Abstract
Glucagonlike peptide-1 receptor (GLP-1R) agonists, which are used to treat type 2 diabetes and obesity, reduce the rates of myocardial infarction and cardiovascular death. GLP-1R has been localized to the human sinoatrial node; however, its expression in ventricular tissue remains uncertain. Here we studied GLP-1R expression in the human heart using GLP-1R–directed antisera, quantitative polymerase chain reaction (PCR), reverse transcription PCR to detect full-length messenger RNA (mRNA) transcripts, and in situ hybridization (ISH). GLP1R mRNA transcripts, encompassing the entire open reading frame, were detected in all four cardiac chambers from 15 hearts at levels approximating those detected in human pancreas. In contrast, cardiac GLP2R expression was relatively lower, and cardiac GCGR expression was sporadic and not detected in the left ventricle. GLP1R mRNA transcripts were not detected in RNA from human cardiac fibroblasts, coronary artery endothelial, or vascular smooth muscle cells. Human Brunner glands and pancreatic islets exhibited GLP-1R immunopositivity and abundant expression of GLP1R mRNA transcripts by ISH. GLP1R transcripts were also detected by ISH in human cardiac sinoatrial node tissue. However, definitive cellular localization of GLP1R mRNA transcripts or immunoreactive GLP-1R protein within human cardiomyocytes or cardiac blood vessels remained elusive. Moreover, validated GLP-1R antisera lacked sufficient sensitivity to detect expression of the endogenous islet or cardiac GLP-1R by Western blotting. Hence, although human cardiac ventricles express the GLP1R, the identity of one or more ventricular cell type(s) that express a translated GLP1R protein requires further clarification with highly sensitive methods of detection.
We assess current methods of detecting GLP-1R expression and apply different techniques in the investigation of GLP-1R expression within distinct chambers and cell types in the human heart.
Incretin hormones, principally glucose-dependent insulinotropic polypeptide and glucagonlike peptide-1 (GLP-1), are secreted from the gut after meal ingestion and amplify glucose-dependent insulin secretion (1). GLP-1 also inhibits glucagon secretion and gastric emptying and promotes satiety; these actions underly the therapeutic use of GLP-1 receptor (GLP-1R) agonists in the treatment of type 2 diabetes (T2D) (2, 3). A single GLP-1R agonist, liraglutide, has also been approved for the treatment of obesity (4); an oral form of semaglutide has entered phase 3 clinical testing (5); and multiple unimolecular agonists that enhance GLP-1 action are under clinical development (6). Hence, there is ongoing interest in the translational mechanisms of GLP-1 action.
The actions of GLP-1 are mediated through a single GLP-1R, a member of the class B family of G protein–coupled receptors (7). The GLP-1R is widely expressed outside the pancreas, including neurons within the enteric, central, and peripheral nervous system; kidney; lung; the immune system; and diverse cell types within the stomach and the small and large bowel (8–10). GLP-1R has also been identified in the cardiovascular system; within blood vessels, vascular smooth muscle cells, and endothelial cells; and in the heart (11, 12).
Consistent with the detection of GLP1R messenger RNA (mRNA) transcripts and protein in human heart and blood vessels, GLP-1R agonists exert multiple actions in the cardiovascular system, including induction of heart rate, vasodilation, and reduction of blood pressure in the setting of hypertension (13). Activation of GLP-1R signaling rapidly improves ventricular function in the context of transient ischemia in preclinical and clinical studies (14) and reduces infarct size in experimental models of ischemia secondary to coronary artery ligation (15, 16). Short-term administration of GLP-1R agonists for a few hours also reduces the extent of reperfusion injury in the myocardium of patients with ST-segment elevation myocardial infarction (17). Nevertheless, the mechanisms and cell types through which GLP-1R signaling reduces injury of the cardiac ventricles remains incompletely understood.
The requirement of regulatory agencies mandating assessment of the cardiovascular safety of newer drugs approved for the treatment of T2D has further enhanced interest in understanding how GLP-1R agonists might regulate cardiovascular function. The first reported outcome study for drugs in this class examined the cardiovascular safety of lixisenatide in human subjects with T2D and recent acute coronary syndrome; lixisenatide did not significantly alter the rate of major adverse cardiovascular events (18). The cardiovascular safety of once-weekly exenatide was also recently established in over 14,000 subjects with T2D, of whom more than 70% had established cardiovascular disease (19). In contrast, two studies examining the cardiovascular safety of liraglutide and semaglutide demonstrated a reduction in the rate of death from cardiovascular causes, nonfatal myocardial infarction, or nonfatal stroke in subjects with T2D (20, 21).
The demonstration that some GLP-1R agonists produce cardioprotective actions in human subjects raises important questions surrounding the cardiovascular biology of the human GLP-1R. Although the expression and function of the GLP-1R in the cardiovascular system has been extensively examined in preclinical studies, the precise localization of GLP1R expression in the human heart remains uncertain. Wallner et al. (22) detected GLP1R mRNA transcripts using reverse transcription polymerase chain reaction (RT-PCR) in RNA from human atrium and ventricle. Exenatide increased contractility in isolated trabeculae from the atrium but not from the majority of ventricular preparations examined (22). Consistent with the actions of GLP-1R agonists to increase heart rate (15), a well-characterized monoclonal antibody detected GLP-1R–immunopositive cells within the sinoatrial node of nonhuman primates and a single human heart (23).
Despite the growing translational interest in understanding sites and mechanisms of GLP-1 action in the human heart, few studies have comprehensively assessed GLP-1R expression in human cardiac tissue. Moreover, detection of the immunoreactive GLP-1R has proven challenging due to the suboptimal sensitivity and specificity of the available reagents (24, 25). To determine the localization of GLP1R expression in the human heart, we studied GLP1R expression in heart samples available from two different human tissue biobanks. We also assessed the sensitivity and specificity of two GLP-1R antisera known to recognize the human GLP-1R. Here we report that GLP1R mRNA transcripts encoding the entire GLP-1R open reading frame were consistently detected in biopsy samples obtained from all four chambers from 15 different human hearts. Despite analysis of histological sections from an independent group of 35 human hearts using a combination of in situ hybridization (ISH) and immunohistochemistry, we were unable to identify the precise cellular localization of human ventricular GLP-1R expression.
Materials and Methods
Human tissue samples
Human heart samples (left atrium, right atrium, left ventricle, right ventricle) for RNA and protein isolation were acquired from the University of Pennsylvania Heart BioBank. Tissues from failing human hearts were obtained from patients receiving heart transplantation at the University of Pennsylvania. The use of human heart tissue for research was approved by the University of Pennsylvania’s Institutional Review Board. Nonfailing heart tissue was obtained from deceased organ donors through the Gift-of-Life donor program in Philadelphia. Prospective informed consent for research use of heart tissue was obtained from all transplant recipients and from the appropriate next of kin for the organ donors. All human tissues received in situ high-potassium cardioplegia at the time of tissue procurement. Conventions for cardiac tissue sampling were as follows: left ventricle—free wall from a noninfarcted region, no septum, and predominantly anterior wall midway between the apex and base; right ventricle—anterior free wall in a region containing minimal epicardial fat; left atrium—free wall, no septum, and no left atrial appendage; right atrium—free wall, no septum, and no right atrial appendage. A description of these human heart samples is provided in Supplemental Table 1 and in Ussher et al. (26). Human cardiac tissues were pulverized in liquid nitrogen and stored at –80°C prior to RNA or protein extraction. Human adipose tissues for RNA isolation were obtained from abdominal wall samples that were removed from patients undergoing breast reconstruction surgery via tram flap and were provided by Dr. John Semple [University Health Network (UHN), Toronto, ON, Canada]. Human intestinal tissue (colon) was obtained from the Mount Sinai Hospital tissue bank after research ethics approval. Human cardiac tissue sections for immunohistochemistry and ISH experiments were obtained from a total of 35 different subjects from the UHN Rapid Autopsy Program as approved by the UHN Research Ethics Board. Supplemental Table 2 provides a list of these human cardiac tissues. Human Brunner glands, pancreas tissue, and cardiac atrial sections were provided by Dr. Catherine Streutker (St. Michael’s Hospital, Department of Surgical Pathology, Toronto, ON, Canada). Human islet samples (>95% purity) were generously provided by Dr. Patrick MacDonald (Alberta Diabetes Institute, University of Alberta, Edmonton, AB, Canada). Human islet donor information is listed in Supplemental Table 3.
Human RNA samples
Human RNA samples were purchased from the following sources: adipose (catalog no. R1234003-10, BioChain, Newark, CA), bone marrow (catalog no. R1234024-10, BioChain; catalog no. HR-704, Zyagen, SanDiego, CA; catalog no. 636591, Takara Bio, Mountain View, CA), coronary artery endothelial cells (catalog no. 300-R10a, Cell Applications Inc., San Diego, CA), human coronary artery smooth muscle cells (catalog no. 350-R10a, Cell Applications Inc.), peripheral blood leukocyte (catalog no. 636592, Takara Bio; catalog no. R1234148-10, BioChain), pericardium (catalog no. R1234133-50, BioChain), liver (catalog no. AM7960, Thermo Fisher, Markham, ON, Canada; catalog no. 540017, Agilent Tech., Santa Clara, CA; catalog no. 636531, Clontech, Mountain View, CA). Human pancreas RNA was kindly provided by Dr. Maria Chrisina Nostro (Department of Physiology, University Health Network, University of Toronto, Toronto, ON, Canada).
Cell lines
Baby hamster kidney (BHK) fibroblast cells stably expressing the human GLP-1 receptor at high, low, and ultra-low levels of expression (BHK clones #12A, #13, and #27, respectively) (27, 28) were provided by Novo Nordisk (Bagsvaerd, Denmark). BHK cells transfected with pcDNA3.1 (Thermo Fisher) were generated as described (29). The human cystic fibrosis pancreatic adenocarcinoma (CFPAC-1) cell line was purchased from American Type Culture Collection (CRL-1918; ATCC, Manassas, VA). Human cardiac fibroblasts were purchased from ScienCell Research Laboratories (San Diego, CA).
RNA isolation and analysis
Total RNA was extracted from tissues or cells using Tri Reagent (Molecular Research Center Inc., Cincinnati, OH). cDNA was synthesized from DNase I–treated (Thermo Fisher) total RNA using random hexamers and Superscript III (Thermo Fisher). Real-time quantitative polymerase chain reaction (qPCR) was carried out using a QuantStudio 5 System and TaqMan Gene Expression Assays (Thermo Fisher). Relative mRNA levels were quantified using the 2−ΔCt method. A list of qPCR primers and assay identification numbers is provided in Supplemental Table 4. Polymerase chain reaction (PCR) products encompassing the majority of the human GLP1R, GCGR, GLP2R, or DPP4 coding regions were amplified from cDNA, separated on an agarose gel, transferred to a nylon membrane, and hybridized with internal human-specific 32P-labeled oligonucleotide probes for detection of GLP1R, GCGR, GLP2R, or DPP4 mRNA transcripts. Blots were exposed to a Storm 860 Phosphor Screen (Biorad, Mississauga, ON, Canada) and visualized using Quantity One imaging software (Bio-Rad). PCR oligonucleotide primer sequences are listed in Supplemental Table 4.
Western blot analyses
Whole tissue (human heart chambers and pancreatic islets) and cell line (BHK fibroblast and CFPAC-1) protein extracts were prepared by homogenization in PBS supplemented with 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and protease inhibitor cocktail (Sigma-Aldrich, Oakville, ON, Canada). Extracts were cleared by centrifugation and then treated for 1 h at 37°C with sample buffer containing β-mercaptoethanol. Protein (30 to 40 μg) was resolved by discontinuous sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electrotransferred onto Amersham Protran nitrocellulose membrane (GE Health Care Life Sciences, Mississauga, ON, Canada) using standard procedures. Immunoprecipitations were performed using 1 mg/mL of cell or tissue protein extract and 2 μg/mL of rabbit polyclonal anti-human GLP-1R antibody [Novus 19400002; Research Resource Identifier (RRID) AB_10004637; Bio-Techne, Oakville, ON, Canada] overnight at 4°C. Immune complexes were captured using Protein A Plus Agarose (Thermo Fisher Scientific) and then treated with sample buffer as described previously prior to sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrotransfer to Protran membrane. Blots were probed at room temperature with antibodies at 1:1000 dilutions in 5% skim milk in Tris-buffered saline containing 0.2% Tween 20. Antigen-antibody complexes were visualized with a secondary antibody conjugated to horseradish peroxidase and an enhanced chemiluminescence kit (SuperSignal WestPico; Thermo Fisher Scientific). Chemiluminescence detection was performed on a Kodak Image Station 4000MM Pro (Carestream Health Canada, Vaughan ON, Canada). The following antibodies to the GLP-1R were used to probe blots: mouse monoclonal 3F52 (DSHB; RRID AB_2618100; University of Iowa, Iowa City, IA), which recognizes the human GLP-1R, and rabbit polyclonal Novus 19400002. The rabbit polyclonal antibodies anti-Akt and anti-Erk1/2 were from Cell Signaling Technology (Whitby, ON, Canada). Antibodies used in this study are listed in Table 1.
Table 1.
Antibodies Used
| Peptide/Protein Target | Antigen Sequence | Name of Antibody | Supplier | Monoclonal or Polyclonal | Dilution Used | RRID |
|---|---|---|---|---|---|---|
| GLP-1R | N/A | Novus 19400002 | Novus Biologicals | Polyclonal | 1:1000 | AB_10004637 |
| GLP-1R | N/A | Novo 3F52 | Novo Nordisk | Monoclonal | 1:1000 | AB_2618100 |
Abbreviations: N/A, not applicable.
GLP-1R immunohistochemistry
Immunohistochemistry was performed on formalin-fixed tissues as described (23). Tissue sections were incubated with the primary GLP-1R antibody (Mab 3F52, 0.83 or 5 μg/mL final concentration) for 2 hours at room temperature and counterstained with Mayer Hematoxylin (Agilent Tech.). Slides were imaged using Leica IM50 Image Manager Software version 1.20 (Leica Microsystems Inc., Concord, ON, Canada). Sections containing human Brunner glands or human pancreas were simultaneously analyzed as positive controls.
ISH
Tissues were cut into 5-μm sections and mounted onto Superfrost Plus (Thermo-Fisher Scientific) glass slides. ISH was performed using the RNAscope 2.5 HD Detection Reagent kit (Advanced Cell Diagnostics Inc., Newark, CA) and human GLP-1R (Hs-GLP1R; catalog no. 519821) or PECAM1 (Hs-PECAM1-No-XMm-01; catalog no. 455931) specific hybridization probes (Advanced Cell Diagnostics Inc.) according to the manufacturer’s instructions and counterstained with Mayer Hematoxylin (Agilent Tech.). Slides were imaged using Leica IM50 Image Manager Software version 1.20 (Leica Microsystems Inc., Concord, ON, Canada). Sections containing human Brunner glands or human pancreas were simultaneously analyzed by ISH as positive controls.
Statistical analyses
Data are presented as mean ± standard error. Statistical significance was determined by two-tailed Student t test using Prism version 5.02 software (GraphPad, San Diego, CA). A P value ≤0.05 was considered statistically significant.
Results
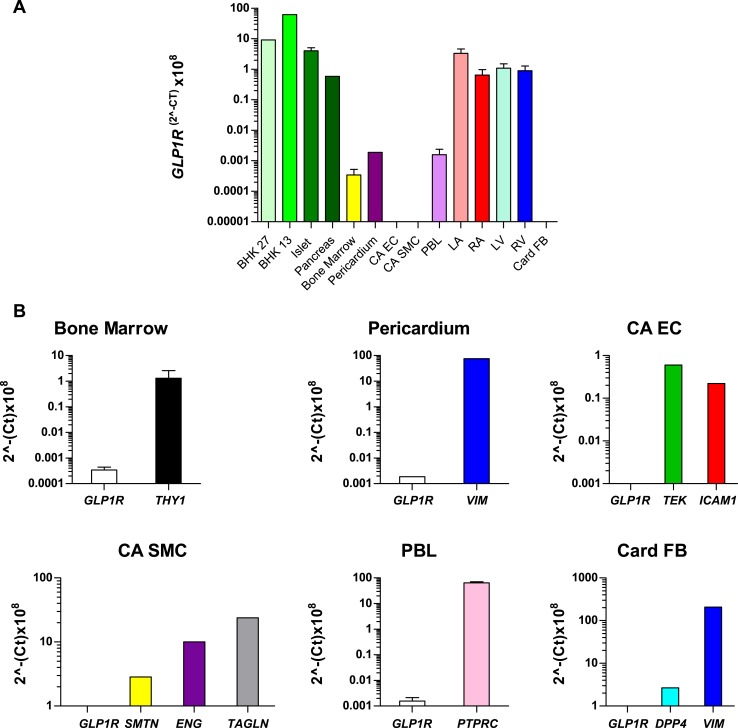
To ascertain the relative levels of GLP1R mRNA transcripts in human tissues, we first quantified GLP1R mRNA transcripts using qPCR (Fig. 1). GLP1R mRNA transcripts were detected in all four chambers of the human heart at levels comparable to those detected in RNA from human pancreas but at lower levels than those detected in RNA from human islets (Fig. 1A). Much lower (100- to 1000-fold less abundant) levels of GLP1R mRNA transcripts were detected in RNA from human bone marrow, peripheral blood leukocytes, and pericardium (Fig. 1A). In contrast, we did not detect GLP1R mRNA transcripts in RNA from human coronary artery endothelial cells, coronary artery vascular smooth muscle cells, or human cardiac fibroblasts (Fig. 1A and 1B and Supplemental Fig. 1).
Figure 1.
GLP1R mRNA transcript levels in the human heart are comparable to those in human pancreas and islets. (A) GLP1R mRNA levels were measured via qPCR analysis in multiple human tissues and in transfected BHK cells that express low levels of the human GLP-1R. For data represented without standard error bars, each single RNA sample was analyzed in duplicate; for isolates depicted with error bars, peripheral blood lymphocyte samples were analyzed in duplicate from two different sources, and at least three different samples were analyzed in duplicate by qPCR for RNAs from islet, bone marrow, left atria (LA), right atria (RA), left ventricle (LV), and right ventricle (RV). (B) qPCR analysis of GLP1R and tissue- or cell-type–specific gene expression in the indicated samples as confirmation of RNA/cDNA integrity. For (A) and (B), data are expressed as cycle threshold (Ct) values because none of the housekeeping genes examined (ACTB, GPI, PSMB4, CHMP2A, and EMC7) exhibited consistent expression levels in all tissues examined. Values are mean ± standard error (where appropriate); n = 1 to 3 samples per tissue. LA samples are from patients P01371, P01430, and P01504. RA samples are from patients P01262, P01371, and P01377. LV samples are from patients P01262, P01430, and P01371. RV samples are from patients P01262, P01371, and P01504 (see Supplemental Table 1 and Figure 4). Islet samples are from donors R177, R199, and R200 (see Supplemental Table 3). CA EC, coronary artery endothelial cells; CA SMC, coronary artery smooth muscle cells; PBL, peripheral blood lymphocytes; Card FB, cardiac fibroblasts.
To assess the qualitative integrity and to verify the origin of the RNA isolated from the human cells studied in Fig. 1A, we analyzed the expression of GLP1R and key genes associated with different cell lineages. We detected mRNA transcripts for THY1 in cells from bone marrow; VIM mRNA transcripts in RNA from pericardium and cardiac fibroblasts; TEK and ICAM1 expression in RNA from endothelial cells; SMTN, ENG, and TAGLN in vascular smooth muscle RNA; and PTPRC transcripts in RNA from peripheral blood leukocytes (Fig. 1B). Collectively, these analyses are consistent with the designated cellular sources of the human RNA samples.
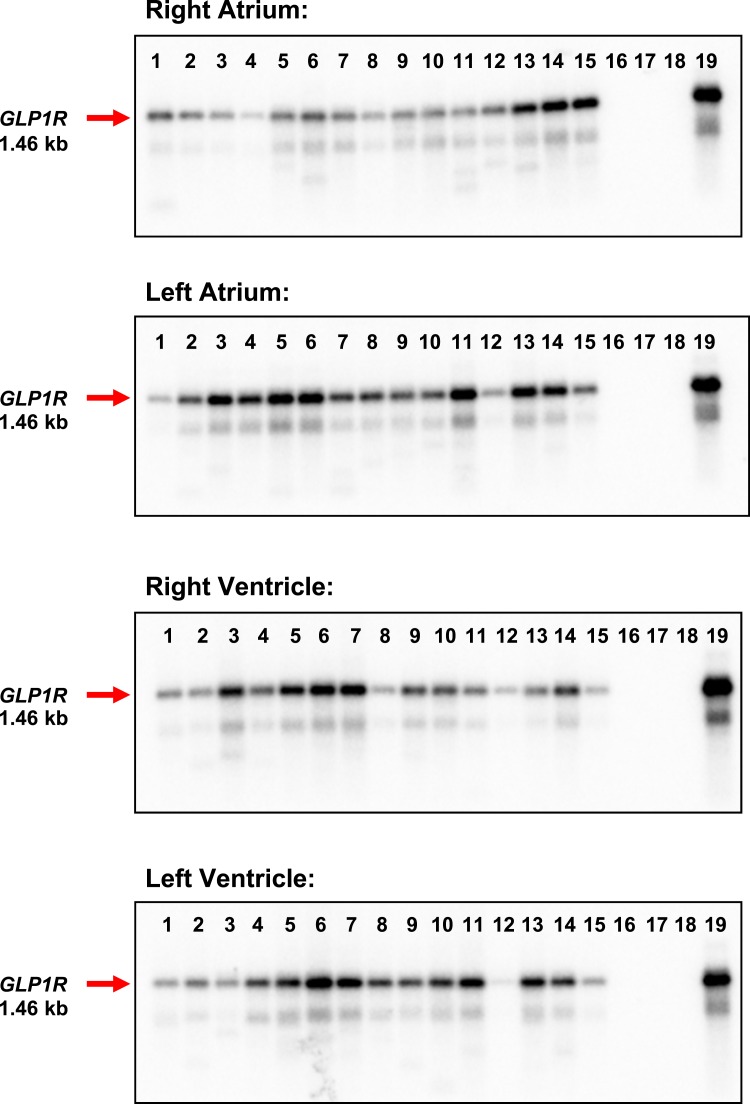
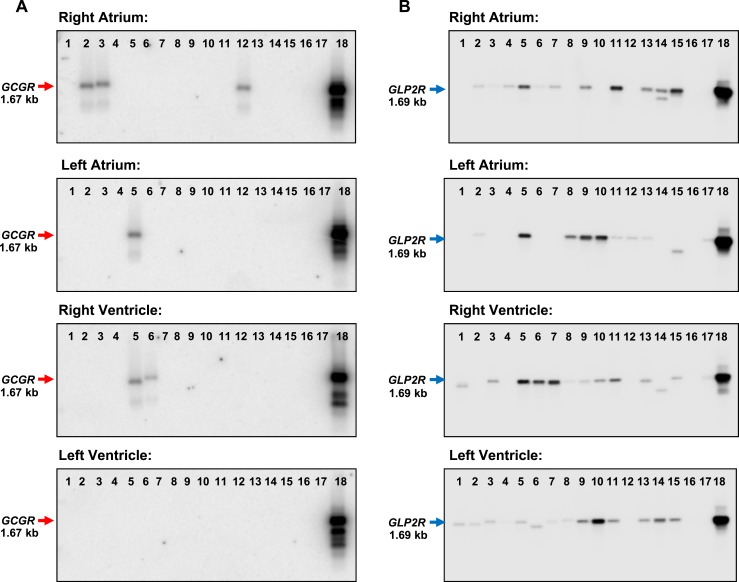
qPCR is sufficiently sensitive to detect very low levels of a partial cDNA fragment but does not provide information about the expression of a full-length mRNA transcript. Accordingly, we assessed GLP1R expression by conventional PCR using primers spanning the entire coding region of the GLP1R mRNA and protein. A full-length GLP1R mRNA transcript was uniformly detected in RNA from all four cardiac chambers in all 15 human hearts examined (Fig. 2). In contrast, mRNA transcripts for the structurally related glucagon receptor (GCGR), which is expressed and functional in the murine adult ventricles (30), were not detected in most of the hearts examined and were absent from the left ventricle (Fig. 3A). Moreover, GLP2R mRNA transcripts were expressed at lower levels compared with GLP1R (P < 0.05 for relative mRNA levels of GLP1R vs GLP2R expression in all four heart chambers; Fig. 4 and data not shown) and were detected in some but not all RNA isolates from all four chambers of the human heart (Fig. 3B).
Figure 2.
A full-length GLP1R is expressed in all four chambers of the human heart. RT-PCR analysis and Southern blotting using an internal GLP1R-specific oligonucleotide probe detected a 1.46-kb GLP1R mRNA transcript (red arrow) in the left and right atria and in the left and right ventricle of all 15 human hearts. Lanes 1 to 15, human heart samples; lane 16, negative control (H2O); lane 17, human cardiac fibroblasts; lane 18, human adipose tissue; lane 19, human pancreas (positive control). See Supplemental Table 1 for a description of the human heart samples.
Figure 3.
GCGR and GLP2R mRNA expression in the human heart. RT-PCR analysis and Southern blotting using an internal oligonucleotide probe specific to each receptor RNA detected a 1.67-kb GCGR mRNA transcript (red arrow, left panels) in a small number of human hearts and a 1.69-kb GLP2R mRNA transcript (blue arrow, right panels) in all four cardiac chambers of several human heart samples. Lanes 1 to 15, human heart samples; lane 16, negative control (H2O); lane 17, human cardiac fibroblasts (GCGR blots) or human adipose tissue (GLP2R blots); lane 18 (positive control), human liver (GCGR blots) or human colon (GLP2R blots). See Supplemental Table 1 for a description of the human heart samples.
Figure 4.
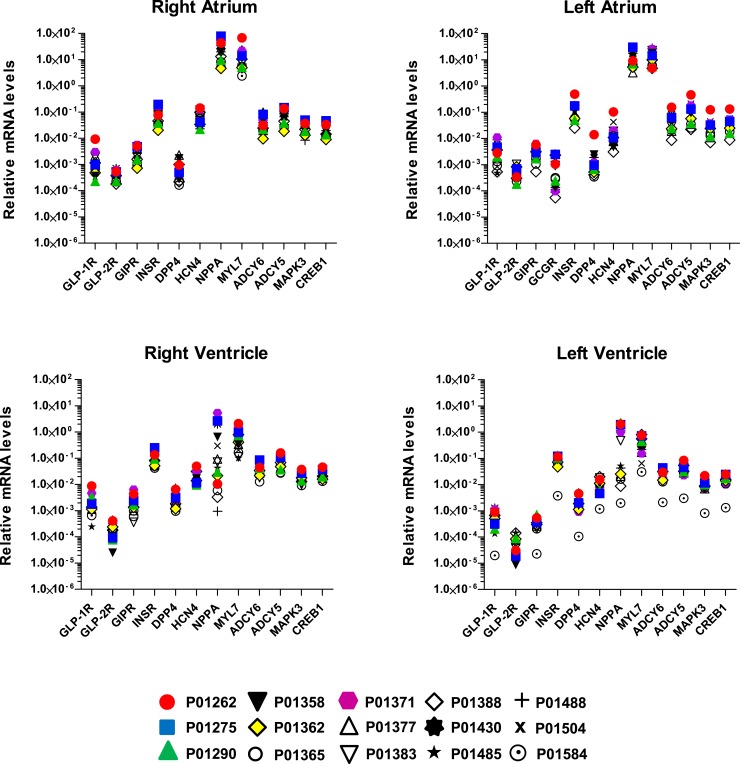
Quantitative gene expression profiles in all four chambers of the human heart. The mRNA levels of 12 or 13 different genes were measured in the four heart chambers from all 15 subjects via qPCR analysis. Data were normalized to β-actin. Each symbol represents analysis of RNA from an individual human heart (see Supplemental Table 1).
To further quantify the relative expression of class B G-protein–coupled receptors in the human heart, we compared levels of GLP1R, GCGR, GIPR, and GLP2R, with mRNA transcripts for INSR, DPP4, HCN4, NPPA, MYL7, ADCY5, ADCY6, MAPK3, and CREB1 in RNA from 15 different human hearts (Fig. 4). The relative expression of the class B G-protein–coupled receptors examined, including GLP1R, was at least 10- to 100-fold lower relative to the abundance of the majority of mRNA transcripts examined, most of which are known to be expressed in cardiomyocytes (Fig. 4). Furthermore, considerable interindividual variation in relative GLP1R expression was detected in all four chambers (Fig. 4). For comparison, the relative expression of the insulin receptor mRNA (INSR), which encodes a transmembrane receptor protein known to be expressed and functional in ventricular cardiac myocytes, was 100- to 1000-fold higher than the level of GLP1R expression in the left ventricle (Fig. 4).
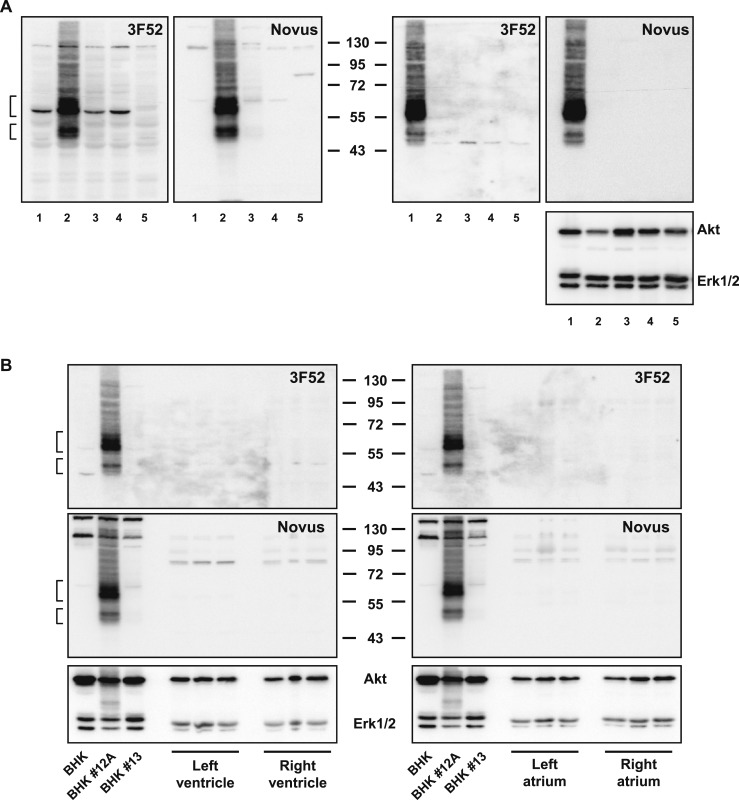
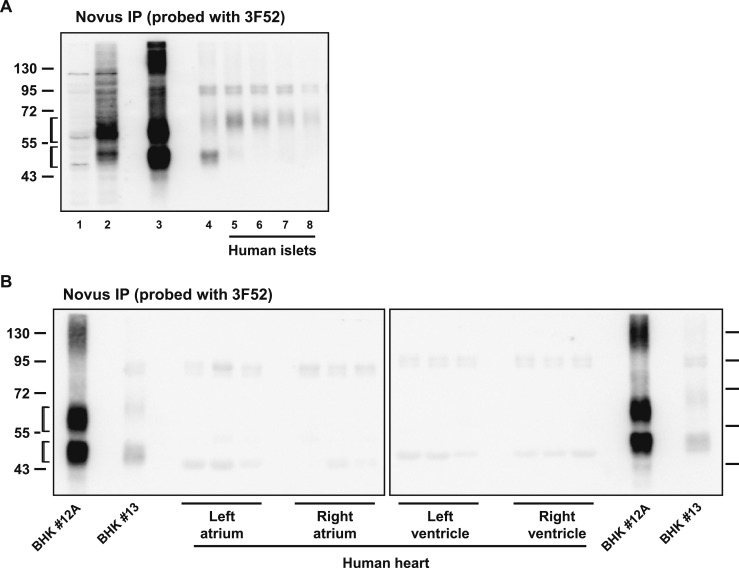
We next assessed whether a combination of immunohistochemistry and ISH would identify specific GLP-1R+ cell types within the human heart. We first examined the suitability of two different GLP-1R antibodies previously shown to detect the human GLP-1R, monoclonal antibody 3F52 (23) and the polyclonal antibody Novus 19400002 (31). Both antibodies detected the GLP-1R protein in Western blot analysis using BHK fibroblasts expressing high levels of a transfected human GLP-1 receptor cDNA (Fig. 5A, left panels, lane 2). In contrast, an immunoreactive GLP-1R was not detected in transfected BHK cells expressing relatively lower levels of transfected GLP-1R mRNA (Fig. 5A, lanes 3 and 4) or in CFPAC-1 cells previously shown to express a functional endogenous human GLP-1R (32) (Fig. 5A, lane 5). Neither antibody was sufficiently sensitive to enable detection of the endogenous GLP-1R in whole tissue protein extracts from isolated human islets (Fig. 5A, right panels) or in protein extracts from all four chambers of the human heart (Fig. 5B). Nonetheless, immunoprecipitation using the Novus antibody followed by Western blot with the 3F52 antibody detected GLP-1R in human islets (Fig. 6A) but not in human heart (Fig. 6B). Moreover, 3F52, but not Novus 19400002, detected the immunoreactive GLP-1R by immunohistochemistry in intestinal sections containing human Brunner glands (Fig. 7A and 7B and data not shown) and in human pancreatic islets (Fig. 7C and 7D).
Figure 5.
(A) Western blot analysis of whole cell or human islet tissue extracts using GLP-1R antibodies against the human GLP-1R, Mab 3F52, or polyclonal Novus. Molecular mass standards (kDa) are indicated in the center. Blots on the left contain whole cell extracts from BHK pcDNA3 (lane 1), BHK clone #12A (lane 2), BHK clone #13 (lane 3), BHK clone #27 (lane 4), and CFPAC-1 cells (lane 5). Blots on the right contain whole cell extracts from BHK clone #12A (lane 1) and whole tissue extracts from human islets #167 (lane 2), #177 (lane 3), #186 (lane 4), and 199 (lane 5). The blot on the far right was immunoblotted with Akt and Erk1/2 antibodies as loading controls. (B) Western blot analysis of whole cell or human cardiac tissue extracts analyzed using the indicated GLP-1R antibody. Molecular mass standards (kDa) are indicated in the center. Immunoblotting with Akt and Erk1/2 antibodies (bottom panels) was used as loading controls. In (A) and (B), the brackets indicate predicted migration positions of immunoreactive GLP-1R protein.
Figure 6.
(A) Whole cell/tissue extracts after immunoprecipitation (IP) and Western blotting with the indicated GLP-1R antibodies. Lanes 1 and 2 are whole cell extracts, and lanes 3 through 8 correspond to immunoprecipitated samples. Lane 1, BHK pcDNA3; lane 2, BHK clone #12A; lane 3, BHK clone #12A; lane 4, BHK clone #13; lane 5, human islet #167; lane 6, human islet #177; lane 7, human islet #186; and lane 8, human islet #199. Molecular mass standards (kDa) are indicated on the left. (B) Whole tissue extracts from human heart chamber samples after IP and Western blotting with the indicated GLP-1R antibodies. Molecular mass standards (kDa) are indicated on the far left and right of the blots. In (A) and (B), the brackets indicate predicted migration positions of immunoreactive GLP-1R protein.
Figure 7.
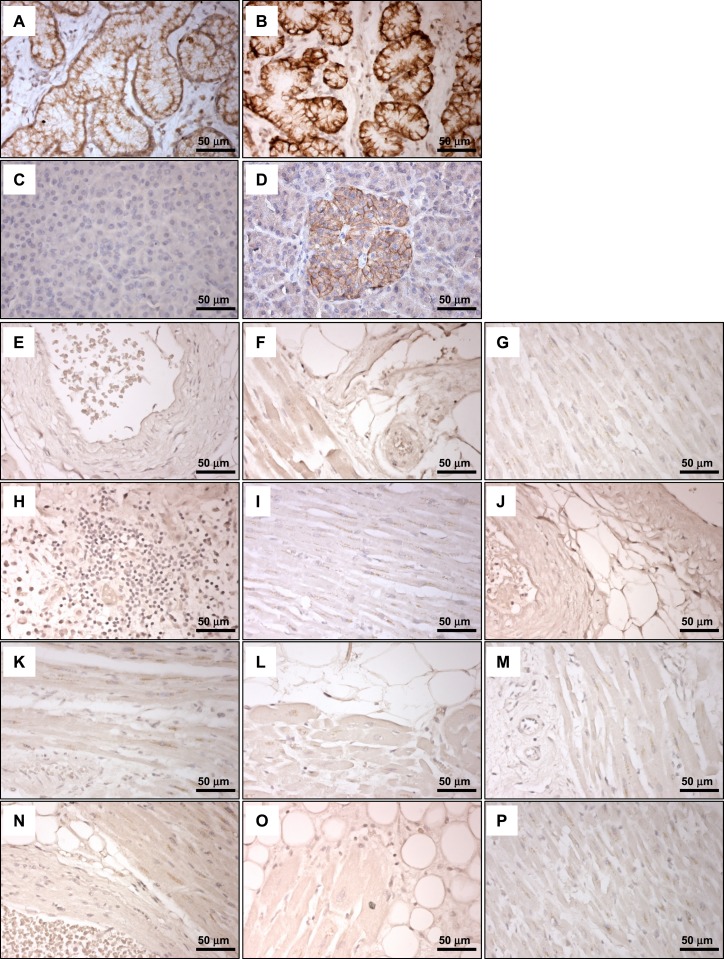
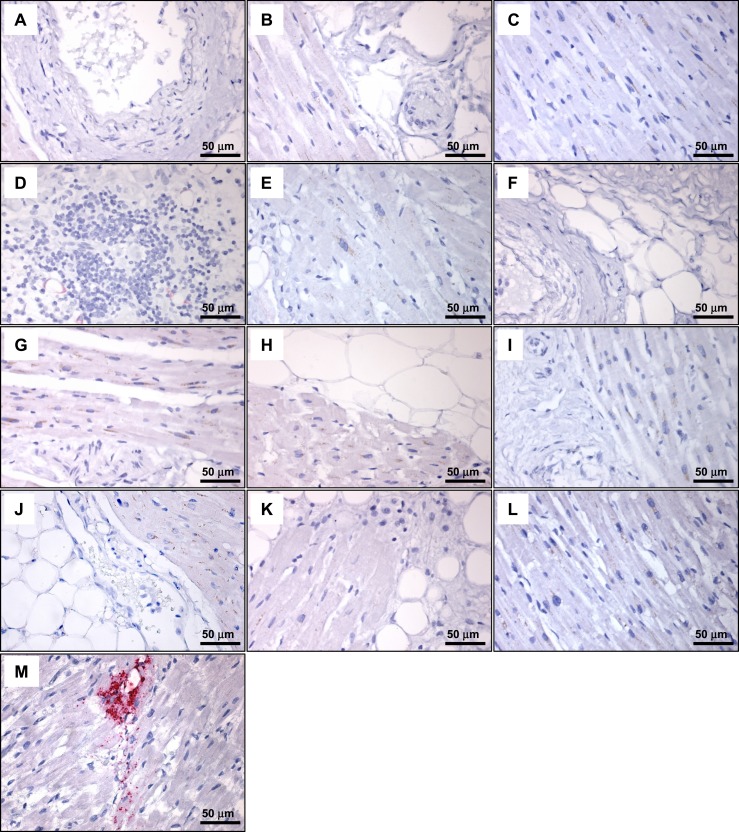
GLP-1R immunohistochemistry with Mab 3F52 does not detect GLP-1R–immunoreactive cells in human ventricular tissue. Immunostaining of hyperplastic human Brunner glands with isotype control antibody (A) and GLP-1R Mab 3F52 (B). The Novus 19400002 antibody did not reliably detect GLP-1R–immunopositive cells in sections containing human Brunner glands (data not shown). Immunostaining of human pancreas with isotype control antibody (C) and GLP-1R Mab 3F52 (D). (E–P) GLP-1R Mab 3F52 immunostaining of human cardiac tissues from individual subjects [(E) 193856; (F) 61704; (G) 70150; (H) 70847; (I) 81858; (J) 164011; (K) 116603; (L) 166469; (M) 179268; (N) 171263; (O) 182651; (P), 52411; see Supplemental Table 2].
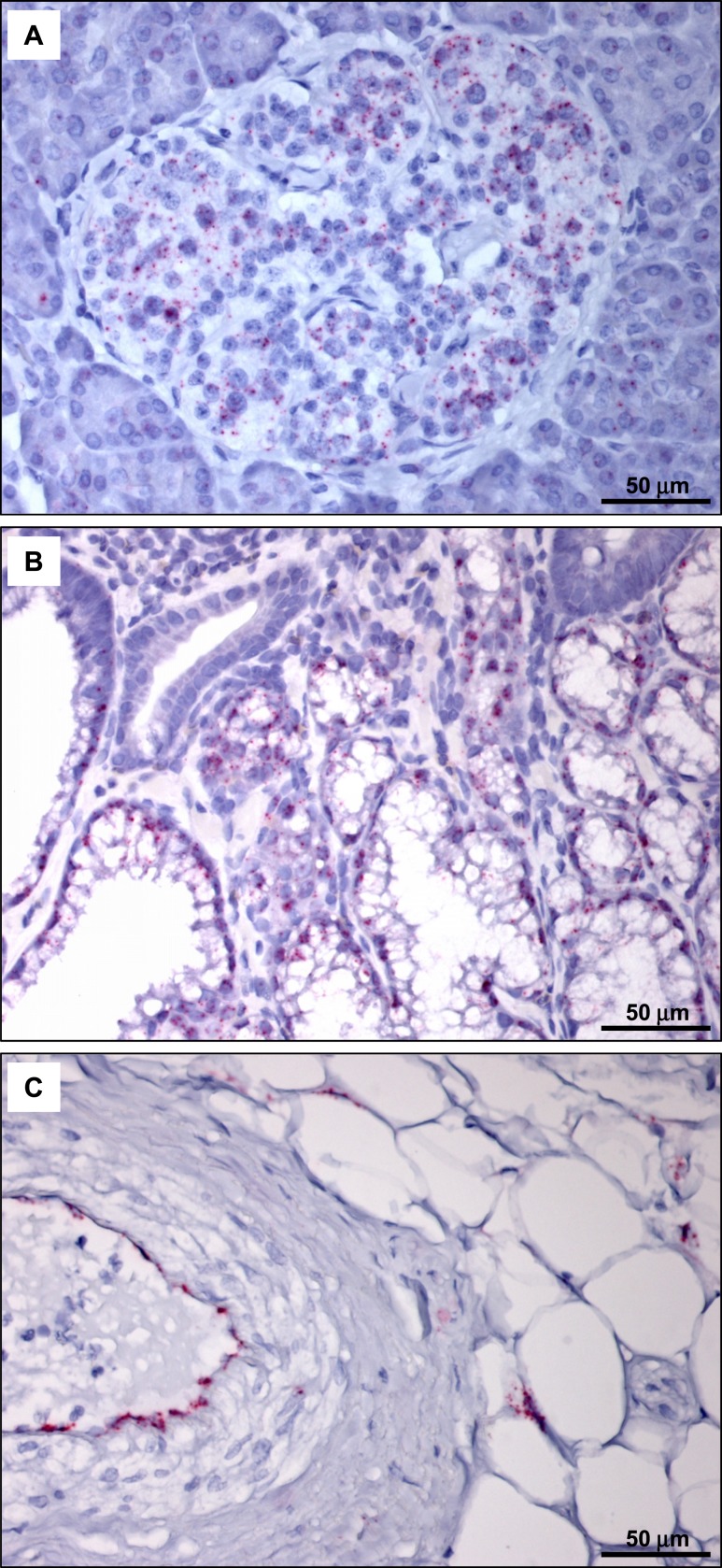
Analysis of multiple histological sections from the ventricles of 35 different human hearts failed to reveal individual GLP-1R–immunopositive cells using the 3F52 GLP-1R antibody (representative staining depicted in Fig. 7E–7P). We next examined whether GLP1R mRNA transcripts could be detected within a specific cardiac cellular subtype, such as cardiac myocytes, blood vessels, fibroblasts, and epicardial fat. We first assessed the sensitivity of our human GLP1R ISH assay, which detected GLP1R expression within human pancreatic islets and Brunner glands (Fig. 8A and 8B). Moreover, the suitability of representative human heart sections for ISH analyses was illustrated by detection of PECAM1 within endothelial cells (Fig. 8C). Although GLP1R+ cells were detected by ISH in atrial tissue from the human sinoatrial node region (Fig. 9M), we were unable to identify individual GLP1R+ cells, including analysis of cardiac myocytes, fat, and blood vessels, in histological sections from 35 human ventricular samples by ISH (representative ISH results are shown in Fig. 9A–9L).
Figure 8.
ISH analysis detects GLP1R mRNA expression in human pancreas (A) and hyperplastic human Brunner glands (B). GLP1R-positive cells are indicated by punctate pink staining. (C) Detection of PECAM1 mRNA expression in human heart (case 164011; Supplemental Table 2) verifies the quality of the tissue and the integrity of the mRNA.
Figure 9.
ISH analysis of GLP1R mRNA expression detects GLP1R-positive cells in human atrial tissue from the sinoatrial node region but not in human ventricular tissue. (A–L) ISH analysis of GLP1R mRNA expression in cardiac tissues from the following cases (see Supplemental Table 2): (A) 193856; (B) 61704; (C) 70150; (D) 70847; (E) 81858; (F) 164011; (G) 116603; (H) 166469; (I) 179268; (J) 171263; (K) 182651; (L) 52411. (M) ISH analysis of GLP1R mRNA expression in tissue from human sinoatrial node region.
Discussion
The demonstration that native GLP-1 and GLP-1R agonists rapidly improve ventricular function in the normoxic and ischemic human heart (14, 15, 33), taken together with the previous detection of GLP-1Rs within human and nonhuman primate atrial tissue (23), implicates one or more direct roles for GLP-1R signaling in the human heart. Despite reports from cardiovascular outcome studies that the GLP-1R agonist liraglutide reduced the rates of cardiovascular death and nonfatal myocardial infarction in human subjects with T2D (21), the precise localization and relative expression of the GLP-1R within human cardiac tissue has not been systematically examined. Here we report the results of our studies examining not only GLP1R but also GCGR and GLP2R expression in all four chambers of the human heart.
Consistent with results presented in publicly available transcriptomic analyses (34), levels of GLP1R mRNA transcripts in human heart were comparable to those detected in RNA from human pancreas. Moreover, our data extend these results by revealing detectable GLP1R mRNA transcripts in all four cardiac chambers. Levels of GLP1R mRNA transcripts were lowest in RNA isolated from the left ventricle. Despite functional expression of the Gcgr in mouse heart, including right and left ventricle (30), we were unable to detect evidence for GCGR expression, the receptor mediating the effects of glucagon, in most of the hearts examined, including in adult human left ventricle. In contrast, mRNA transcripts for the structurally related GLP2R, the receptor transducing actions of GLP-2, were extremely low yet were detected in most ventricular samples from the human hearts studied.
The relatively similar abundance of GLP1R mRNA transcripts in all four chambers of the human heart reported here differs from the expression pattern previously reported in rats and mice. Glp1r expression has been detected predominantly in the atria of normotensive and hypertensive mice (30, 35), whereas Glp1r mRNA transcripts were expressed at very low levels in the mouse cardiac ventricles (30) and were not detected in rat ventricular tissue or ventricular cardiomyocytes (36). In contrast, GLP1R mRNA transcripts were detected at comparable levels in ventricular and atrial RNA from the human heart in this study, although levels were generally lower in RNA isolated from the left ventricle. Species specificity of GLP1R expression has also been demonstrated in the thyroid, with detectable expression of the Glp1r in the rodent C-cell and thyroid (37) and extremely low-level or undetectable GLP1R expression in the nonneoplastic human thyroid (27). Furthermore, our current analyses of atrial GLP1R expression did not routinely include RNA isolated from the human sinoatrial node, a region of the human heart that expresses abundant GLP1R mRNA and protein (23). Hence, our findings reinforce the importance of considering species-specific differences and variation in receptor expression arising from differential RNA sampling when interpreting relative GLP-1R expression data from preclinical and human studies.
GLP-1R agonists acutely increased myocardial blood flow in human subjects with diabetes without coronary artery disease (38), and infusion of native GLP-1 rapidly augmented myocardial blood flow in human subjects without diabetes (39). Moreover, an immunoreactive GLP-1R has been reported in studies of human coronary artery endothelial cells and in rat kidney vascular smooth muscle cells (40, 41). Similarly, GLP-1 directly induced elastic fiber formation in studies of human cardiac fibroblasts (42). In contrast to reports of vascular Glp1r-directed reporter expression in murine coronary arteries (43), we were unable to detect expression of canonical GLP1R mRNA transcripts in RNA from human coronary artery endothelial cells, human coronary artery vascular smooth muscle cells, or human cardiac fibroblasts. Hence, the extent to which these noncardiomyocyte cell types may, under some circumstances, express the GLP-1R within regions of the human heart requires further confirmation.
In the course of assessing the suitability of two different antisera for detection of the cardiac GLP-1R, we were unable to detect immunoreactive GLP-1R by conventional Western blotting using extracts from several different preparations of human islets. In contrast, both antisera readily detected the human GLP-1R in Western blot analysis of hGLP-1R–transfected fibroblasts. Although a putative endogenous GLP-1R protein has been detected in multiple cell lines and tissue extracts, including islets and heart, by many groups using Western blot analysis (44–47), the sensitivity and specificity of the majority of antisera used for these studies has not been uniformly verified (24, 25, 31). Because the β-cell is among the cell types exhibiting the most robust expression of the endogenous GLP-1R, our inability to detect expression of the endogenous GLP-1R in conventional Western blotting of human islets provides a further note of caution regarding the accuracy of multiple reports of GLP-1R protein expression using Western blotting and native tissue extracts.
In summary, we detected low-level expression of the full-length GLP1R mRNA transcript in all four chambers of the human heart. In contrast, mRNA transcripts encoding the structurally related receptors for peptides coencoded by the same proglucagon precursor (e.g., glucagon and GLP-2) were not detected (GCGR) or were detected at even lower levels in a less uniform manner (GLP2R). Despite a previous report detecting GLP1R mRNA transcripts in human ventricular myocytes by RT-PCR (22), a combination of ISH and immunocytochemistry was not successful in localizing GLP-1R expression to individual cell types within the dozens of human heart sections studied here.
Caveats and limitations
These studies have several limitations. It may be important that our PCR analyses were carried out on RNA obtained from cardiac tissue subjected to cardioplegia followed by rapid freezing prior to RNA preparation. In contrast, although the human heart samples we obtained for histological analysis from the Rapid Autopsy Program were fixed relatively quickly (often within hours) to minimize degradation of RNA and protein, we cannot exclude the possibility that failure to localize GLP1R RNA and protein within specific cells in the heart may reflect instability of the RNA transcript or protein, compromising detection. Furthermore, although we analyzed the tissue sections by immunohistochemistry or ISH within several days to 2 weeks after generation of the slides, it is possible that the sensitivity of GLP1R/GLP-1R detection falls off rapidly over time in paraffin sections, further hampering GLP1R transcript or GLP-1R protein localization.
It seems likely, based on available murine data (43), that the cardiac GLP-1R may be expressed at very low levels in scattered cells, perhaps within a subset of nerves or blood vessels, including endothelial cells (48), or adjacent to valves not contained within the several dozen heart sections examined here. A complementary possibility limiting detection of the GLP-1R protein is that GLP1R mRNA transcripts may not be uniformly translated into a detectable GLP-1R protein in all cardiac cells expressing the GLP1R mRNA. Alternatively, cardiac-specific posttranslational modifications of the GLP-1R may compromise detection of the immunoreactive protein using the 3F52 antibody. Furthermore, the relative level of GLP1R expression within individual cardiac cells may be very low, precluding definitive identification by ISH. Hence, further studies, perhaps using cell enrichment techniques, ideally from heart samples rapidly obtained from well-preserved hearts, may be required to enable more sensitive and precise identification of the cellular sites of GLP-1R expression in human cardiac tissue. Nevertheless, our studies highlight the consistent detection of low-level GLP1R expression within human ventricular tissue, findings with potential relevance for understanding how GLP-1R agonists directly affect cardiac function in vivo.
Supplementary Material
Acknowledgments
The authors thank Charles Pyke and Pia Mortensen (Novo Nordisk Inc.) for guidance in the use of immunocytochemistry and ISH to detect GLP-1R expression and Miriam Cnop, Université Libre de Bruxelles Center for Diabetes Research, for helpful discussions.
Financial Support: These studies were supported by CIHR Foundation Grant 154321, the Canada Research Chairs Program, the Banting and Best Diabetes Centre Novo Nordisk Chair in Incretin Biology, and a grant from Novo Nordisk in support of the cardiovascular mechanisms of GLP-1 action (all to D.J.D.).
Author Contributions: L.L.B., B.Y., X.C., and E.E.M. carried out the experiments. D.J.D., T.P.C., and K.B.M. planned the experiments. J.B., C.J.S., L.L.B., B.Y., and D.J.D. reviewed human tissue blocks and histology. L.L.B. and D.J.D. wrote the first draft of the manuscript. D.J.D. is the guarantor of the data and takes full responsibility for the data in this manuscript. All authors reviewed and edited the manuscript prior to submission.
Current Affiliation: E. E. Mulvihill’s current affiliation is the University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada.
Disclosure Summary: D.J.D. has served as an advisor or consultant within the past 12 months to Arisaph Pharmaceuticals Inc., Intarcia Therapeutics, Merck Research Laboratories, Novo Nordisk Inc., and Pfizer Inc. Neither D.J.D. or his family members hold stock directly or indirectly in any of these companies. The remaining authors have nothing to disclose.
Glossary
Abbreviations:
- BHK
baby hamster kidney
- CFPAC-1
cystic fibrosis pancreatic adenocarcinoma
- GLP-1
glucagonlike peptide-1
- GLP-1R
glucagonlike peptide-1 receptor
- ISH
in situ hybridization
- PCR
polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- RT-PCR
reverse transcription polymerase chain reaction
- T2D
type 2 diabetes
References
- 1. Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132(6):2131–2157. [DOI] [PubMed] [Google Scholar]
- 2. Campbell JE, Drucker DJ. Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab. 2013;17(6):819–837. [DOI] [PubMed] [Google Scholar]
- 3. Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368(9548):1696–1705. [DOI] [PubMed] [Google Scholar]
- 4. Astrup A, Rössner S, Van Gaal L, Rissanen A, Niskanen L, Al Hakim M, Madsen J, Rasmussen MF, Lean ME; NN8022-1807 Study Group . Effects of liraglutide in the treatment of obesity: a randomised, double-blind, placebo-controlled study. Lancet. 2009;374(9701):1606–1616. [DOI] [PubMed] [Google Scholar]
- 5. Davies M, Pieber TR, Hartoft-Nielsen ML, Hansen OKH, Jabbour S, Rosenstock J. Effect of oral semaglutide compared with placebo and subcutaneous semaglutide on glycemic control in patients with type 2 diabetes: a randomized clinical trial. JAMA. 2017;318(15):1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sadry SA, Drucker DJ. Emerging combinatorial hormone therapies for the treatment of obesity and T2DM. Nat Rev Endocrinol. 2013;9(7):425–433. [DOI] [PubMed] [Google Scholar]
- 7. Thorens B. Expression cloning of the pancreatic β cell receptor for the gluco-incretin hormone glucagon-like peptide 1. Proc Natl Acad Sci USA. 1992;89(18):8641–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullock BP, Heller RS, Habener JF. Tissue distribution of messenger ribonucleic acid encoding the rat glucagon-like peptide-1 receptor. Endocrinology. 1996;137(7):2968–2978. [DOI] [PubMed] [Google Scholar]
- 9. Wismann P, Barkholt P, Secher T, Vrang N, Hansen HB, Jeppesen PB, Baggio LL, Koehler JA, Drucker DJ, Sandoval DA, Jelsing J. The endogenous preproglucagon system is not essential for gut growth homeostasis in mice. Mol Metab. 2017;6(7):681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campos RV, Lee YC, Drucker DJ. Divergent tissue-specific and developmental expression of receptors for glucagon and glucagon-like peptide-1 in the mouse. Endocrinology. 1994;134(5):2156–2164. [DOI] [PubMed] [Google Scholar]
- 11. Wei Y, Mojsov S. Tissue-specific expression of the human receptor for glucagon-like peptide-I: brain, heart and pancreatic forms have the same deduced amino acid sequences. FEBS Lett. 1995;358(3):219–224. [DOI] [PubMed] [Google Scholar]
- 12. Pujadas G, Drucker DJ. Vascular biology of glucagon receptor superfamily peptides: mechanistic and clinical relevance. Endocr Rev. 2016;37(6):554–583. [DOI] [PubMed] [Google Scholar]
- 13. Ussher JR, Drucker DJ. Cardiovascular actions of incretin-based therapies. Circ Res. 2014;114(11):1788–1803. [DOI] [PubMed] [Google Scholar]
- 14. Read PA, Khan FZ, Dutka DP. Cardioprotection against ischaemia induced by dobutamine stress using glucagon-like peptide-1 in patients with coronary artery disease. Heart. 2012;98(5):408–413. [DOI] [PubMed] [Google Scholar]
- 15. Drucker DJ. The cardiovascular biology of glucagon-like peptide-1. Cell Metab. 2016;24(1):15–30. [DOI] [PubMed] [Google Scholar]
- 16. Noyan-Ashraf MH, Momen MA, Ban K, Sadi AM, Zhou YQ, Riazi AM, Baggio LL, Henkelman RM, Husain M, Drucker DJ. GLP-1R agonist liraglutide activates cytoprotective pathways and improves outcomes after experimental myocardial infarction in mice. Diabetes. 2009;58(4):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lønborg J, Vejlstrup N, Kelbæk H, Bøtker HE, Kim WY, Mathiasen AB, Jørgensen E, Helqvist S, Saunamäki K, Clemmensen P, Holmvang L, Thuesen L, Krusell LR, Jensen JS, Køber L, Treiman M, Holst JJ, Engstrøm T. Exenatide reduces reperfusion injury in patients with ST-segment elevation myocardial infarction. Eur Heart J. 2012;33(12):1491–1499. [DOI] [PubMed] [Google Scholar]
- 18. Pfeffer MA, Claggett B, Diaz R, Dickstein K, Gerstein HC, Køber LV, Lawson FC, Ping L, Wei X, Lewis EF, Maggioni AP, McMurray JJ, Probstfield JL, Riddle MC, Solomon SD, Tardif JC, Investigators E; ELIXA Investigators . Lixisenatide in patients with type 2 diabetes and acute coronary syndrome. N Engl J Med. 2015;373(23):2247–2257. [DOI] [PubMed] [Google Scholar]
- 19. Holman RR, Bethel MA, Mentz RJ, Thompson VP, Lokhnygina Y, Buse JB, Chan JC, Choi J, Gustavson SM, Iqbal N, Maggioni AP, Marso SP, Öhman P, Pagidipati NJ, Poulter N, Ramachandran A, Zinman B, Hernandez AF, Group ES; EXSCEL Study Group . Effects of once-weekly exenatide on cardiovascular outcomes in type 2 diabetes. N Engl J Med. 2017;377(13):1228–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marso SP, Bain SC, Consoli A, Eliaschewitz FG, Jódar E, Leiter LA, Lingvay I, Rosenstock J, Seufert J, Warren ML, Woo V, Hansen O, Holst AG, Pettersson J, Vilsbøll T; SUSTAIN-6 Investigators . Semaglutide and cardiovascular outcomes in patients with type 2 diabetes. N Engl J Med. 2016;375(19):1834–1844. [DOI] [PubMed] [Google Scholar]
- 21. Marso SP, Daniels GH, Brown-Frandsen K, Kristensen P, Mann JFE, Nauck MA, Nissen SE, Pocock S, Poulter NR, Ravn LS, Steinberg WM, Stockner M, Zinman B, Bergenstal RM, Buse JB LEADER Steering CommitteeLEADER Trial Investigators . Liraglutide and cardiovascular outcomes in type 2 diabetes. N Engl J Med. 2016;375(4):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wallner M, Kolesnik E, Ablasser K, Khafaga M, Wakula P, Ljubojevic S, Thon-Gutschi EM, Sourij H, Kapl M, Edmunds NJ, Kuzmiski JB, Griffith DA, Knez I, Pieske B, von Lewinski D. Exenatide exerts a PKA-dependent positive inotropic effect in human atrial myocardium: GLP-1R mediated effects in human myocardium. J Mol Cell Cardiol. 2015;89(Pt B):365–375. [DOI] [PubMed] [Google Scholar]
- 23. Pyke C, Heller RS, Kirk RK, Ørskov C, Reedtz-Runge S, Kaastrup P, Hvelplund A, Bardram L, Calatayud D, Knudsen LB. GLP-1 receptor localization in monkey and human tissue: novel distribution revealed with extensively validated monoclonal antibody. Endocrinology. 2014;155(4):1280–1290. [DOI] [PubMed] [Google Scholar]
- 24. Panjwani N, Mulvihill EE, Longuet C, Yusta B, Campbell JE, Brown TJ, Streutker C, Holland D, Cao X, Baggio LL, Drucker DJ. GLP-1 receptor activation indirectly reduces hepatic lipid accumulation but does not attenuate development of atherosclerosis in diabetic male ApoE(-/-) mice. Endocrinology. 2013;154(1):127–139. [DOI] [PubMed] [Google Scholar]
- 25. Pyke C, Knudsen LB. The glucagon-like peptide-1 receptor: or not? Endocrinology. 2013;154(1):4–8. [DOI] [PubMed] [Google Scholar]
- 26. Ussher JR, Campbell JE, Mulvihill EE, Baggio LL, Bates HE, McLean BA, Gopal K, Capozzi M, Yusta B, Cao X, Ali S, Kim M, Kabir MG, Seino Y, Suzuki J, Drucker DJ. Inactivation of the glucose-dependent insulinotropic polypeptide receptor improves outcomes following experimental myocardial infarction. Cell Metab. 2017;S1550–4131(17)30671-X. [DOI] [PubMed]
- 27. Bjerre Knudsen L, Madsen LW, Andersen S, Almholt K, de Boer AS, Drucker DJ, Gotfredsen C, Egerod FL, Hegelund AC, Jacobsen H, Jacobsen SD, Moses AC, Mølck AM, Nielsen HS, Nowak J, Solberg H, Thi TD, Zdravkovic M, Moerch U. Glucagon-like peptide-1 receptor agonists activate rodent thyroid C-cells causing calcitonin release and C-cell proliferation [published correction appears in Endocrinology. 2012;153(2):1000]. Endocrinology. 2010;151(4):1473–1486. [DOI] [PubMed] [Google Scholar]
- 28. Knudsen LB, Hastrup S, Underwood CR, Wulff BS, Fleckner J. Functional importance of GLP-1 receptor species and expression levels in cell lines. Regul Pept. 2012;175(1-3):21–29. [DOI] [PubMed] [Google Scholar]
- 29. Baggio LL, Huang Q, Brown TJ, Drucker DJ. Oxyntomodulin and glucagon-like peptide-1 differentially regulate murine food intake and energy expenditure. Gastroenterology. 2004;127(2):546–558. [DOI] [PubMed] [Google Scholar]
- 30. Ali S, Ussher JR, Baggio LL, Kabir MG, Charron MJ, Ilkayeva O, Newgard CB, Drucker DJ. Cardiomyocyte glucagon receptor signaling modulates outcomes in mice with experimental myocardial infarction. Mol Metab. 2014;4(2):132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drucker DJ. Incretin action in the pancreas: potential promise, possible perils, and pathological pitfalls. Diabetes. 2013;62(10):3316–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koehler JA, Drucker DJ. Activation of GLP-1 receptor signaling does not modify the growth or apoptosis of human pancreatic cancer cells. Diabetes. 2006;55:1369–1379. [DOI] [PubMed] [Google Scholar]
- 33. Read PA, Hoole SP, White PA, Khan FZ, O’Sullivan M, West NE, Dutka DP. A pilot study to assess whether glucagon-like peptide-1 protects the heart from ischemic dysfunction and attenuates stunning after coronary balloon occlusion in humans. Circ Cardiovasc Interv. 2011;4(3):266–272. [DOI] [PubMed] [Google Scholar]
- 34. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim M, Platt MJ, Shibasaki T, Quaggin SE, Backx PH, Seino S, Simpson JA, Drucker DJ. GLP-1 receptor activation and Epac2 link atrial natriuretic peptide secretion to control of blood pressure. Nat Med. 2013;19(5):567–575. [DOI] [PubMed] [Google Scholar]
- 36. Wohlfart P, Linz W, Hübschle T, Linz D, Huber J, Hess S, Crowther D, Werner U, Ruetten H. Cardioprotective effects of lixisenatide in rat myocardial ischemia-reperfusion injury studies. J Transl Med. 2013;11(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamada C, Yamada Y, Tsukiyama K, Yamada K, Udagawa N, Takahashi N, Tanaka K, Drucker DJ, Seino Y, Inagaki N. The murine glucagon-like peptide-1 receptor is essential for control of bone resorption. Endocrinology. 2008;149(2):574–579. [DOI] [PubMed] [Google Scholar]
- 38. Gejl M, Søndergaard HM, Stecher C, Bibby BM, Møller N, Bøtker HE, Hansen SB, Gjedde A, Rungby J, Brock B. Exenatide alters myocardial glucose transport and uptake depending on insulin resistance and increases myocardial blood flow in patients with type 2 diabetes. J Clin Endocrinol Metab. 2012;97(7):E1165–E1169. [DOI] [PubMed] [Google Scholar]
- 39. Subaran SC, Sauder MA, Chai W, Jahn LA, Fowler DE, Aylor KW, Basu A, Liu Z. GLP-1 at physiological concentrations recruits skeletal and cardiac muscle microvasculature in healthy humans. Clin Sci (Lond). 2014;127(3):163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nyström T, Gutniak MK, Zhang Q, Zhang F, Holst JJ, Ahrén B, Sjöholm A. Effects of glucagon-like peptide-1 on endothelial function in type 2 diabetes patients with stable coronary artery disease. Am J Physiol Endocrinol Metab. 2004;287(6):E1209–E1215. [DOI] [PubMed] [Google Scholar]
- 41. Jensen EP, Poulsen SS, Kissow H, Holstein-Rathlou NH, Deacon CF, Jensen BL, Holst JJ, Sorensen CM. Activation of GLP-1 receptors on vascular smooth muscle cells reduces the autoregulatory response in afferent arterioles and increases renal blood flow. Am J Physiol Renal Physiol. 2015;308(8):F867–F877. [DOI] [PubMed] [Google Scholar]
- 42. Qa’aty N, Wang Y, Wang A, Mao S, Vincent M, Husain M, Hinek A. The antidiabetic hormone glucagon-like peptide-1 induces formation of new elastic fibers in human cardiac fibroblasts after cross-activation of IGF-IR. Endocrinology. 2015;156(1):90–102. [DOI] [PubMed] [Google Scholar]
- 43. Richards P, Parker HE, Adriaenssens AE, Hodgson JM, Cork SC, Trapp S, Gribble FM, Reimann F. Identification and characterisation of glucagon-like peptide-1 receptor expressing cells using a new transgenic mouse model. Diabetes. 2014;63:1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakamura Y, Kanai T, Saeki K, Takabe M, Irie J, Miyoshi J, Mikami Y, Teratani T, Suzuki T, Miyata N, Hisamatsu T, Nakamoto N, Yamagishi Y, Higuchi H, Ebinuma H, Hozawa S, Saito H, Itoh H, Hibi T. CCR2 knockout exacerbates cerulein-induced chronic pancreatitis with hyperglycemia via decreased GLP-1 receptor expression and insulin secretion. Am J Physiol Gastrointest Liver Physiol. 2013;304(8):G700–G707. [DOI] [PubMed] [Google Scholar]
- 45. Artinian SB, Al Lafi SM, Boutary SS, Bitar KM, Zwainy NS, Bikhazi AB. Assessment of glucagon-like peptide-1 analogue and renin inhibitor on the binding and regulation of GLP-1 receptor in type 1 diabetic rat hearts. Exp Diabetes Res. 2011;2011:489708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu YS, Huang ZW, Wang L, Liu XX, Wang YM, Zhang Y, Zhang M. Sitagliptin alleviated myocardial remodeling of the left ventricle and improved cardiac diastolic dysfunction in diabetic rats. J Pharmacol Sci. 2015;127(3):260–274. [DOI] [PubMed] [Google Scholar]
- 47. Zhou J, Livak MF, Bernier M, Muller DC, Carlson OD, Elahi D, Maudsley S, Egan JM. Ubiquitination is involved in glucose-mediated downregulation of GIP receptors in islets. Am J Physiol Endocrinol Metab. 2007;293(2):E538–E547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaur H, Carvalho J, Looso M, Singh P, Chennupati R, Preussner J, Günther S, Albarrán-Juárez J, Tischner D, Classen S, Offermanns S, Wettschureck N. Single-cell profiling reveals heterogeneity and functional patterning of GPCR expression in the vascular system. Nat Commun. 2017;8:15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.