Abstract
We have identified a retroviral envelope gene with a complete, intact open reading frame (ORF) in 20 species of spiny-rayed fishes (Acanthomorpha). The taxonomic distribution of the gene, “percomORF”, indicates insertion into the ancestral lineage >110 Ma, making it the oldest known conserved gene of viral origin in a vertebrate genome. Underscoring its ancient provenence, percomORF exists as an isolated ORF within the intron of a widely conserved host gene, with no discernible proviral sequence nearby. Despite its remarkable age, percomORF retains canonical features of a retroviral glycoprotein, and tests for selection strongly suggest cooption for a host function. Retroviral envelope genes have been coopted for a role in placentogenesis by numerous lineages of mammals, including eutherians and marsupials, representing a variety of placental structures. Therefore percomORF’s presence within the group Percomorpha—unique among spiny-finned fishes in having evolved placentation and live birth—is especially intriguing.
Keywords: endogenous retrovirus, viral gene cooption, evolution.
Vertebrate genomes include vast numbers of sequences derived from the infection of germ cells by retroviruses, which integrate into host DNA as a normal part of their replication cycle (Jern and Coffin 2008; Johnson 2015). Most of these endogenous retrovirus (ERV) sequences are overwritten by mutations over time, but occasionally an ERV gene is coopted for a function that benefits the host, and becomes fixed in the population. While investigating the origins of a sequence that had been published as potentially representing a remnant of a filovirus in the genome of the stickleback (Gasterosteus aculeatus) (Belyi et al. 2010), we found that the sequence is actually part of an ORF encoding a complete and intact retroviral envelope glycoprotein. Retroviral glycoproteins stud the surface of the virion and mediate receptor binding and membrane fusion, allowing entry of the viral cargo into the target cell.
The UCSC Genome Browser (Speir et al. 2016) indicates orthologs of the stickleback sequence in two species of pufferfishes, Takifugu rubripes and Tetraodon nigroviridis, and BLAST searches of the NCBI databases with the full-length translated sequence recovered the ORF from an additional 17 species of spiny-rayed fishes (supplementary table S1, Supplementary Material online). Although no matches were found within the EST database, a search of the NCBI Sequence Read Archive (SRA) revealed an exact match to the stickleback sequence among RNA transcripts expressed in embryonic tissue at 3 days post-fertilization (supplementary methods S1, Supplementary Material online). All species belong to the crown group known as the Percomorpha, thus we named the ORF “percomORF”.
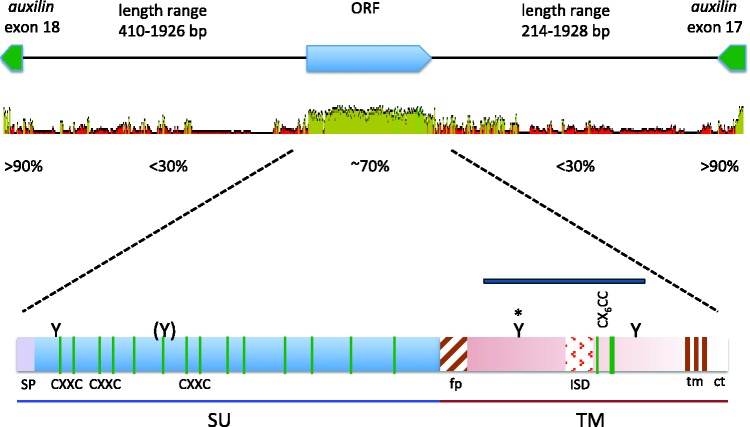
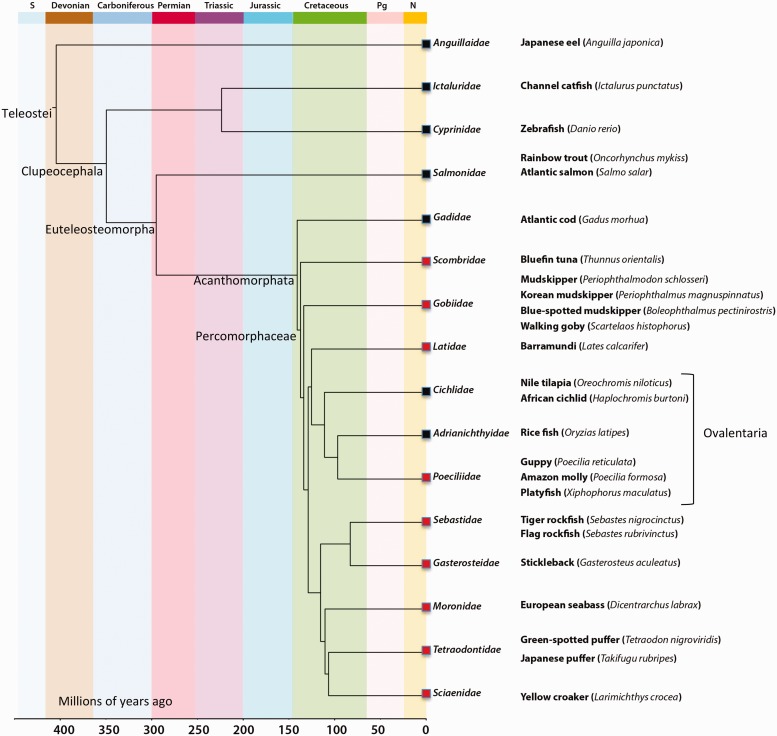
Analysis of the flanking regions revealed that percomORF is positioned within an intron of a gene encoding an auxilin-like protein, DNAJC6, that is widely and highly conserved among vertebrates. We were able to trace the DNAJC6 coding region across 18 exons in Xiphophorus maculatus, and percomORF lies within the last intron, between exons 17 and 18, in opposite orientation to DNAJC6 (fig. 1). This distinctive location allowed us to establish that percomORF is in a syntenic position in all 20 species and thus arose from a single event in a common ancestor. We also examined the corresponding regions in other teleost lineages to generate a presence/absence profile (fig. 2). Its presence in multiple lineages of Percomorphaceae indicates that percomORF originated no more recently than 109–140 Ma, the estimated range for the Percomorphaceae divergence (Near et al. 2012; Betancur-R et al. 2013).
Fig. 1.
Top, genomic position of percomORF. The orientation of the coding regions for auxilin exons and percomORF, and the varying lengths of the ORF-flanking intron sequences are shown along the top. Below is a graph representing the average pairwise percent nucleotide identity of each region, generated using GENEIOUS v. 6.0.4, available from http://www.geneious.com. Dark green indicates 100% identity across all sequences at a given site; green, >30%, identity; red, <30% identity. Height of graph at each position conveys the fraction of sequences with an identical nucleotide at that position. Bottom, conserved features of PercomORF. SU and TM subunits are indicated at the bottom of the figure; green vertical lines represent cysteines, with approximate positions of the CXXC motifs in SU, and the CX6CC motif in TM indicated; fusion peptide (fp), immunosuppressive domain (ISD), and transmembrane region (tm) are shown as patterned boxes; SP, signal peptide; Y, predicted N-glycan site, with parentheses at partially conserved site, and asterisk marking the “heptad stutter”; ct, cytoplasmic tail. The blue bar above TM represents the partial sequence from the stickleback that was included in a study on filoviruses in mammalian genomes (Belyi et al. 2010).
Fig. 2.
Estimated age of percomORF. Time-calibrated phylogeny scaled to the geological time scale, adapted from Near et al. (2012) and based on 9 nuclear genes and 36 fossil age constraints. Orders in which percomORF or an empty site was found are indicated by red boxes and black boxes, respectively. Lineages that are part of the Ovalentaria clade are indicated. The S, Pg, and N (top) signify the Silurian, Paleogene, and Neogene geological periods.
Despite its ancient age, percomORF encodes all of the typical features of a functional retroviral envelope glycoprotein, with no stop codons interrupting the predicted reading frame in any of the species, implying maintenance of function for the benefit of the host, or exaptation. For example, the maintenance of receptor-binding function is suggested by 14 cysteines positionally conserved in the region corresponding to the surface (SU) subunit (fig. 1). Cysteines in SU typically help the protein fold into a globular conformation that functions in receptor binding (Chiou et al. 1992; van Anken et al. 2008). Likewise, percomORF retains motifs associated with membrane fusion such as: (1) a run of basic residues typical of a canonical furin cleavage site by which SU and the fusion subunit (TM) are cleaved to generate a fusion-ready trimer of heterodimers and (2) a stretch of hydrophobic residues at the N-terminus of TM, consistent with the fusion peptide. The CX6CC motif in the region coding for TM is typical of retroviruses whose subunits are joined by a covalent disulfide bond (Pinter et al. 1997; Henzy and Coffin 2013), and these motifs are usually found in conjunction with an isomerase domain (CXXC) in SU, which has been shown to mediate bond dissociation upon receptor binding (Wallin et al. 2004). Interestingly, percomORF encodes three such motifs (fig. 1). In addition to the conservation of motifs associated with function, exaptation is further supported by dN/dS analysis (supplementary table S2, Supplementary Material online). Under various evolutionary models, we found that between 189 and 232 of the 422 codons analyzed have dN/dS values that are consistent with a regime of strong purifying selection (average dN/dS: 0.25), while only a handful of positions (0–5) have dN/dS ratios suggesting positive selection.
One feature of the stickleback sequence that suggested it may be a filoviral glycoprotein is 32% amino acid sequence identity with Reston ebolavirus glyco-protein over a region in the TM subumit encoding 89 residues (fig. 1, blue bar) (Belyi et al. 2010). However, although filovirus glycoproteins are known to share structural features with retroviral glycoproteins (Gallaher 1996; Weissenhorn et al. 1998; Jeffers et al. 2002), several motifs encoded by percomORF are specific to retroviruses. These are (1) isomerase motifs in SU, which are common in retroviruses with covalently-bonded subunits (Pinter et al. 1997; Wallin et al. 2004; Henzy and Coffin 2013) but have not been found in filoviruses; (2) a fusion peptide that is N-terminal, whereas filoviral fusion peptides are internal and flanked by cysteines and (3) a motif (QNRAALD) in the immunosuppressive domain (ISD) that is typical of nonmammalian retroviruses rather than filoviruses (Henzy and Johnson 2013). A consensus sequence generated from an alignment of all 20 orthologs retains these motifs (supplementary fig. S1, Supplementary Material online), consistent with their presence in the ancestral retroviral protein.
Another feature that suggested that the stickleback sequence may be a filoviral gene is its existence in isolation from other viral genes (Belyi et al. 2010). Indeed, all of the percomORF loci we identified contained no discernible retroviral genes within a 10-kb flanking window, consistent with previous findings (Belyi et al. 2010). Whereas retroviruses insert a DNA copy of the entire viral genome (known as a “provirus”) into host DNA as a normal part of replication, other endogenous virus sequences are thought to arise through the interaction of viral mRNA with a retrotransposon such as a LINE-1 element (Horie et al. 2010; Katzourakis and Gifford 2010; Taylor et al. 2010), and thus exist as isolated genes. However, at a different locus we found a bona fide retroviral provirus in the Japanese eel (GenBank: KI305800.1) with a predicted Env protein that has 50% amino acid identity with the original partial sequence (compared with only 32% identity seen with Reston ebolavirus), serving as additional evidence for the retroviral origin of percomORF. Notably, we did not find any degenerate sequences more closely related to percomORF than that of the Japanese eel, likely reflecting divergence of percomORF’s evolutionary trajectory as a host gene from that of the originating retrovirus.
The presence of percomORF as an isolated gene may reflect its age. Notably, the Fv-1 gene of mice, like percomORF, is present in some species as an isolated gene with no discernible proviral sequence flanking it (Schlecht-Louf et al. 2010). An analysis of Fv-1 orthologous sites suggests that the locus has undergone multiple insertions, deletions, and rearrangements in various lineages, which may have caused the loss of proviral sequence in mice while the gene itself was maintained under selection (GR Young and JP Stoye, personal communication). The location of percomORF within the intron of DNAJC6 allowed us to compare the flanking sequence among the 20 orthologs. Similar to Fv-1, the regions flanking percomORF are very poorly conserved both in sequence and in length (fig. 1), consistent with loss of proviral sequence due to degeneration of neutral sites over a long period of evolution. By the same reasoning, it is likely that any markings of noncanonical integration, such as the inverted repeats that accompany LINE-1-mediated insertion, would be erased by mutations. Notably, the much “younger” ERVWE1 locus—a co-opted env gene originating from a retroviral insertion in the genome of a hominoid ancestor 25–40 Ma—is part of a provirus in which the gag and pol genes have acquired multiple inactivating mutations (Mallet et al., 2004).
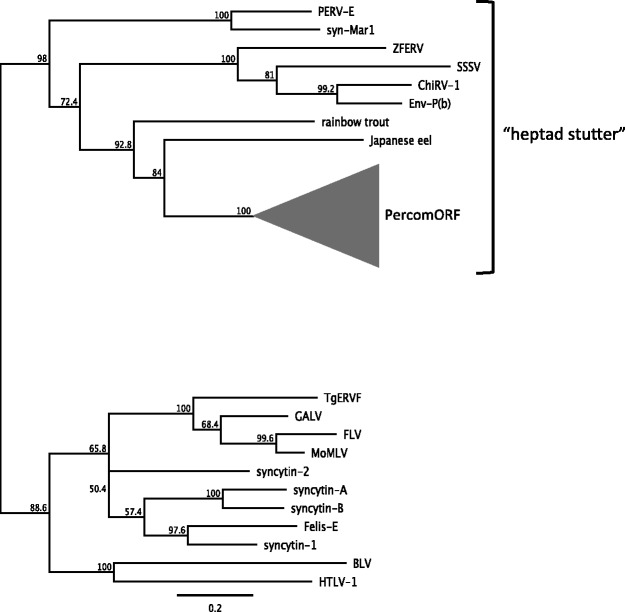
Among the features that percomORF does share with filovirus glycoproteins is a rigidly conserved predicted N-glycan site in the N-terminal portion of TM (fig. 1 and supplementary fig. S1, Supplementary Material online). N-glycans at this position have been found to disrupt the register of the heptad repeat of the fusion subunit, causing a loosening of the coiled-coil region, or “stutter” (Igonet et al. 2011; Higgins et al. 2014), which has been associated with viruses that infect via an endocytic pathway (Igonet et al. 2011). However this feature is typical of nonmammalian retroviruses (Henzy and Johnson 2013), as well as filoviruses (Koellhoffer et al. 2012), snake arenaviruses (Higgins et al. 2014; Koellhoffer et al. 2014), influenza virus, and SARS coronavirus (Supekar et al. 2004). On phylogenetic trees, gammaretrovirus-like env sequences form two large clades that reflect the presence or absence of the predicted heptad stutter. The PercomORF TM forms a well-supported branch within the heptad stutter superclade (fig. 3), which also includes a syncytin from squirrel-related rodents, MAR-1 (Redelsperger et al. 2014). Basal to the PercomORF clade is the TM sequence from the Japanese eel, possibly representing the original viral lineage from which percomORF arose.
Fig. 3.
Relationship of PercomORF to other gammaretroviral-like Env proteins. A neighbor-joining tree was generated (midpoint-rooted) using GENEIOUS v. 6.0.4, based on an amino acid alignment spanning the TM coding region, excluding the cytoplasmic tail. PercomORF sequences (collapsed into a clade represented by the gray triangle) cluster with the group of gammaretroviral-like Env proteins that carry a “heptad stutter” in the N-terminal heptad repeat. The lower clade consists of gammaretroviral sequences that do not include the stutter. All of the sequences besides PercomORF occur in the context of proviruses.
Exapted retroviral glycoproteins described in the literature have been associated with several functions (Jern and Coffin 2008; Lavialle et al. 2013; Johnson 2015; Malfavon-Borja and Feschotte 2015). Fv-4, for example, is a partial retroviral glycoprotein sequence in mice that expresses a defective envelope protein (Ikeda and Sugimura 1989) that binds to its cognate receptor, downregulating or blocking its expression (Kai et al. 1986) and thereby protecting the host from infection by related retroviruses (Limjoco et al. 1993). Other examples exploit the fusogenicity of the glycoprotein. Mammalian “syncytins”, for example, are maintained in various lineages for their ability to fuse syncytiotrophoblast cells during formation of the placenta (Lavialle et al. 2013), and appear to contribute to myoblast fusion in mice (Redelsperger et al. 2016). Syncytin-mediated cell–cell fusion is mechanistically analogous to membrane fusion during entry of the virus into the target cell. Thus, syncytins must maintain the fusion-mediating domains of retroviral Env proteins in addition to receptor-binding regions. The maintenance of fusion-related motifs in percomORF suggests that it could provide a fusogenic function. Moreover, the predicted cytoplasmic tail (CT) of percomORF is unusually short (supplementary fig. S1, Supplementary Material online) compared with that of related retroviral glycoproteins. The C-terminal 16 amino acids of the CT of murine leukemia viruses (MLVs) is cleaved by a viral protease to activate fusion (Yang and Compans 1997), and in HIV-1, truncation of the CT has been shown to increase cell–cell fusion in vitro (Kolchinsky et al. 2001). Therefore, a truncated CT in percomORF could represent selection for fusogenicity.
The divergence of the Percomorpha was associated with an expansion in the diversity of reproductive modes. Although the majority of its members are lecithotrophic (deriving nutrients from the yolk), the Percomorpha are the only group within the Acanthomorpha that includes taxa that have evolved placentation and viviparity (Wourms 1981). Specifically, most of the taxa with these traits belong to the clade “Ovalentaria”, which means “sticky eggs”, referring to the demersal eggs with adhesive filaments that are produced by taxa of this group (Wainwright et al. 2012). Ovalentaria are known to have very complex and highly derived reproductive strategies involving peculiar chorionic morphology (Reznick et al. 2007), and have experienced the evolution and loss of placentas on numerous occasions (Reznick et al. 2002; Pollux et al. 2009). While the majority of percomORF-positive species from this limited study are not viviparous, reflecting the composition of the Percomorpha group as a whole, it is intriguing to speculate on a role for percomORF in the dynamic innovation in reproductive strategies that characterizes this group, possibly contributing to syncytia formation in placenta, “sticky eggs”, or another chorionic feature.
In conclusion, percomORF is to our knowledge the oldest intact gene of viral—older by as much as 50 My than syncytin-Car1, which entered the germline of carnivores an estimated 60–85 Ma (Cornelis et al. 2012). The maintenance of an intact ORF and canonical functional motifs, its expression in stickleback embryos, together with dN/dS analyses strongly suggest that percomORF has been coopted by the host species. The role of similar retroviral env genes in chorionic structures and placenta formation in mammals, combined with percomORF’s association with the only group of fish in which placentation evolved leads naturally to speculation on a syncytin-like role for percomORF. However, follow-up studies involving a wider range of taxa and morphological data; expression profiles in a variety of tissue types and developmental stages; and assays for fusogenicity of the protein, are needed to assess its role in the evolution of ray-finned fishes.
Supplementary Material
Supplementary methods S1, tables S1 and S2 and figure S1 are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grant AI083118 (W.E.J.) and by start-up funds provided by Boston College.
Footnotes
Associate editor: Thomas Leitner
References
- Belyi VA, Levine AJ, Skalka AM. 2010. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6:e1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R, Broughton RE, Wiley EO, Carpenter K, López JA, Li C, Holcroft NI, Arcila D, Sanciangco M, Cureton Ii JC, et al. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou SH, Freed EO, Panganiban AT, Kenealy WR. 1992. Studies on the role of the V3 loop in human immunodeficiency virus type 1 envelope glycoprotein function. AIDS Res Hum Retroviruses 8:1611–1618. [DOI] [PubMed] [Google Scholar]
- Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Véron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl. Acad Sci U S A. 109:E432–E441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher WR. 1996. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 85:477–478. [DOI] [PubMed] [Google Scholar]
- Henzy JE, Coffin JM. 2013. Betaretroviral envelope subunits are noncovalently associated and restricted to the mammalian class. J Virol. 87:1937–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzy JE, Johnson WE. 2013. Pushing the endogenous envelope. Philos Trans R Soc Lond B Biol Sci. 368:20120506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CD, Malashkevich VN, Almo SC, Lai JR. 2014. Influence of a heptad repeat stutter on the pH-dependent conformational behavior of the central coiled-coil from influenza hemagglutinin HA2. Proteins 82:2220–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, et al. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igonet S, Vaney M-C, Vonrhein C, Vonhrein C, Bricogne G, Stura EA, Hengartner H, Eschli B, Rey FA. 2011. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc Natl Acad Sci U S A. 108:19967–19972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Sugimura H. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 63:5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers SA, Sanders DA, Sanchez A. 2002. Covalent modifications of the Ebola virus glycoprotein. J Virol. 76:12463–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet. 42:709–732. [DOI] [PubMed] [Google Scholar]
- Johnson WE. 2015. Endogenous retroviruses in the genomics era. Annu Rev Virol. 2:135–159. [DOI] [PubMed] [Google Scholar]
- Kai K, Sato H, Odaka T. 1986. Relationship between the cellular resistance to Friend murine leukemia virus infection and the expression of murine leukemia virus-gp70-related glycoprotein on cell surface of BALB/c-Fv-4wr mice. Virology 150:509–512. [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6:e1001191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellhoffer JF, Dai Z, Malashkevich VN, Stenglein MD, Liu Y, Toro R, S Harrison J, Chandran K, DeRisi JL, Almo SC, et al. 2014. Structural characterization of the glycoprotein GP2 core domain from the CAS virus, a novel arenavirus-like species. J Mol Biol. 426:1452–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellhoffer JF, Malashkevich VN, Harrison JS, Toro R, Bhosle RC, Chandran K, Almo SC, Lai JR. 2012. Crystal structure of the Marburg virus GP2 core domain in its postfusion conformation. Biochemistry (Mosc.) 51:7665–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 75:2041–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of “syncytins”, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 368:20120507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limjoco TI, Dickie P, Ikeda H, Silver J. 1993. Transgenic Fv-4 mice resistant to Friend virus. J Virol. 67:4163–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfavon-Borja R, Feschotte C. 2015. Fighting fire with fire: endogenous retrovirus envelopes as restriction factors. J Virol. 89:4047–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci. U S A. 101:1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A. 109:13698–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Kopelman R, Li Z, Kayman SC, Sanders DA. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 71:8073–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollux BJA, Pires MN, Banet AI, Reznick DN. 2009. Evolution of placentas in the fish family Poeciliidae: an empirical study of macroevolution. Annu Rev Ecol Evol Syst. 40:271–289. [Google Scholar]
- Redelsperger F, Cornelis G, Vernochet C, Tennant BC, Catzeflis F, Mulot B, Heidmann O, Heidmann T, Dupressoir A. 2014. Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the Rodentia squirrel-related clade. J Virol. 88:7915–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelsperger F, Raddi N, Bacquin A, Vernochet C, Mariot V, Gache V, Blanchard-Gutton N, Charrin S, Tiret L, Dumonceaux J, et al. 2016. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLOS Genet. 12:e1006289.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D, Meredith R, Collette BB. 2007. Independent evolution of complex life history adaptations in two families of fishes, live-bearing halfbeaks (zenarchopteridae, beloniformes) and poeciliidae (cyprinodontiformes). Evol Int J Org Evol. 61:2570–2583. [DOI] [PubMed] [Google Scholar]
- Reznick DN, Mateos M, Springer MS. 2002. Independent origins and rapid evolution of the placenta in the fish genus Poeciliopsis. Science 298:1018–1020. [DOI] [PubMed] [Google Scholar]
- Schlecht-Louf G, Renard M, Mangeney M, Letzelter C, Richaud A, Ducos B, Bouallaga I, Heidmann T. 2010. Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc Natl Acad Sci U S A. 107:3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS, et al. 2016. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 44:D717–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar VM, Bruckmann C, Ingallinella P, Bianchi E, Pessi A, Carfí A. 2004. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc Natl Acad Sci U S A. 101:17958–17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Leach RW, Bruenn J. 2010. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol Biol. 10:193.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Anken E, Sanders RW, Liscaljet IM, Land A, Bontjer I, Tillemans S, Nabatov AA, Paxton WA, Berkhout B, Braakman I. 2008. Only five of 10 strictly conserved disulfide bonds are essential for folding and eight for function of the HIV-1 envelope glycoprotein. Mol Biol Cell 19:4298–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright PC, Smith WL, Price SA, Tang KL, Sparks JS, Ferry LA, Kuhn KL, Eytan RI, Near TJ. 2012. The evolution of pharyngognathy: a phylogenetic and functional appraisal of the pharyngeal jaw key innovation in labroid fishes and beyond. Syst Biol. 61:1001–1027. [DOI] [PubMed] [Google Scholar]
- Wallin M, Ekström M, Garoff H. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 23:54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 2:605–616. [DOI] [PubMed] [Google Scholar]
- Wourms JP. 1981. Viviparity: the maternal-fetal relationship in fishes. Am Zool. 21:473–515. [Google Scholar]
- Yang C, Compans RW. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 71:8490–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.