) | 10 (83.3) | Adeno | .005 | 9.81 (2.02 to 47.69) | |
| B | 37 (3.6) | A1/2 | 981 (96.4) | Control | – | – |
| B | 17 (2.0) | 841 (98.0) | CIN3 | .04 | 0.54 (0.32 to 0.95) | |
| B | 2 (3.7) | 52 (96.3) | SCC | .98 | 1.02 (0.24 to 4.35) | |
| B | 0 (0.0) | 47 (100.0) | AIS | – | – | |
| B | 1 (9.1) | 10 (90.9) | Adeno | .36 | 2.65 (0.33 to 21.26) | |
| C | 15 (1.5) | A1/2 | 981 (98.5) | Control | – | – |
| C | 27 (3.1) | 841 (96.9) | CIN3 | .02 | 2.10 (1.11 to 3.97) | |
| C | 2 (3.7) | 52 (96.3) | SCC | .23 | 2.52 (0.56 to 11.29) | |
| C | 1 (2.1) | 47 (97.9) | AIS | .75 | 1.39 (0.18 to 10.76) | |
| C | 0 (0.0) | 10 (100.0) | Adeno | – | – | |
| D2 | 5 (0.5) | A1/2 | 981 (99.5) | Control | – | – |
| D2 | 21 (2.4) | 841 (97.6) | CIN3 | .001 | 4.90 (1.84 to 13.05) | |
| D2 | 2 (3.7) | 52 (96.3) | SCC | .02 | 7.55 (1.43 to 39.82) | |
| D2 | 7 (13.0) | 47 (87.0) | AIS | 2.3E-08 | 29.22 (8.94 to 95.51) | |
| D2 | 7 (41.2) | 10 (58.8) | Adeno | 1.5E-13 | 137.34 (37.21 to 506.88) | |
| D3 | 33 (3.3) | A1/2 | 981 (96.7) | Control | – | – |
| D3 | 60 (6.7) | 841 (93.3) | CIN3 | 7.0E-04 | 2.12 (1.37 to 3.28) | |
| D3 | 9 (14.8) | 52 (85.2) | SCC | 4.6E-05 | 5.15 (2.34 to 11.31) | |
| D3 | 25 (34.7) | 47 (65.3) | AIS | 1.2E-19 | 15.81 (8.71 to 28.71) | |
| D3 | 20 (66.7) | 10 (33.3) | Adeno | 8.5E-22 | 59.45 (25.81 to 136.98) |
* P values were calculated using a two-sided logistic regression model. Adeno = adenocarcinomas; AIS = adenocarcinoma in situ; CI = confidence interval; CIN3 = cervical intraepithelial neoplasia grade 3, without AIS; OR = odds ratio; SCC = squamous cell carcinomas.
Variant Lineage Risk by Race and Ethnicity
Self-reported race/ethnicity was different among HPV16 variant lineages ( P < .001) ( Table 1 ). Women with an A4, B, C, or D2/D3 variant lineage infection had statistically significantly different race distributions compared with women with an A1/A2 variant infection ( Supplementary Figure 2 , available online). White women with the A1/A2 variant lineages had a higher risk of CIN3+ compared with all other women (OR = 1.35, 95% CI = 1.13 to 1.61, P = .001) ( Table 4 ). Especially when compared with African-American women with an A1/A2 variant lineage infection, white women with an A1/A2 variant had a higher risk of CIN3 + (OR = 1.71, 95% CI = 1.15 to 2.54, P = .008) (data not shown). These P values remain statistically significant after a Bonferroni correction for multiple tests. There was a suggestion that Asian women with the A4 (Asian) sublineage had a higher risk of CIN3 + (OR = 2.19, 95% CI = 0.64 to 7.5, P = .21) and that Hispanic women with the D2/D3 sublineages had a higher risk of CIN3 + (OR = 1.70, 95% CI = 0.77 to 3.75, P = .19) compared with the other racial/ethnic groups ( Table 4 ). The increased risk of AIS and adenocarcinoma associated with the D2/D3 variant lineages compared with A1/A2 was stronger in Hispanic women (OR for adenocarcinoma = 165.60, 95% CI = 19.06 to 1438.50, P = 3.6x10 −06 ) compared with white women (OR for adenocarcinoma = 64.25, 95% CI = 22.83 to 180.85, P = 3.2x10 −15 ) ( Supplementary Table 1 , available online).
Table 4.
Risk of CIN3+ for selected racial groups stratified by HPV16 variant lineage
| HPV16 variant | Tested racial group | No. (%) | Reference racial group | No. (%) | Status | P * | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| A1/2 | White | 506 (51.6) | All others | 475 (48.4) | Control | – | – |
| 560 (58.9) | 390 (41.1) | CIN3+ | .001 | 1.35 (1.13 to 1.61) | |||
| A4 | Asian | 12 (60.0) | All others | 8 (40.0) | Control | – | – |
| 23 (76.7) | 7 (23.3) | CIN3+ | .21 | 2.19 (0.64 to 7.50) | |||
| D2/3 | Hispanic | 10 (26.3) | All others | 28 (73.7) | Control | – | – |
| 57 (37.7) | 94 (62.3) | CIN3+ | .19 | 1.70 (0.77 to 3.75) |
* P values were calculated using a two-sided logistic regression model. CI = confidence interval; CIN3 + = cervical intraepithelial neoplasia grade 3 and AIS and all cancers; OR = odds ratio.
Discussion
We report the largest study to date of HPV16 variant lineages and risk of cervical precancer/cancer in 3200 women, based on HPV16 whole-genome sequencing and precise HPV16 sublineage classification. We further evaluated risk modification by race/ethnicity. Smaller studies have shown that HPV16 variants influence risk of persistence and progression; however, most have combined all “European” and all “non-European” lineages and grouped infection outcomes as “CIN2+” because of small numbers ( 11 ). We substantially refined and extended these initial associations by evaluating all HPV16 variant sublineages individually by specific histologic outcomes. We showed that the non-A HPV16 lineages (B/C/D) as a group were associated with a higher risk of precancer and cancer compared with the A lineages, but, importantly, through our large sample size we found that this is misleading and the summary conceals important qualitative heterogeneity in both viral lineages and disease outcomes, which masked very strong and specific associations.
In particular, we first showed that the variant lineages that are often grouped are not homogeneous in regards to pathogenicity. The “European” variant lineages are not uniform; the A4 sublineage was linked to an increased risk of cancer compared with the A1/A2 clade. The “African” variants also displayed heterogeneity for disease outcomes, with the B variant lineage being associated with a statistically significantly reduced risk of CIN3+ compared with the A1/A2 sublineages, while the C lineage conferred statistically significantly elevated risk. The D lineages were associated with a substantially higher risk of precancer/cancer compared with the A1/A2 sublineages; because of small numbers, we could not assess D1 or D4 with precision.
In terms of severity of histologic outcomes, we again found important heterogeneity. In general, inclusion of CIN2 in the precancer outcome group was shown to be misleading because, except for one lineage, none of the variant lineages was associated with an increased risk of CIN2. Risks for cancer were even stronger than risks for CIN3. We were underpowered to study invasive cancers thoroughly although the OR estimates were so large that we were able nonetheless to observe major risk relationships.
We observed markedly increased risk of adenocarcinomas associated specifically with the D2/D3 and A4 sublineages; this provides strong evidence for a link between viral genomics and histologic outcome. An association between D lineages and adenocarcinoma was reported in some smaller studies ( 21 , 23–25 ) but not others ( 26 , 27 ). One case series had also suggested a higher frequency of the D lineages in AIS compared with CIN3 ( 24 ). Our study identified a strong association between D2/D3 sublineages and both AIS and adenocarcinomas. The A4 sublineage has previously been associated with an increased risk of squamous precancer and cancer ( 39 ); with modest numbers, we only observed an increased risk for cancer, and specifically only adenocarcinomas.
Given the magnitude of these associations, HPV16 genomic variation may partly determine the pathogenesis of cervical cancer, but the mechanisms are not at all clear. The A4 and D2/D3 variant lineages may have a greater ability to infect glandular epithelial cells. By analogy, closely related genotypes HPV16 and HPV31 utilize differing initial cell entry routes ( 40 , 41 ). Therefore, it is possible that HPV16 entry and processing in glandular vs squamous cells is variant specific ( 24 ). Or, variants might differ in their effects on carcinogenesis postinfection. The basis of this important relationship between specific HPV16 variants and histological subtypes warrants further study.
One study found that the D variant lineages had 3.3- and 2.8-fold increased oncogene P97 promoter activity when compared with the A lineages, and the B and C lineages had comparable P97 promoter activity with the A lineages ( 42 ). We speculate that transcriptional differences may provide a potential mechanism of how HPV genomic variations may in part determine the pathogenesis of an HPV16 infection leading to cancer. Figure 1 depicts the sublineage-specific nucleotide and amino acid differences in each of the HPV16 sublineages, and, of interest, the sublineages associated with a strong increased risk of cancer, A4, D2, and D3, have most of their amino acid changes within the E1, E2, and L2 ORFs. In differentiated host epithelial cells, both the viral DNA helicase E1 and the viral replication/transcription factor E2 are required for productive replication of the HPV genome ( 43 , 44 ). Interestingly, an earlier study suggested that HPV genomes with a disrupted E1 ORF may have some replication advantage over those with an intact E1 ORF ( 45 ). However, viral variant lineages are defined by highly correlated nucleotide changes across their genomes, previously termed “lineage fixation” ( 46 ), which makes linking function to one of these changes or regions difficult and requires further functional evaluation before speculating more about a specific mechanistic model.
Our data shows that race/ethnicity is associated with infection with specific HPV16 variant lineages, and the risk of precancer and cancer for specific HPV16 variant lineages varies by a women’s race/ethnicity. Our data suggests that if a woman’s self-reported race/ethnicity matches that of the origin of the infecting HPV16 virus, there is an increased risk of precancer/cancer. That is, white women with an A1/A2 variant infection have an increased risk of CIN3+ compared with women of other races infected with an A1/A2 variant. Similarly, there was a suggestion that Asian women and Hispanic women had increased risks associated with the A4 (Asian) variant and D2/D3 (Asian-American) variants, respectively, compared with the other races. We further showed that the association between the D2/D3 sublineages and glandular lesions was stronger in Hispanic women compared with the other races.
Our findings corroborate and extend a previous report that showed white women were more likely to have CIN3 than were African American women, given an HPV16 European variant infection ( 14 ). The mechanism for these observed differences in outcomes is unclear, but the virus-host interaction may be related to some aspect of the host’s immune response; it is possible that an HPV16 lineage that has co-evolved with a particular human race has an advantage in evading immune surveillance and persisting, thus increasing risk of precancer and cancer.
A limitation of our study, even with our large sample size, is that the population was primarily a white, non-Hispanic population, so we had limited numbers of the other racial/ethnic groups represented. Thus, the findings for race-specific differences in risk associated with HPV16 variant lineages warrant further evaluation in a more diverse population; in particular, we had too few African Americans with B and C lineages in our study to assess this relationship. Self-reported race is only a surrogate of genetic ancestry; future studies are needed to evaluate human genetic ancestry with genetic markers to determine more precise ancestry relationships with HPV16 variant lineages. A strength of our study is the approach using whole-genome analyses that ensures proper assignment to HPV16 variant lineages as not all regions of the genome have similar variant-informative content ( 47 ). As a major conclusion of our investigation, we are encouraged to analyze finer details of HPV phylogenetic branchings and gene sequences, including non–lineage specific SNPs. We are expanding to whole-genome sequencing of specimens from cases and controls containing HPV16-related types in the alpha-9 species group (eg, HPV31) to find additional clues to the unique carcinogenicity of HPV16.
In summary, our study of over 3200 women infected with HPV16 shows that HPV16 variant lineages have considerable risk differences for precancer and cancer, and specific variant sublineages strongly influence HPV16 carcinogenicity and histologic outcome. Our results suggest that a finer level of grouping of HPV16 variant lineage/sublineage classifications and disease outcomes is warranted; it is not valid to group heterogeneous lineages/sublineages (eg, B and C as the “African variants”) and outcomes (ie, most lineages are not associated with an altered risk of CIN2). We provide compelling evidence that specific HPV16 variant sublineages, D2/D3 and A4, are strongly associated with glandular lesions, and, given the difficulty in screening for these lesions using cytology, a highly specific HPV variant test could conceivably have clinical and prevention implications.
Funding
This study was funded by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health. RB was supported in part by the National Cancer Institute (CA78527).
Notes
The study funder had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
References
- 1. Schiffman M, Herrero R, DeSalle R , et al. . The carcinogenicity of human papillomavirus types reflects viral evolution . Virology. 2005. ; 337 : 76 – 84 . [DOI] [PubMed] [Google Scholar]
- 2. Bouvard V, Baan R, Straif K , et al. . A review of human carcinogens—Part B: biological agents . Lancet Oncol. 2009. ; 10 : 321 – 322 . [DOI] [PubMed] [Google Scholar]
- 3. Forman D, de Martel C, Lacey CJ , et al. . Global Burden of Human Papillomavirus and Related Diseases . Vaccine. 2012. ; 30(Suppl 5) : F12 – F23 . [DOI] [PubMed] [Google Scholar]
- 4. Bernard H-U, Burk RD, Chen Z , et al. . Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments . Virology. 2010. ; 401 : 70 – 79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCredie M, Sharples K, Paul C , et al. . Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study . Lancet Oncol. 2008. ; 9 : 425 – 434 . [DOI] [PubMed] [Google Scholar]
- 6. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer . Lancet. 2007. ; 370 : 890 – 907 . [DOI] [PubMed] [Google Scholar]
- 7. Burk RD, Chen Z, Van Doorslaer K. Human Papillomaviruses: Genetic Basis of Carcinogenicity . Public Health Genomics. 2009. ; 12 : 281 – 290 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Sanjose S, Quint WGV, Alemany L , et al. . Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study . Lancet Oncol. 2010. ; 11 : 1048 – 1056 . [DOI] [PubMed] [Google Scholar]
- 9. Guan P, Howell-Jones R, Li N , et al. . Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer . Int J Cancer. 2012. ; 131 : 2349 – 2359 . [DOI] [PubMed] [Google Scholar]
- 10. Cullen M, Boland JF, Schiffman M , et al. . Deep sequencing of HPV16 genomes: A new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection . Papillomavirus Res . 2015. ; 1 : 3 – 11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burk RD, Harari A, Chen Z. Human papillomavirus genome variants . Virology. 2013. ; 445 : 232 – 243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hildesheim A, Schiffman M, Bromley C , et al. . Human Papillomavirus Type 16 Variants and Risk of Cervical Cancer . J Natl Cancer Inst. 2001. ; 93 : 315 – 318 . [DOI] [PubMed] [Google Scholar]
- 13. Pientong C, Wongwarissara P, Ekalaksananan T , et al. . Association of human papillomavirus type 16 long control region mutation and cervical cancer . Virol J. 2013. ; 10 : 30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xi LF, Koutsky LA, Hildesheim A , et al. . Risk for High-Grade Cervical Intraepithelial Neoplasia Associated with Variants of Human Papillomavirus Types 16 and 18 . Cancer Epidemiol Biomarkers Prev. 2007. ; 16 : 4 – 10 . [DOI] [PubMed] [Google Scholar]
- 15. Schiffman M, Rodriguez AC, Chen Z , et al. . A Population-Based Prospective Study of Carcinogenic Human Papillomavirus Variant Lineages, Viral Persistence, and Cervical Neoplasia . Cancer Res. 2010. ; 70 : 3159 – 3169 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cornet I, Gheit T, Iannacone MR , et al. . HPV16 genetic variation and the development of cervical cancer worldwide . Br J Cancer. 2013. ; 108 : 240 – 244 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gheit T, Cornet I, Clifford GM , et al. . Risks for Persistence and Progression by Human Papillomavirus Type 16 Variant Lineages Among a Population-Based Sample of Danish Women . Cancer Epidemiol Biomarkers Prev. 2011. ; 20 : 1315 – 1321 . [DOI] [PubMed] [Google Scholar]
- 18. Zehbe I, Voglino G, Delius H, Wilander E, Tommasino M. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms . Lancet. 1998. ; 352 : 1441 – 1442 . [DOI] [PubMed] [Google Scholar]
- 19. Zuna RE, Moore WE, Shanesmith RP , et al. . Association of HPV16 E6 variants with diagnostic severity in cervical cytology samples of 354 women in a US population . Int J Cancer. 2009. ; 125 : 2609 – 2613 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sichero L, Ferreira S, Trottier H , et al. . High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18 . Int J Cancer. 2007. ; 120 : 1763 – 1768 . [DOI] [PubMed] [Google Scholar]
- 21. Berumen J, Ordoñez RM, Lazcano E , et al. . Asian-American Variants of Human Papillomavirus 16 and Risk for Cervical Cancer: a Case–Control Study . J Natl Cancer Inst. 2001. ; 93 : 1325 – 1330 . [DOI] [PubMed] [Google Scholar]
- 22. Freitas LB, Chen Z, Muqui EF , et al. . Human Papillomavirus 16 Non-European Variants Are Preferentially Associated with High-Grade Cervical Lesions . PLoS ONE. 2014. ; 9 : e100746 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burk RD, Terai M, Gravitt PE , et al. . Distribution of Human Papillomavirus Types 16 and 18 Variants in Squamous Cell Carcinomas and Adenocarcinomas of the Cervix . Cancer Res. 2003. ; 63 : 7215 – 7220 . [PubMed] [Google Scholar]
- 24. Quint KD, de Koning MNC, van Doorn L-J, Quint WGV, Pirog EC. HPV genotyping and HPV16 variant analysis in glandular and squamous neoplastic lesions of the uterine cervix . Gynecol Oncol. 2010. ; 117 : 297 – 301 . [DOI] [PubMed] [Google Scholar]
- 25. Rabelo-Santos SH, Villa LL, Derchain SF , et al. . Variants of human papillomavirus types 16 and 18: Histological findings in women referred for atypical glandular cells or adenocarcinoma in situ in cervical smear . Int J Gynecol Pathol. 2006. ; 25 : 393 – 397 . [DOI] [PubMed] [Google Scholar]
- 26. De Boer MA, Peters LAW, Aziz MF , et al. . Human papillomavirus type 18 variants: Histopathology and E6/E7 polymorphisms in three countries . Int J Cancer. 2005. ; 114 : 422 – 425 . [DOI] [PubMed] [Google Scholar]
- 27. Lizano M, De la Cruz-Hernández E, Carrillo-García A , et al. . Distribution of HPV16 and 18 intratypic variants in normal cytology, intraepithelial lesions, and cervical cancer in a Mexican population . Gynecol Oncol. 2006. ; 102 : 230 – 235 . [DOI] [PubMed] [Google Scholar]
- 28. Xi LF, Kiviat NB, Hildesheim A , et al. . Human Papillomavirus Type 16 and 18 Variants: Race-Related Distribution and Persistence . J Natl Cancer Inst. 2006. ; 98 : 1045 – 1052 . [DOI] [PubMed] [Google Scholar]
- 29. Lopera EA, Baena A, Florez V , et al. . Unexpected inverse correlation between Native American ancestry and Asian American variants of HPV16 in admixed Colombian cervical cancer cases . Infect Genet Evol. 2014. ; 28 : 339 – 348 . [DOI] [PubMed] [Google Scholar]
- 30. Junes-Gill K, Sichero L, Maciag PC , et al. . Human papillomavirus type 16 variants in cervical cancer from an admixtured population in Brazil . J Med Virol. 2008. ; 80 : 1639 – 1645 . [DOI] [PubMed] [Google Scholar]
- 31. Castle P, Shaber R, LaMere B , et al. . Human papillomavirus (HPV) genotypes in women with cervical precancer and cancer at Kaiser Permanente Northern California . Cancer Epidemiol Biomarkers Prev. 2011. ; 20 : 946 – 953 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bernard H-U. Coevolution of papiliomaviruses with human populations . Trends Microbiol. 1994. ; 2 : 140 – 143 . [DOI] [PubMed] [Google Scholar]
- 33. Bernard H-U, Calleja-Macias IE, Dunn ST. Genome variation of human papillomavirus types: Phylogenetic and medical implications . Int J Cancer. 2006. ; 118 : 1071 – 1076 . [DOI] [PubMed] [Google Scholar]
- 34. Yamada T, Manos MM, Peto J , et al. . Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective . J Virol. 1997. ; 71 : 2463 – 2472 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burk RD, Ho GYF, Beardsley L , et al. . Sexual Behavior and Partner Characteristics Are the Predominant Risk Factors for Genital Human Papillomavirus Infection in Young Women . J Infect Dis. 1996. ; 174 : 679 – 689 . [DOI] [PubMed] [Google Scholar]
- 36. Castle PE, Schiffman M, Gravitt PE , et al. . Comparisons of HPV DNA detection by MY09/11 PCR methods . J Med Virol. 2002. ; 68 : 417 – 23 . [DOI] [PubMed] [Google Scholar]
- 37. Van Doorslaer K, Tan Q, Xirasagar S , et al. . The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis . Nucleic Acids Res. 2013. ; 41 : D571 – D578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models . Bioinformatics. 2006. ; 22 : 2688 – 2690 . [DOI] [PubMed] [Google Scholar]
- 39. Chopjitt P, Ekalaksananan T, Pientong C , et al. . Prevalence of human papillomavirus type 16 and its variants in abnormal squamous cervical cells in Northeast Thailand . Int J Infect Dis. 2009. ; 13 : 212 – 219 . [DOI] [PubMed] [Google Scholar]
- 40. Day PM, Lowy DR, Schiller JT. Papillomaviruses infect cells via a clathrin-dependent pathway . Virology. 2003. ; 307 : 1 – 11 . [DOI] [PubMed] [Google Scholar]
- 41. Smith JL, Campos SK, Ozbun MA. Human Papillomavirus Type 31 Uses a Caveolin 1- and Dynamin 2-Mediated Entry Pathway for Infection of Human Keratinocytes . J Virol. 2007. ; 81 : 9922 – 9931 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kämmer C, Warthorst U, Torrez-Martinez N, Wheeler CM, Pfister H. Sequence analysis of the long control region of human papillomavirus type 16 variants and functional consequences for P97 promoter activity . J Gen Virol. 2000. ; 81 : 1975 – 1981 . [DOI] [PubMed] [Google Scholar]
- 43. McBride AA. Chapter 4 Replication and Partitioning of Papillomavirus Genomes . In: Advances in Virus Research . Academic Press; . 2008. ; 155 – 205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mori S, Kusumoto-Matsuo R, Ishii Y, Takeuchi T, Kukimoto I. Replication interference between human papillomavirus types 16 and 18 mediated by heterologous E1 helicases . Virol J. 2014. ; 11 : 11 – 11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fu L, Terai M, Matsukura T, Herrero R, Burk RD. Codetection of a Mixed Population of candHPV62 Containing Wild-Type and Disrupted E1 Open-Reading Frame in a 45-Year-Old Woman with Normal Cytology . J Infect Dis. 2004. ; 190 : 1303 – 1309 . [DOI] [PubMed] [Google Scholar]
- 46. Chen Z, Terai M, Fu L , et al. . Diversifying Selection in Human Papillomavirus Type 16 Lineages Based on Complete Genome Analyses . J Virol. 2005. ; 79 : 7014 – 7023 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith B, Chen Z, Reimers L , et al. . Sequence Imputation of HPV16 Genomes for Genetic Association Studies . PLoS ONE. 2011. ; 6 : e21375 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.