Abstract
Quantitative proteomics of postmortem human brain can identify dysfunctional proteins that contribute to neurodegenerative disorders like Alzheimer disease (AD) and frontotemporal dementia. Similar studies in chronic traumatic encephalopathy (CTE) are limited, therefore we hypothesized that proteomic sequencing of CTE frontal cortex brain homogenates from varying CTE pathologic stages may provide important new insights into this disorder. Quantitative proteomics of control, CTE and AD brains was performed to characterize differentially expressed proteins, and we identified over 4000 proteins in CTE brains, including significant enrichment of the microtubule associated protein tau. We also found enrichment and pathologic aggregation of RNA processing factors as seen previously in AD, supporting the previously recognized overlap between AD and CTE. In addition to these similarities, we identified CTE-specific enrichment of proteins which increase with increasing severity of CTE pathology. NADPH dehydrogenase quinone 1 (NQO1) was one of the proteins which showed significant enrichment in CTE and also correlated with increasing CTE stage. NQO1 demonstrated neuropathologic correlation with hyperphosphorylated tau in glial cells, mainly astrocytes. These results demonstrate that quantitative proteomic analysis of CTE postmortem human brain can identify disease relevant findings and novel cellular pathways involved in CTE pathogenesis.
Keywords: Alzheimer disease, Chronic traumatic encephalopathy, Insoluble proteome, Microtubule associated protein tau, NADPH dehydrogenase quinone 1 (NQO1), Proteomics, U1-70k
INTRODUCTION
Neurodegenerative diseases often involve the abnormal accumulation of proteins in neurons and glia (1, 2). The accumulations can be identified through immunohistochemical analysis as well as with high throughput discovery approaches like proteomic sequencing. Previous studies utilizing quantitative proteomics have reported both known and novel disease relevant findings in Alzheimer disease (AD), frontotemporal dementia and Parkinson disease (PD) (3–14), however, proteomic investigations of postmortem human brain from subjects with chronic traumatic encephalopathy (CTE) are lacking. Understanding proteins that aggregate in CTE could provide much needed insight into disease mechanisms.
CTE is a neurodegenerative disease thought to be caused by a prolonged history of repetitive head impacts. Recent consensus criteria has described CTE as a distinct tauopathy with a pathognomonic lesion consisting of an abnormal perivascular accumulation of phosphorylated tau within neurons and astrocytes in an irregular, patchy distribution concentrated at the depth of the cortical sulci (15). Other supportive features include the aggregation of phosphorylated TAR DNA binding protein 43 (TDP-43) as well as certain gross anatomical features like a cavum septum pellucidum and atrophy of deep gray matter structures. A significant number of later stage cases also have typical Alzheimer pathology including β-amyloid-positive plaques (16). There are likely other important proteins that aggregate and contribute to CTE.
Although ongoing prospective studies will likely provide much needed insight concerning the scope of CTE, more information is needed on the aggregation mechanisms driving the disease. One approach is to use proteomic sequencing of the detergent insoluble brain fraction to characterize those proteins that aggregate in CTE as these proteins are likely to be dysfunctional and may contribute to disease progression. We previously used this approach in AD postmortem brain (4, 13, 14). In addition to classic disease associated proteins like tau and β-amyloid, we identified novel AD associated insoluble proteins including multiple RNA processing factors like U1-70k, U1-A and SmD (13, 14). We also found evidence for RNA processing dysfunction, and further investigations suggested that these proteins may somehow link tau and amyloid pathologies. Therefore, a similar approach with CTE insoluble proteome could provide important clues about disease pathogenesis.
A potential limitation in using large scale proteomics to study CTE is whether or not proteomic sequencing will provide an accurate reflection of the ongoing neurodegenerative process. Early stage CTE cases have relatively sparse accumulations of pathology and this may make it difficult to capture disease relevant changes (17). Another possible concern is that pathologic overlap with AD in the later stage cases may interfere with identifying important CTE-specific changes.
We performed the first large-scale quantitative protein sequencing study of CTE to assess the insoluble proteome in a group of CTE brains encompassing a range of pathological severity. Control and AD cases were sequenced for comparison. We identified expected enrichment of insoluble tau along with novel proteins, including NADPH dehydrogenase quinone 1 (NQO1), that demonstrated progressive enrichment through stages of CTE pathology. Findings were validated with immunohistochemical staining and novel changes were correlated with tau pathology. These studies demonstrated a unique signature in the insoluble proteome that distinguishes CTE from both AD and control brains. Importantly, we also observed enrichment of RNA processing factors in CTE brains, which were similar to changes we had previously observed in AD (13), suggesting possible mechanistic overlap. Overall, this study confirmed that proteomic analysis can provide important new insights into CTE pathogenesis and support the importance of conducting larger scale studies.
MATERIALS AND METHODS
Neuropathological Characteristics of Cases
Brains with a history of repetitive head injury from the VA-BU-CLF Brain Bank were neuropathologically evaluated for changes of CTE using recently established NINDS consensus criteria for CTE (15) as well as criteria for AD, PD, dementia with Lewy bodies, frontotemporal lobe degeneration, or motor neuron disease (Supplementary Data Table S1). To diagnose CTE, the criteria requires at least 1 perivascular hyperphosphorylated (ptau) tau lesion that consists of ptau aggregates inside of neurons, astrocytes, and cell processes around a small vessel (CTE lesion). CTE is divided into 4 stages wherein the localization and severity of ptau deposition defines the stage. In stage I, sparse, isolated perivascular foci of ptau are observed in the frontal cortex. In stage II, ptau lesions are found in multiple cortical regions. In stage III, extensive ptau pathology is found in medial temporal lobe structures like the hippocampus and amygdala. In stage IV, there is wide spread ptau involvement in all brain structures. Three cases utilized in this study were labeled as having overlapping stages (i.e., I–II, II–III) due to the difficulty in selecting a clear stage II or stage III neuropathologic diagnosis. Cases with CTE that were used here did not meet diagnostic criteria for AD, PD, dementia with Lewy bodies, frontotemporal lobe degeneration, or motor neuron disease. A team of board certified neuropathologists (T.D.S., B.R.H., A.C.M.) provided semiquantitative scores of the presence of Aβ neuritic plaques (CERAD score), TDP-43 pathology, and α-synuclein aggregates that are detailed in Supplementary Data Table S1. Scores denote 0 = normal, 1 = mild, 2 = moderate, 3 = severe. Additional brains from nonathlete controls were obtained from the Boston University Alzheimer’s Disease Center Brain Bank (Supplementary Data Table S1). Controls were selected based on a lack of history of contact sport play and lack of neurodegenerative diseases. Additional control, CTE and AD cases were obtained from the Emory Alzheimer’s Disease Research Center brain bank (Supplementary Data Table S1). IRB approval for brain donation was obtained through the Boston University Alzheimer’s Disease and CTE Center (ADC), the Edith Nourse Rogers Memorial Veterans Hospital (Bedford, MA) and Emory University (Atlanta, GA). Next of kin provided written consent for participation and brain donation.
Antibodies
Primary antibodies included U1-70 K (EM439; custom rabbit polyclonal), SmD (50940, Abcam, Cambridge, MA), tau (AT8 clone, Pierce, Rockford, IL), NQO1 (34173, Abcam; 3187 Cell Signaling), glial fibrillary acidic protein ([GFAP], MAB360, Millipore, Illerica, MA), IBA1 (019-19741, Wako), CD31 (Clone JC/70 A; ThermoFisher, Waltham, MA), NRF2 (62352, Abcam), O4 (MAB1326, Novus Biologicals, Littleton, CO), NG2 (MAB5384, Millipore), Olig2 (AB33427, Abcam), and parvalbumin (PA1-933 ThermoFisher). Secondary antibodies included Alexa 488 and Alexa 568 (Jackson ImmunoResearch, West Grove, PA), cyanine 3 tyramide (SAT704A001EA, Perkin Elmer, Wellesley, MA) and biotinylated goat antimouse and antirabbit (Vector Laboratories, Burlingame, CA).
Immunohistochemistry and Immunofluorescence of Postmortem Human Brain
Brain tissue for immunostaining was processed by fixation in periodate-lysine-paraformaldehyde or paraformaldehyde and stored at 4 °C. A tissue block from the dorsolateral frontal cortex was taken perpendicular to the superior frontal sulcus, and either embedded in paraffin and cut at 20 μm or submerged in 30% sucrose for generating 50-µm cryosections. Immunohistochemistry of paraffin sections was performed as previously described (18). Immunohistochemistry and dual immunofluorescence on cryopreserved free-floating 50-µm sections was performed as previously described (13, 19–21).
Digital Microscopy and Analysis
NQO1 and AT8 immunostained slides were scanned and digitized at 20× magnification using the Aperio ScanScope (Leica, Buffalo Grove, IL) as previously described (18). The gray matter at the depth of the cortical sulci (defined as the bottom third of 2 connecting gyri) was selected and highlighted in ImagesScope (Leica). NQO1-positive cells were counted by hand while AT8 area was quantified using Leica image analysis software. The Aperio-positive pixel count algorithm (version 9) was set to recognize and quantify the total area of-positive AT8 tau stain. All quantifications were standardized to area measured and presented as density per analyzed area.
Statistics
Statistical analysis was performed with SPSS version 20.0 (IBM Inc., Armonk, New York) and Prism v6 (Graphpad Software, La Jolla, CA). NQO1 and AT8 density were log transformed to normalize for regression analysis. A one-way ANOVA was used to compare NQO1 density among controls and CTE groups. For multiple linear regressions, age at death was included to control for age specific effects.
Preparation of Detergent Insoluble Fraction and LC-MS/MS With Tandem Mass Tag Quantification
Brain detergent insoluble fractions were generated as described previously from fresh frozen human lateral prefrontal cortex (13, 14). All attempts were made to utilize the same region for each brain. In brief, approximately 200 mg of frozen human frontal cortex was dounce homogenized (buffer: 50 mM HEPES pH 7.0, 250 mM sucrose, 1 mM EDTA, and 1× HALT (ThermoFisher) protease inhibitor cocktail) followed by the addition of N-lauroylsarcosine (final 1% w/v) and NaCl (final 0.5 M). The sample was sonicated followed by ultracentrifugation at 180,000g. The pellet was solubilized in 8 M urea buffer and protein concentrations were determined (BCA method, ThermoFisher). Proteins were digested with trypsin and LC-MS/MS was performed as previously described except isobaric tandem mass tags ([TMTs]; ThermoFisher) were added to peptides to allow for multiplexing and quantification (22). Four batches of TMT peptide mixtures (10-plex) were prepared and mixed such that batch-confounding by disease status or CTE stage was avoided, and each peptide batch was offline fractionated via ERLIC solid phase for liquid chromatography. ERLIC fractions (n = 11) for each batch were run on a Fusion Orbitrap (ThermoFisher) mass spectrometer. At least 1 TMT-labeled channel in each batch was comprised of a multi-brain mixture representing all cases analyzed as a standard by which abundances across batches could be normalized. Analysis of MS/MSspectra for identification, and MS3 multi-notch accumulated reporter ion intensities was performed as previously reported, using Proteome Discover v2.1 (ThermoFisher) (4, 22, 23). Raw data was searched against peptides generated from the April 2015 Uniprot complete nonredundant FASTA database (90,411 entries). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PX007695 (24) and summarized in Supplementary Data Table S2. For quantification, we included all proteins that were identified and quantified in at least 3 of the 4 batches and excluded proteins with >1 value missing in an individual CTE stage. Normalized log2 reporter ion intensity ratio versus the global multi-brain mixture was used to compare protein expression across cases and batches. The Student t-test (2-tailed) was utilized to calculate p values and significance for the differentially enriched or depleted proteins. Fold change comparison was calculated by subtracting the control log2 values from the CTE or AD log2 values.
RESULTS
Proteomic Analysis of CTE Detergent Insoluble Proteome
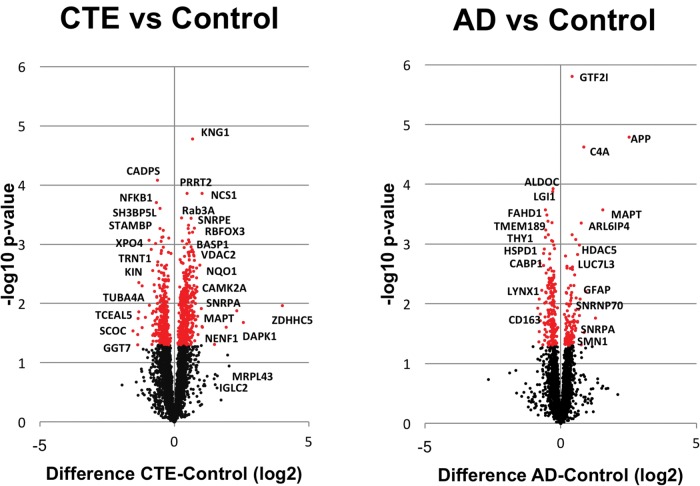
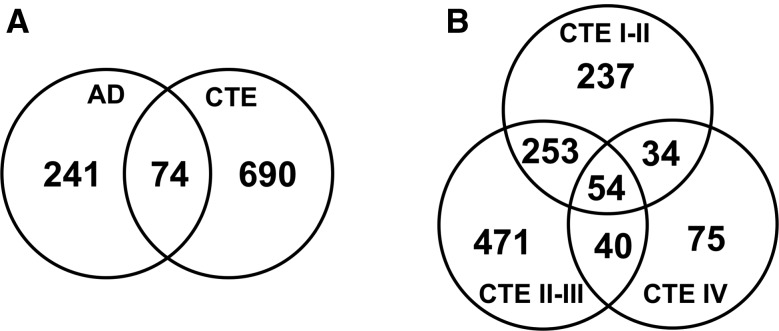
We have previously identified significant disease related protein changes in AD and other neurodegenerative disorders with LC-MS/MS of brain detergent insoluble fraction (13, 14). We were interested exploring this technique in cases of CTE, but the pathology is often irregular and patchy, particularly in earlier stages. We therefore used a similar approach for discovery of proteomic changes in CTE with using TMT labeling and off-line fractionation to improve the number of proteins identified and quantified. TMT labeling was performed on brain detergent insoluble fractions in CTE (n = 11) and control cases (n = 6). AD cases (n = 8) were also included since AD pathology often coexists with CTE pathology, especially in later stage subjects (Supplementary Data Table S1). We identified over 4,000 unique proteins, and, compared to controls, we found significantly increased/reduced expression (p < 0.05) in 317 proteins for AD (n = 8) and 764 proteins for CTE (n = 11) (Supplementary Data Table S2). Volcano plots (Fig. 1) show the distributions of proteins that were significantly enriched and reduced in CTE (n = 11) and AD (n = 8) cases as compared to controls. These plots provide perspective on numbers of significant proteins as well as the comparative significance of the expression change. To demonstrate stage specific changes, CTE cases were further divided into respective stages in the volcano plots shown in Supplementary Data Figure S1. Although there was overlap between the AD and CTE insoluble proteomes as well as between CTE stages (Fig. 2; Supplementary Data Table S3), there were also proteins unique to each neurodegenerative disease. As expected, we identified enrichment of the microtubule associated protein tau in both AD and CTE. β-amyloid enrichment was identified in AD but not in the overlapping protein list with CTE, likely due to the inclusion of lower stage CTE cases which do not typically have β-amyloid pathology. TDP-43 and α-synuclein were not identified in either the AD/CTE or CTE stage overlapping proteins (Supplementary Data Table S3).
FIGURE 1.
Volcano plots all insoluble brain proteins sequenced from CTE (n = 11) and AD (n = 8) as compared to control cases (n = 6). The plot shows protein abundance, either enriched or reduced (log2-fold-change), against the t-statistic (−log10(p-value)), which is a measure of significance. Red dots are significant differentially expressed proteins (p < 0.05).
FIGURE 2.
Venn diagrams of significantly reduced and enriched insoluble proteins as compared to controls. (A) AD (n = 8) and CTE (n = 11). (B) CTE I–II (n = 3), CTE II–III (n = 4) and CTE IV (n = 4).
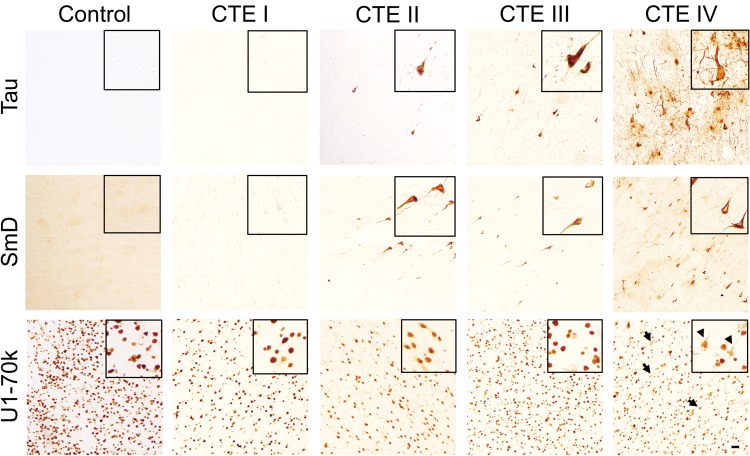
Interestingly, we also identified enrichment of RNA processing factors (Supplementary Data Tables S2, S3, and Fig. S1), including U1-70k. Enrichment of U1-70k and other members of the U1snRNP complex were previously identified in the insoluble proteome of AD in association with neurofibrillary tangle-like aggregates (13). We performed immunohistochemical staining of CTE postmortem brain to assess for similar pathologies and identified tangle-like aggregates of both U170k and SmD in CTE cases (Fig. 3). The SmD pathology followed more closely with density of tau pathology staining with increasing CTE stage, whereas U1-70k aggregates were sparse in earlier CTE stages as compared to CTE stage IV. The results from these studies demonstrate the ability of LC-MS/MS proteomics to identify known pathological features (e.g., tau) and reveal a previously unknown shared pathology in CTE and AD. The limited overlap in the profile of differentially expressed proteins further distinguish CTE and AD as distinct pathologic entities.
FIGURE 3.
Immunohistochemistry of RNA processing factors in CTE. Human frontal cortex 50-µm free-floating sections from control (n = 6) and CTE (n = 11) cases with increasing CTE stage was immunostained with Tau (AT8), SmD, and U1-70k. Sections were from dorsolateral prefrontal cortex and images shown were from depths of the sulci. Inset figures provide a magnified view from each figure panel. Black arrows designate U1-70k-positive tangle-like aggregates in the CTE stage IV case. Scale bar: 100 µm.
Proteomic Changes With Increasing CTE Pathologic Stages
Four stages have been described in CTE based on the overall density and distribution of tau pathology within the brain (15, 17). Previous work has suggested that increased pathologic burden of tau was associated with the development of dementia in CTE (18). While the correlation between tau and symptom severity has not yet been firmly established, increasing pathology with higher stage is likely to reflect pathophysiological processes that may provide clues about disease pathogenesis and progression. As previously noted, there is reason for concern that random sampling from earlier stage CTE cases might fail to capture disease related changes. To evaluate this possibility, we divided the CTE cohort into 3 groups based on CTE stage. Three instead of 4 groups were utilized due to the small number of cases. When compared to control brains, we identified unique groups of differentially expressed proteins in the detergent insoluble proteome of each of the 3 groups (CTE I–II: 578; CTE II–III: 818; CTE IV: 203) as well as shared protein changes across the different stages (Fig. 2; Supplementary Data Table S3). Notably, tau was significantly enriched in the CTE II–III and CTE IV groups, but not in the CTE I–II group. This finding is consistent with neuropathological observations that show more limited tau pathology in these lower CTE stages (17). Interestingly, our proteomics results did not reveal enrichment of β-amyloid in any of the CTE groups despite the previously reported overlap of β-amyloid pathology between AD and late stage cases (16). We suspect this was due to the small number of cases and variability in β-amyloid deposition.
The top 25 most enriched proteins in the CTE IV subgroup included 4 proteins that demonstrated progressive enrichment with increasing stage across the 3 CTE subgroups. These included NADPH quinone dehydrogenase 1 (NQO1), neuronal calcium sensor 1 (NCS1), leucine-rich repeat, tropomodulin, proline-rich region containing (RLTPR), and calmodulin 2 (CALM2) (Table). There were also 3 other proteins (kininogen 1 [KNG1]), mitochondrial ribosomal protein L32 (MRPL32), and voltage dependent anion channel 3 [VDAC3]) that demonstrated significant enrichment in all groups, but did not show progressive enrichment with increasing pathological stage. When considering all the CTE cases as a single group, 4 of these proteins (NCS1, NQO1, MRPL32, and CALM2) remained in the top 25 most enriched insoluble proteins. These experiments demonstrate novel stage-specific changes identified by analyzing insoluble proteome changes across CTE severity.
TABLE.
CTE Insoluble Brain Proteome
| CTE (I–II) vs CTL | CTE (II–III) vs CTL | CTE (IV) vs CTL | |
|---|---|---|---|
| 1 | COL1A1 | NENF | ITM2B |
| 2 | KRT16 | IGLC2 | PAK4 |
| 3 | COX7C | IGHG1 | ZAK |
| 4 | BANF1 | IGHA1 | NQO1* |
| 5 | MRPL32* | IGKC | C19orf66 |
| 6 | EWSR1 | KRT9 | NCS1* |
| 7 | FTH1 | EFHD2 | CYP27A1 |
| 8 | PHF5A | ERAL1 | NAA50 |
| 9 | ARHGEF25 | NCS1* | KNG1* |
| 10 | KRT10 | ABHD11 | MRPL32* |
| 11 | EGFL7 | NQO1* | MAPT |
| 12 | FGD1 | KRT1 | NDUFC1 |
| 13 | FBXO41 | PDS5A | PKIG |
| 14 | GPC1 | GPM6B | DCAF5 |
| 15 | GFM2 | CSTB | RLTPR* |
| 16 | ERAL1 | NAV3 | CRYZ |
| 17 | DCAF5 | C19orf66 | ATP6V0C |
| 18 | CAMK2A | GTPBP10 | RNF34 |
| 19 | RPL10A | LAMA5 | PGAM2 |
| 20 | RPL12 | CAMK2A | SYNPR |
| 21 | NCS1* | LAMA1 | CALM2* |
| 22 | MTIF2 | LAMC3 | DNAJB2 |
| 23 | SNRPB | PALD1 | CRTC2 |
| 24 | BSN | RPL12 | VDAC3* |
| 25 | FARS2 | RAB15 | DNAJC5 |
| 45-VDAC3* | 34-CALM2* | ||
| 66-CALM2* | 60-RLTPR* | ||
| 86-KNG1* | 73-MRPL32* | ||
| 106-RLTPR* | 79-VDAC3* | ||
| 119-NQO1* | 110-KNG1* |
The top 25 significantly enriched proteins are shown across increasing CTE stages. Seven of the top 25 stage IV proteins are also enriched in the first 2 groups. If the protein is not in the top 25, it is listed below the table with the position number on the list. CTE, chronic traumatic encephalopathy; CTL, control.
Seven proteins of interest.
Abnormal Accumulation of Insoluble Protein Target
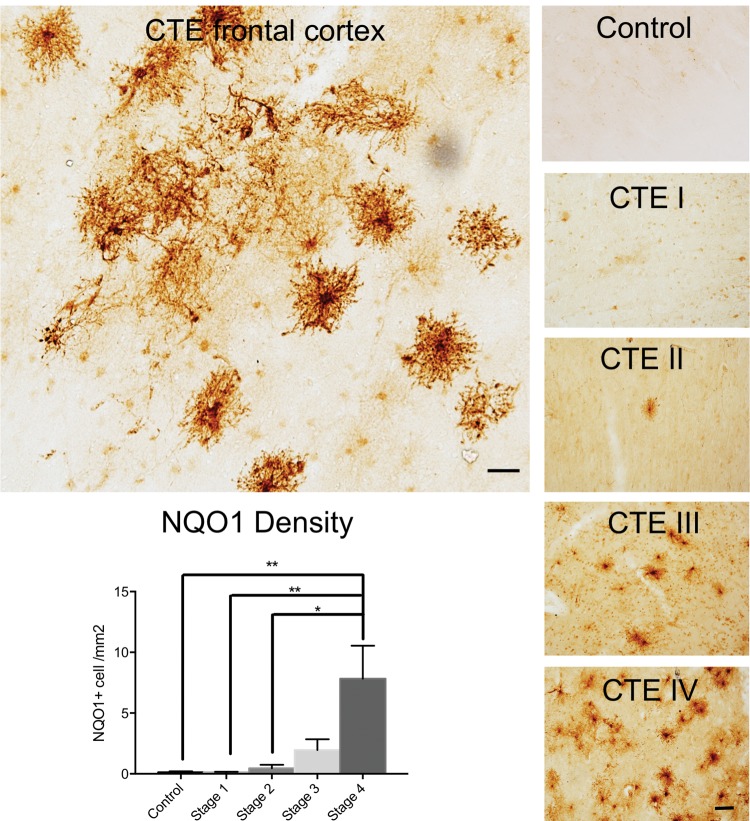
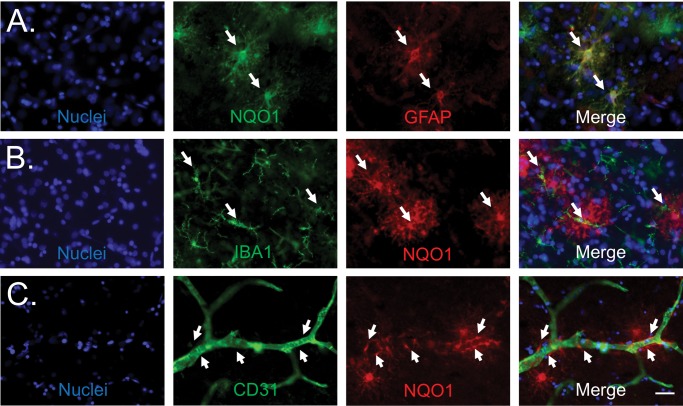
Results from sequencing the insoluble proteome of CTE cases suggested previously unknown disease related changes. We sought to further evaluate the proteomic changes observed in CTE cases, including tissue localization by immunohistochemistry, to better understand the cellular basis for altered protein expression. Of the proteins that were enriched in the stage IV CTE cases, NQO1 demonstrated the largest relative increase in accumulation with advancing CTE pathology from stage I to IV. NQO1 was also the most enriched of the 54 shared differentially expressed insoluble proteins in CTE across all stages (Fig. 2; Supplementary Data Table S3) and was not enriched in the AD insoluble proteome. Additionally, previously published reports suggest that NQO1 may play a role in neurodegeneration (25–28). Since proteins in the detergent insoluble proteome are known to aggregate (tau, amyloid, RNA processing factors), we performed immunohistochemical labeling of NQO1 to assess possible aberrant localization or accumulation in CTE and AD brains. CTE cases demonstrated large fluffy cells with NQO1 immunostaining, and this staining pattern became more abundant with increasing CTE stage (Fig. 4). A majority of AD cases (54%, 12/22) did not contain any NQO1-positive cells. Thirty-two percent (7/12) of AD cases demonstrated occasional and 14% (3/22) demonstrated more frequent NQO1-positive cells (Supplementary Data Fig. S2). Morphologically, the cells had the appearance of reactive astrocytes or possibly activated microglia. In double-labeling experiments, many of the NQO1-positive cells were also positive for GFAP, a marker of astrocytes, but there was no overlap with the microglial marker IBA1 (Fig. 5). NQO1-positive cellular processes frequently contacted microvasculature as seen in dual labeling experiments with CD31, further identifying these cells as astrocytes. There were NQO1-positive cells that did not have astrocytic markers, however we were unable to identify the cell subtype, even after assessing with additional antibodies to oligodendrocytes (oligodendrocyte transcription factor 2 [olig2] and neural/glial antigen 2 [NG2], oligodendrocyte precursor cells [NG2 and O4] and interneurons [parvalbumin; data not shown]). Given the paucity of the NQO1-positive cells in control cases, dual labeling figures are also not shown. We also evaluated nuclear factor E2-related factor 2 (NRF2), an NQO1 transcription factor, in order to determine whether the increased NQO1 immunostaining was the result of increased cellular production of NRF2 in response to oxidative injury. Dual immunofluorescence staining of CTE brain showed that only rare NQO1-positive cells demonstrated NRF2 immunostaining while the majority of NQO1-positive cells did not have clear NRF2 colabeling (Supplementary Data Fig. S3).
FIGURE 4.
NQO1 immunostaining in CTE brain. The upper left panel shows NQO1 labeling in human frontal cortex of a stage IV CTE case. Scale bar: 50 µm. The 5 panels on the right show representative examples of increasing NQO1 pathology with increasing CTE stage (control n = 6 and CTE n = 11) in frontal cortex 50-µm free-floating sections. Scale bar: 100 µm. The bottom left panel provides quantitation of NQO1 density from paraffin sections of frontal cortex with increasing CTE stage (Control n = 9, CTE I n = 8, CTE II n = 8, CTE III n = 9, CTE IV n = 11) as a validation cohort. One whole sulcus was utilized for measuring NQO1 density in each case.
FIGURE 5.
NQO1-positive cells localize to blood vessels and GFAP but not IBA 1. (A) NQO1 (green), GFAP (red), Hoechst (blue). White arrows: NQO1-positive cells with GFAP staining (yellow color on merge panel). (B) IBA1 (green), NQO1 (red), Hoechst (blue). White arrows: IBA1-positive microglia adjacent to but not colocalizing with NQO1-positive cells. (C) CD31 (green), NQO1 (red), and Hoechst (blue). White arrows: foot processes from NQO1-positive cells. Sections utilized were from the depths of sulci from dorsolateral prefrontal cortex. Scale bar: 50 µm.
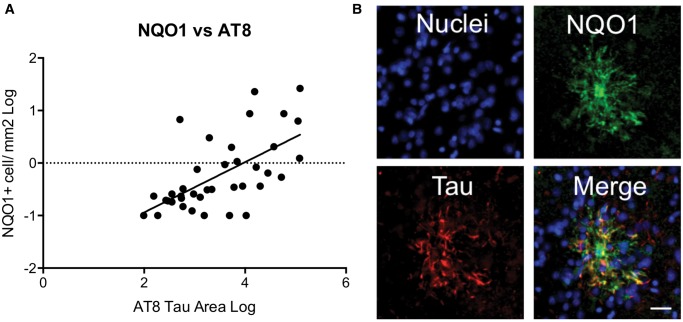
The density of NQO1 stained cells significantly correlated with hyperphosphorylated tau immunostaining and NQO1 colocalized with tau immunostaining within these cells (Fig. 6). Almost all NQO1-positive cells also contained the abnormal accumulation of hyperphosphorylated tau (Supplementary Data Fig. S4) Later stage CTE cases are typically from older individuals than earlier stage cases. Therefore, to determine if the NQO1-tau relationship was age related, we performed a multiple linear regression analysis which showed that NQO1 significantly predicted AT8 tau density (β = 0.604, p < 0.001) independently of age (β = 0.073, p = 0.589). These immunohistochemical studies validate findings from our proteomics studies and suggest that CTE is associated with aberrant expression of NQO1 in astrocytes and that NQO1 abnormalities correlate with the accumulation of tau pathology in increasing stages of CTE. Elevated NRF2 levels may be driving the NQO1 expression in some of the cells.
FIGURE 6.
Correlation of NQO1 and tau in CTE frontal cortex. (A) NQO1 correlation with hyperphosphorylated tau density in cases with CTE. (B) Immunofluorescence labeling of nuclei Hoechst (blue), NQO1 (green), and tau (red) in a single large NQO1-positive cell. Yellow color in the merge image designates colocalization. Scale bar: 50 µm.
DISCUSSION
This study used proteomic sequencing to identify disease related changes in CTE postmortem brain. Given prior success characterizing aggregation prone proteins (4, 6, 29), we sequenced over 4,000 proteins from the CTE detergent insoluble proteome with TMT sample labeling and mass spectrometry. We identified expected insoluble proteins like tau as well as other novel proteins like the RNA processing factors that previously were only associated with AD (13, 14). Homology between the CTE and AD insoluble proteomes was <25%, further confirming that CTE was a different pathologic entity. Despite the relatively sparse pathology in early CTE stages, we identified proteins uniquely changing in each stage as well as proteins demonstrating increased enrichment with higher CTE stages. An example of this was NQO1, which demonstrated greater insolubility and pathologic immunostaining with increasing stage. NQO1 pathology also correlated with tau immunostaining independent of age and colocalized with tau in astrocytes. Together, these data confirmed that mass spectrometry can identify novel CTE proteomic changes associated with disease.
The identification of insoluble tau was expected given that aggregates of hyperphosphorylated tau are a main pathological hallmark of AD and CTE. Neurofibrillary tangles were present in both diseases, however, there were also distinct CTE tau pathological traits including the perivascular accumulation and deep sulcal localization. The identification of RNA processing factors was also not entirely surprising since the overlap between CTE and AD pathology has been well described (16). Mechanistically, this could suggest that RNA processing is disrupted in CTE such that all pre-mRNAs are not properly converted into functional mRNA, leading to aberrant protein expression that may contribute to disease pathogenesis. Similarly, the RNA processing factor U1-70 K contains low complexity domains that may contribute to abnormal protein aggregation, including the formation of neurofibrillary tangles (29). SmD does not contain low complexity domains, however, it is also robustly associated with neurofibrillary tangle pathology. It is unclear if domains in SmD or the low complexity domains in U1-70 K contribute to the development of AD or CTE pathology, however, studies are currently in progress to test this hypothesis.
CTE cases can also have β-amyloid pathology, especially in the older age groups and later CTE stages (16). Data suggested that coexisting amyloid pathology may indeed function to accelerate the CTE tau pathology at an earlier age although the converse action of CTE pathology accelerating amyloid pathology may occur as well (16). For this cohort, we did not identify significantly enriched amyloid, even in the late-stage CTE cases. This could speak to inherent variability within the tissues, however, we previously identified enriched fragments of amyloid precursor protein in a smaller CTE cohort of late stage cases (data not shown) suggesting that it can be done. Increasing overall case numbers and stratifying based on coexisting pathologies (like β-amyloid) may improve sensitivity for detecting protein changes in future studies.
Although the proteomic data suggested that insoluble enrichment of NQO1 was specific for the CTE cases, we also identified a few AD cases which contained NQO1-positive cells. However, the distribution of immunostaining was not as robust as seen in the CTE cases, as most of the AD cases had no significant NQO1 immunostaining or only occasional NQO1-positive cells. This finding is not surprising given that there is overlap between CTE and AD pathology, especially in the later stage cases that are also inherently older. Fully characterizing prior history of CTE risk factors can also be difficult as it is possible that the small number of AD cases with more robust NQO1 staining may have had a prior history of repetitive head injury, despite attempts to capture this information via questionnaires. Further studies investigating the driving force behind the NQO1 enrichment will be helpful in clarifying its role in neurodegenerative diseases.
This study was limited by relatively small numbers of CTE cases, especially when dividing the cohort to query the different CTE stages. Even with this limitation, we demonstrated the feasibility of tracking enriched proteins through increasing CTE stages. As proof of concept, we selected one of these proteins to validate, NQO1. NQO1 was the most enriched of all the overlapping insoluble proteins and demonstrated increasing enrichment with CTE stage. NQO1 was also interesting from a biological perspective because it is involved in the reduction of reactive oxygen species. In addition, studies have suggested that NQO1 may play a role in AD, however, the results from these studies are not conclusive (25). The enrichment of NQO1 in the insoluble proteome and the further identification of NQO1 immunostaining with hyperphosphorylated tau, could suggest that NQO1 is forming pathological aggregates that reduce its capacity to function. As a result, the cells could be more susceptible to reactive oxygen species and oxidative injury and therefore prone to degenerate, perhaps contributing to disease pathogenesis in CTE. Alternatively, increased NQO expression in these cells may actually be a protective cellular response, and represent the cell’s attempt to reduce oxidative injury in cells that are being damaged by the abnormal tau aggregations or by oxidative injury in the surrounding parenchyma. The increased NRF2 immunostaining in a subpopulation of the NQO1-positive cells would support the later hypothesis that the cells are attempting to upregulate protective mechanisms to reduce oxidative injury. Similarly, there were NRF2-positive cells, possibly neurons or interneurons, that have NRF2 expression near the NQO1-positive cells (Supplementary Data Fig. S3), and these cells may also be contributing to the NQO1 expression levels. If additional studies support this finding, recent interest in targeting the NRF2/NQO1 pathway and repurposing NRF2 activators as a therapeutic strategy for AD and PD may also prove useful for CTE (26, 28, 30).
Another challenge with validating NQO1 was confirming the identity of the NQO1-positive cells. The GFAP dual staining and cellular processes contacting microvasculature supported that NQO1-positive cells were astrocytes, however, a population of cells was negative for GFAP and other markers. Although these could be astrocytes without robust GFAP expression, another study with transcriptomic data suggested that NQO1 expression was present in oligodendrocyte precursor cells (OPC) (31). Morphologically this could be consistent, however, we were unable to confirm this hypothesis despite trying multiple dual-labeling experiments with OPC markers. Either way, the contribution of glial cells to the neurodegenerative cascade is becoming increasingly recognized. Additional studies including disease modeling may be helpful in clarifying how NQO1 contributes to the disease process.
Overall, we confirmed that analysis of the CTE insoluble proteome could generate relevant findings even when analyzing aggregate data across all CTE stages. We also demonstrated that proteins can be followed through increasing CTE stage and validated with an orthogonal approach. In addition to the RNA processing factors and NQO1, we identified a wealth of many other proteins to explore in the context of CTE progression even with this small cohort. Larger case numbers will likely improve the ability to detect more changes, reduce false positives, and also provide further opportunity for more sophisticated systems biology analysis. The ultimate goal is that this approach will provide additional mechanistic insight into CTE pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the use of resources and facilities at the Edith Nourse Rogers Memorial Veterans Hospital (Bedford, MA) as well as all the individuals whose participation and contributions made this work possible.
Compliance with ethical standards: IRB approval for brain donation was obtained through the Boston University Alzheimer’s Disease and CTE Center (ADC), the Edith Nourse Rogers Memorial Veterans Hospital, Bedford, MA, the Emory Alzheimer’s Disease Research Center and Emory University, Atlanta, GA.
Availability of data and materials: The datasets generated and analyzed during the current study are available on ProteomeXchange (Project accession: PXD007694) and summarized in Supplementary Data Table S2.
REFERENCES
- 1. Taylor JP, Hardy J, Fischbeck KH.. Toxic proteins in neurodegenerative disease. Science 2002; 296:1991–5 10.1126/science.1067122 [DOI] [PubMed] [Google Scholar]
- 2. Ross CA, Poirier MA.. Protein aggregation and neurodegenerative disease. Nat Med 2004; 10:S10–7 [DOI] [PubMed] [Google Scholar]
- 3. Dammer EB, Fallini C, Gozal TM, et al. Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One 2012; 7:e38658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donovan LE, Higginbotham L, Dammer EB, et al. Analysis of a membrane-enriched proteome from postmortem human brain tissue in Alzheimer's disease. Proteomics Clin Appl 2012; 6:201–11 10.1002/prca.201100068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seyfried NT, Dammer EB, Swarup V, et al. A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer's disease. Cell Syst 2017; 4:60–72 10.1016/j.cels.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seyfried NT, Gozal YM, Donovan LE, et al. Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J Proteome Res 2012; 11:2721–38 10.1021/pr2010814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Licker V, Kovari E, Hochstrasser DF, et al. Proteomics in human Parkinson's disease research. J Proteomics 2009; 73:10–29 10.1016/j.jprot.2009.07.007 [DOI] [PubMed] [Google Scholar]
- 8. Musunuri S, Wetterhall M, Ingelsson M, et al. Quantification of the brain proteome in Alzheimer's disease using multiplexed mass spectrometry. J Proteome Res 2014; 13:2056–68 10.1021/pr401202d [DOI] [PubMed] [Google Scholar]
- 9. Begcevic I, Kosanam H, Martinez-Morillo E, et al. Semiquantitative proteomic analysis of human hippocampal tissues from Alzheimer's disease and age-matched control brains. Clin Proteomics 2013; 10:5. 10.1186/1559-0275-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agresta AM, De Palma A, Bardoni A, et al. Proteomics as an innovative tool to investigate frontotemporal disorders. Proteomics Clin Appl 2016; 10:457–69 10.1002/prca.201500090 [DOI] [PubMed] [Google Scholar]
- 11. Licker V, Turck N, Kovari E, et al. Proteomic analysis of human substantia nigra identifies novel candidates involved in Parkinson's disease pathogenesis. Proteomics 2014; 14:784–94 10.1002/pmic.201300342 [DOI] [PubMed] [Google Scholar]
- 12. Steger M, Tonelli F, Ito G, et al. Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 2016; 5. doi: 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai B, Hales CM, Chen PC, et al. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc Natl Acad Sci U S A 2013; 110:16562–7 10.1073/pnas.1310249110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hales CM, Dammer EB, Deng Q, et al. Changes in the detergent-insoluble brain proteome linked to amyloid and tau in Alzheimer's Disease progression. Proteomics 2016; 16:3042–53 10.1002/pmic.201600057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKee AC, Cairns NJ, Dickson DW, et al. The first NINDS/NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. Acta Neuropathol 2016; 131:75–86 10.1007/s00401-015-1515-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stein TD, Montenigro PH, Alvarez VE, et al. Beta-amyloid deposition in chronic traumatic encephalopathy. Acta Neuropathol 2015; 130:21–34 10.1007/s00401-015-1435-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKee AC, Stein TD, Nowinski CJ, et al. The spectrum of disease in chronic traumatic encephalopathy. Brain 2013; 136:43–64 10.1093/brain/aws307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cherry JD, Tripodis Y, Alvarez VE, et al. Microglial neuroinflammation contributes to tau accumulation in chronic traumatic encephalopathy. Acta Neuropathol Commun 2016; 4:112. 10.1186/s40478-016-0382-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hales CM, Dammer EB, Diner I, et al. Aggregates of small nuclear ribonucleic acids (snRNAs) in Alzheimer's disease. Brain Pathol 2014; 24:344–51 10.1111/bpa.12133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hales CM, Seyfried NT, Dammer EB, et al. U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer's disease due to autosomal dominant genetic mutations and trisomy 21. Mol Neurodegener 2014; 9:15. 10.1186/1750-1326-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herskowitz JH, Feng Y, Mattheyses AL, et al. Pharmacologic inhibition of ROCK2 suppresses amyloid-beta production in an Alzheimer's disease mouse model. J Neurosci 2013; 33:19086–98 10.1523/JNEUROSCI.2508-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gokhale A, Hartwig C, Freeman AH, et al. The proteome of BLOC-1 genetic defects identifies the Arp2/3 actin polymerization complex to function downstream of the schizophrenia susceptibility factor dysbindin at the synapse. J Neurosci 2016; 36: 12393–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dammer EB, Duong DM, Diner I, et al. A neuron enriched nuclear proteome isolated from human brain. J Proteome Res 2013; 12:3193–206 10.1021/pr400246t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vizcaino JA, Csordas A, del-Toro N, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 2016; 44:D447–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raina AK, Templeton DJ, Deak JC, et al. Quinone reductase (NQO1), a sensitive redox indicator, is increased in Alzheimer's disease. Redox Rep 1999; 4:23–7 10.1179/135100099101534701 [DOI] [PubMed] [Google Scholar]
- 26. Simoni E, Serafini MM, Caporaso R, et al. Targeting the Nrf2/amyloid-beta liaison in Alzheimer's disease: a rational approach. ACS Chem Neurosci 2017; 8:1618–27 10.1021/acschemneuro.7b00100 [DOI] [PubMed] [Google Scholar]
- 27. Cao H, Wang L, Chen B, et al. DNA demethylation upregulated Nrf2 expression in Alzheimer's disease cellular model. Front Aging Neurosci 2015; 7:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lastres-Becker I, Garcia-Yague AJ, Scannevin RH, et al. Repurposing the NRF2 activator dimethyl fumarate as therapy against synucleinopathy in Parkinson's disease. Antioxid Redox Signal 2016; 25:61–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Diner I, Hales CM, Bishof, et al. Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. J Biol Chem 2014; 289:35296–313 10.1074/jbc.M114.562959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson DA, Johnson JA.. Nrf2: a therapeutic target for the treatment of neurodegenerative diseases. Free Radic Biol Med 2015; 88:253–67 10.1016/j.freeradbiomed.2015.07.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34:11929–47 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.