While bronchiolitis is considered a relatively homogenous viral infection, these results provide novel evidence to support the emerging hypothesis that bronchiolitis due to respiratory syncytial virus only or rhinovirus only may have different mechanisms that involve a complex interplay between virus, microbiome, and host.
Keywords: Bronchiolitis, respiratory syncytial virus, rhinovirus, metabolomics, microbiome
Abstract
Background
Bronchiolitis, the leading cause of hospitalization among infants in the United States, is most commonly caused by respiratory syncytial virus (RSV), followed by rhinovirus (RV). Conventional perception is that bronchiolitis is a single entity, albeit with different viral etiologies and degrees of severity.
Methods
We conducted a cross-sectional study of nasopharyngeal aspirates from 106 infants hospitalized with bronchiolitis due to either RSV only (80 patients) or RV only (26 patients). We performed metabolomics analysis and 16S ribosomal RNA gene sequencing on all samples and metagenomic sequencing on 58 of 106 samples.
Results
Infants with RSV-only and RV-only infections had significantly different nasopharyngeal metabolome profiles (P < .001) and bacterial metagenome profiles (P < .05). RSV-only infection was associated with metabolites from a range of pathways and with a microbiome dominated by Streptococcus pneumoniae. By contrast, RV-only infection was associated with increased levels of essential and nonessential N-acetyl amino acids and with a high relative abundance of Haemophilus influenzae. These co-occurring species were associated with driving the bacterially derived metabolic pathways. Multi-omic analysis showed that both the virus and the microbiome were significantly associated with metabolic function in infants hospitalized with bronchiolitis.
Conclusion
Although replication of these findings is necessary, they highlight that bronchiolitis is not a uniform disease between RSV and RV infections, a result with future implications for prevention and treatment.
Bronchiolitis, a lower respiratory tract infection in infants, is an important public health problem worldwide and the leading cause of US infant hospitalizations [1]. Respiratory syncytial virus (RSV) and rhinovirus (RV) account for approximately 85% of the viruses causing severe bronchiolitis [2]. Although different viruses cause bronchiolitis, the American Academy of Pediatrics recommends that clinicians not test children with bronchiolitis for viruses, since treatment does not vary by viral etiology [3]. Furthermore, owing to the lack of clinical trials demonstrating a consistently beneficial responses to pharmacotherapy, the 2014 American Academy of Pediatrics clinical practice guideline discusses bronchiolitis as a single, relatively homogenous entity [4, 5].
Recently, however, we identified distinct clinical phenotypes of bronchiolitis, signaling that bronchiolitis probably is a heterogeneous condition [6]. Furthermore, compared with children with RSV bronchiolitis, children with RV bronchiolitis have a shorter hospital length of stay [2], are significantly more likely to be treated with systemic corticosteroids [7], and have an increased risk of childhood asthma [7, 8]. Although indicative of heterogeneity, these associations between viral etiology and outcomes do not provide insight into the underlying pathobiology of bronchiolitis, which involves complex interplay between the virus, microbiome, and host. Thus, we hypothesized that RSV and RV infections would be associated with underlying metabolic differences, as manifested by differences in the airway microbiome and metabolome. To test this hypothesis, we used metabolomic and metagenomic approaches to investigate nasopharyngeal aspirates from infants hospitalized with bronchiolitis. This integrated multi-omic approach examined molecular differences between RSV and RV pathobiology while accounting for both microbial and host factors.
METHODS
Study Design, Setting, and Participants
The study design, setting, participants, and methods of data collection are from the 35th Multicenter Airway Research Collaboration (MARC-35) [9]. Briefly, using a standardized protocol, site investigators at 17 sites across 14 US states enrolled infants hospitalized with a diagnosis of bronchiolitis from an attending physician. Bronchiolitis was defined on the basis of the American Academy of Pediatrics guidelines, as follows: acute respiratory illness with some combination of rhinitis, cough, tachypnea, wheezing, crackles, and retractions [3]. To make the timing of sample collection more synchronous, we excluded infants with previous enrollment, those who were transferred to a participating hospital >24 hours after the original hospitalization, and those for whom consent to participate was provided >24 hours after hospitalization. We also excluded those with known heart-lung disease, immunodeficiency, immunosuppression, or gestational age of <32 weeks. All patients were treated at the discretion of the treating physician. The institutional review board at each of the participating hospitals approved the study. Written informed consent was obtained from the parent or legal guardian.
We selected MARC-35 samples for metabolomic analysis to examine the severity of illness [10] and to determine the presence of RSV-only versus RV-only infections. Given the cost of this testing and the volume of sample required, we not only limited the overall sample size for testing, but also selected nasopharyngeal samples with sufficient volume for metabolomic analysis and metagenomic shotgun whole-genome sequencing (WGS). For the present analysis, we analyzed 106 samples with RSV-only or RV-only infections. Of the 106 samples, all had 16S ribosomal RNA gene sequencing data, and 58 had metagenomic shotgun WGS data. Detailed descriptions of the sample preparation, data acquisition, and processing and of the statistical analysis may be found in the Supplementary Materials.
Sample Collection and Testing
Sample Collection
Nasopharyngeal aspirates were collected by trained site investigators and stored at −80°C, using a standardized protocol [2]. All samples were handled and processed identically.
Virologic Testing
Viral genomic load was determined as previously described [11]. A cycle threshold (Ct) of <40 was considered positive. The Ct provides a semiquantitative measure of viral load, with an inverse linear relationship between viral load and Ct. Thus, the inverse of the Ct (ie, viral load) was used for analysis.
Metabolomic Profiling
Metabolomic analysis was performed by Metabolon (Durham, NC), using ultra-performance liquid chromatography–tandem mass spectrometry in both positive and negative ionization modes. Metabolon processed the samples in a masked fashion and a random order. Sample preparation was performed as described previously [12].
Bacterial 16S Ribosomal RNA Gene Profiling
Bacterial genomic DNA was extracted from the nasopharyngeal samples by using the MOBIO PowerSoil DNA Isolation Kit (Mo Bio Laboratories; Carlsbad, CA) as described previously [13]. The sequencing methods were adapted from those developed for the National Institutes of Health–Human Microbiome Project [14], using the V4 region (515F and 806R [15]), and were sequenced on the MiSeq platform (Illumina; San Diego, CA), using the 2 × 250–bp paired-end protocol. Resulting reads were quality filtered using strict merging criteria, and biom tables were rarified to 1500 reads/sample.
Metagenomic Shotgun WGS
Metagenomic shotgun WGS was available for 58 nasopharyngeal samples [10]. Individual libraries constructed from each sample were sequenced using the 2 × 100–bp paired-end read protocol on the HiSeq platform (Illumina). The process of quality filtering, trimming, and demultiplexing was performed using a pipeline developed at Baylor College of Medicine.
Statistical Analyses
The primary exposure was RSV-only or RV-only infection, and the primary outcome was the nasopharyngeal airway metabolome. Orthogonal partial least squares discriminant analysis (OPLS-DA) was performed using MetaboAnalyst 3.0 and was validated using 2000 random permutations [16]. Significant metabolites were determined using the 2-tailed Welch t test, with the false-discovery rate (FDR) algorithm used to adjust for multiple comparisons [17]. Linear regression models adjusted for 7 potential confounding variables (ie, age, sex, preterm birth, use of antibiotics, use of systemic corticosteroid therapy before hospitalization, positive pressure ventilation [PPV], and hospital site) and were corrected with the FDR algorithm. MixOmics [18] was implemented in R, version 3.3 [19], based on sparse partial least squares regression analysis, and was performed in canonical mode with LASSO penalization. FishTaco (functional shifts taxonomic contributors) was used to determine which species was associated with “reducing” or “driving” the significantly altered functions [20], and model-based integration of metabolite observations and species abundances (MIMOSA) was used to determine biologically feasible correlations between the shotgun WGS bacterial Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs and the resulting metabolites of likely bacterial origin [21].
RESULTS
Study Population
Nasopharyngeal metabolomic and microbiome data were generated from 106 infants enrolled in the MARC-35 study, a 17-center cohort study of 1016 infants hospitalized with bronchiolitis [9]. Compared with the nonanalytic MARC-35 cohort (910 patients), the 106 patients in the current analytic cohort were younger and more likely to require PPV, defined as continuous PPV and/or endotracheal intubation (Supplementary Table 1). The 106 infants in the analytic cohort had a median age at hospitalization of 3 months (interquartile range, 1–5 months), and 61% were male. Eighty (75%) were infected with RSV only, and 26 (25%) were infected with RV only (ie, no other coinfecting virus was detected among 16 viruses tested). Patient characteristics did not differ significantly between the 2 virus groups, with the exception of a higher proportion of non-Hispanic white infants and a greater proportion of infants using PPV in the RSV-only group (Table 1).
Table 1.
Characteristics of 106 Infants Hospitalized With Either Respiratory Syncytial Virus (RSV)–Only or Rhinovirus (RV)–Only Bronchiolitis
| Characteristic | RSV Only (n = 80) | RV Only (n = 26) |
P |
|---|---|---|---|
| Age, mo | 2.7 (1–5) | 2.7 (1.4–7.9) | .64 |
| Male sex | 47 (59) | 18 (69) | .34 |
| Race/ethnicity | <.001 | ||
| Non-Hispanic white | 43 (54) | 11 (42) | |
| Non-Hispanic black | 11 (14) | 6 (23) | |
| Hispanic | 21 (26) | 8 (31) | |
| Other | 5 (6) | 1 (4) | |
| Maternal smoking during pregnancy | 5 (6) | 4 (15) | .18 |
| Prematurity (gestation wk 32–37 ) | 16 (20) | 7 (27) | .46 |
| Cesarean section delivery | 29 (36) | 10 (38) | .87 |
| Postnatal smoke exposure at home | 9 (12) | 0 (0) | .97 |
| Siblings, no. | 1 (1–2) | 1 (1–2) | .54 |
| History of antibiotic use prior to enrollment | 22 (28) | 8 (31) | .75 |
| History of corticosteroid use prior to enrollment | 11 (14) | 3 (12) | .77 |
| Positive pressure ventilation use | 16 (23) | 0 (0) | .03 |
Data are median value (interquartile range) or no. (%) of infants.
Infants With RSV-Only or RV-Only Infection Have Significantly Different Metabolomic Profiles
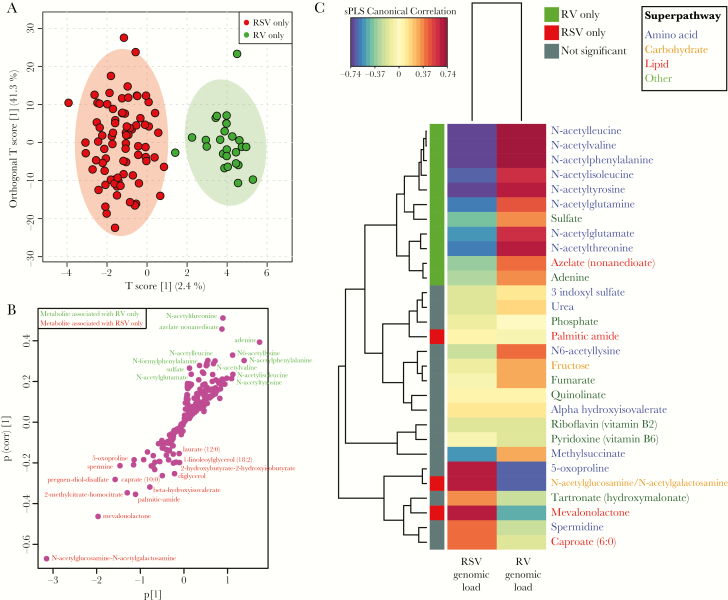
Metabolomic analysis detected 254 metabolites. When comparing infants with RSV-only bronchiolitis to those with RV-only bronchiolitis, the metabolomic profiles clustered distinctly on the basis of viral etiology (Figure 1A). This difference was validated using 2000 permutations (P < .001). Since the use of PPV was significantly greater in infants with RSV-only infection and may influence the metabolites (Table 1), we removed the infants who underwent PPV from the analysis and still found significant differences in the metabolomic profiles between infants with RSV-only infection and those with RV-only infection (P < .001). Metabolites associated with RSV-only infection were from a range of superpathways, including carbohydrate metabolism (N-acetylglucosamine-N-acetylgalactosamine), lipid metabolism (mevalonolactone), amino acid metabolism (5-oxoproline), and energy metabolism (2-methylcitrate-homocitrate; Figure 1B). By contrast, metabolites associated with RV-only infection consisted primarily of N-acetyl metabolites from the amino acid superpathway, specifically, N-acetylthreonine, N-acetylleucine, N-acetylisokeucine, N-acetylphenyalanine, N-acetylvaline, N-acetylglutamate, N-acetyltyrosine, and N-acetyllysine (Figure 1B).
Figure 1.
Nasopharyngeal airway metabolomic profiles in infants with respiratory syncytial virus (RSV)–only or rhinovirus (RV)–only bronchiolitis. Analysis includes 106 infants and 254 detected metabolites. A, Orthogonal partial least squares discriminant analysis (OPLS-DA) score scatterplot of samples classified according to RSV-only (red) or RV-only (green) infection. Each dot represents the metabolomic profile of 1 infant. Ellipses show 95% confidence intervals. Metabolomic profiles were significantly different between RSV and RV infections (permutation P < .001). B, OPLS-DA S-plot of metabolites, where each dot represents a metabolite. Metabolites in red are discriminatory in RSV-only infection, and metabolites in green are discriminatory in RV-only infection. C, Heat map showing the correlations of the viral genomic load (measured as the inverse cycle threshold) of RSV and RV infections with the metabolomic profiles. Only the top 25 (of all 254 metabolites) most strongly correlated metabolites are shown. Canonical correlations are based on sparse partial least squares (sPLS). Colored squares on the left column denote metabolites significantly increased in RSV-only infection (red), significantly increased in RV-only infection (green), and not significant increased (gray), as per Table 2.
Of the 254 detected metabolites, 31 significantly differed between RSV-only and RV-only infections by relative intensity, with significant differences for 25 metabolites remaining after adjustment for 7 covariates, including race/ethnicity and PPV use (FDR P < .05; Table 2). These associations between the qualitative viral differences (ie, viral presence or absence) and all 254 detected metabolites were also supported by correlations between quantitative viral differences (ie, genomic load of RSV and RV, based on inverse Ct values) and all detected metabolites (Figure 1C). In other words, not only were the metabolites different on the basis of infecting virus, but also the amount of virus correlated with increased metabolite intensity. Of the samples with detectable virus, the median viral load Ct was 23.0 (interquartile range, 21.5–26.4) for RSV and 26.4 (interquartile range, 24.6–28.1) for RV.
Table 2.
Metabolites Associated With Respiratory Syncytial Virus (RSV) or Rhinovirus (RV) Among Infants Hospitalized with RSV-Only or RV-Only Bronchiolitis
| Virus, Metabolite | Subpathway(s) | Superpathway | KEGG Compound | P | |
|---|---|---|---|---|---|
| Unadjusted | Adjusteda | ||||
| RSV | |||||
| N-acetylglucosamine-N- acetylgalactosamine | Aminosugar metabolism | Carbohydrate | … | <.001 | .002 |
| Mevalonolactone | Mevalonate metabolism | Lipid | … | <.001 | .02 |
| Cystine | Methionine, cysteine, SAM, and taurine metabolism | Amino acid | C00491 | .033 | .004 |
| Palmitic-amide | Fatty acid, amide | Lipid | … | .001 | .004 |
| 2-methylcitrate-homocitrate | TCA cycle | Energy | … | .001 | .016 |
| Caprate (10:0) | Medium chain fatty acid | Lipid | C01571 | .018 | .024 |
| Diglycerol | Chemical | Xenobiotics | … | .025 | .044 |
| Beta-hydroxyisovalerate | Leucine, isoleucine, and valine metabolism | Amino acid | … | .015 | .054 |
| Pregnen-diol-disulfate | Steroid | Lipid | … | .018 | .060 |
| Laurate (12:0) | Medium chain fatty acid | Lipid | C02679 | .046 | .081 |
| RV | |||||
| N-acetylthreonine | Glycine, serine, and threonine metabolism | Amino acid | … | <.001 | .002 |
| N-acetyltyrosine | Phenylalanine and tyrosine metabolism | Amino acid | … | <.001 | .002 |
| N6-acetyllysine | Lysine metabolism | Amino acid | C02727 | .001 | .002 |
| Adenine | Purine metabolism, adenine containing | Nucleotide | C00147 | .000 | .002 |
| Azelate (nonanedioate) | Fatty acid, dicarboxylate | Lipid | C08261 | .000 | .002 |
| N-acetylleucine | Leucine, isoleucine, and valine metabolism | Amino acid | C02710 | .001 | .002 |
| Glucuronate | Aminosugar metabolism | Carbohydrate | C00191 | .005 | .002 |
| N-acetylvaline | Leucine, isoleucine, and valine metabolism | Amino acid | … | .001 | .002 |
| N-acetylphenylalanine | Phenylalanine and tyrosine metabolism | Amino acid | C03519 | .001 | .002 |
| N-acetylisoleucine | Leucine, isoleucine, and valine metabolism | Amino acid | … | .003 | .014 |
| 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) | Phospholipid metabolism | Lipid | … | .040 | .014 |
| N-acetyltaurine | Methionine, cysteine, SAM, and taurine metabolism | Amino acid | … | .017 | .017 |
| Sulfate | Chemical | Xenobiotics | C00059 | .011 | .024 |
| Glycerol | Glycerolipid metabolism | Lipid | C00116 | .017 | .024 |
| N-acetylglutamate | Glutamate metabolism | Amino acid | C00624 | .005 | .031 |
| 4-hydroxyphenylpyruvate | Phenylalanine and tyrosine metabolism | Amino acid | C01179 | .011 | .031 |
| Glycerol 3-phosphate | Glycerolipid metabolism | Lipid | C00093 | .017 | .031 |
| N-formylphenylalanine | Phenylalanine and tyrosine metabolism | Amino acid | .011 | .031 | |
| Gluconate | Food component/plant | Xenobiotics | C00257 | .045 | .038 |
| N-acetylarginine | Urea cycle; arginine and proline metabolism | Amino acid | C02562 | .014 | .060 |
| Folate | Folate metabolism | Cofactors and vitamins | C00504 | .034 | .131 |
aLinear regression models adjusting for age, race/ethnicity, history of prematurity, history of antibiotic use prior to the enrollment, use of systemic corticosteroids during the prehospitalization visit, use of positive pressure ventilation, and hospital site.
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes; SAM, S-adenyl methionine; TCA, tricarboxylic acid.
Infants With RSV-Only or RV-Only Infections Have Significantly Different Bacterial Functional Potential Primarily Associated With Streptococcus pneumoniae or Haemophilus influenzae, Respectively
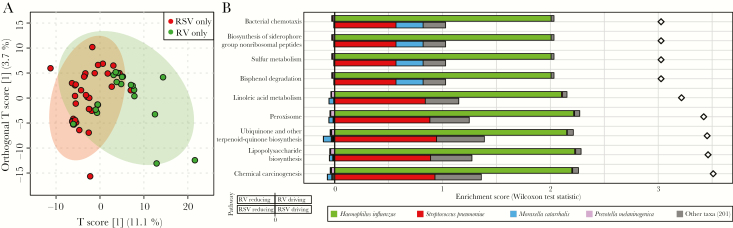
Since RSV and RV infect an airway colonized with bacteria, we conducted metagenomic shotgun WGS to determine the functional potential of the microbiome (ie, the presence but not necessarily expression of genes in microbial genomes). From the 106 nasopharyngeal samples, 58 were included in the metagenomic analyses (39 from infants in the RSV-only group and 19 from infants in the RV-only group). There were no demographic differences between the RSV-only and RV-only groups in the metagenomic subset, although infants with RSV-only infection were more likely to undergo PPV (Supplementary Table 2). Infants with either RSV-only or RV-only infection had significantly different metagenomic (metabolomic potential) profiles (permutation P = .043; Figure 2).
Figure 2.
Metabolic potential of the bacterial community among infants with respiratory syncytial virus (RSV)–only or rhinovirus (RV)–only infection. The analysis includes metagenomic subset of 58 infants and 3883 Kyoto Encyclopedia of Genes and Genomes orthologs (KOs). A, Orthogonal partial least squares discriminant analysis grouped by RSV-only (red) or RV-only (green) infection. Each dot represents the metagenomic profile of 1 infant. Metagenomic profiles were significantly different between RSV-only and RV-only infections (permutation P = .043). B, FishTaco analysis determined the contribution of bacterial species to metabolic pathways (determined from KOs) that were significantly different between RSV-only and RV-only infections. Plots are divided by the 9 significant pathways. In each plot, the bars with negative enrichment (left of the line) show the association between bacterial species reducing function, and bars with positive enrichment (right of the line) show the association between bacterial species driving function. For each pathway, RV-only samples are on the top row, and RSV-only samples are the bottom row.
We performed further analysis of the metagenomic data by using FishTaco, which determines taxa that are associated with “reducing” or “driving” functional shifts [20]. This analysis showed that 9 bacterially derived metabolic pathways were significantly different between infants with RSV-only infection and those with RV-only infection (Figure 2B). Among infants with RSV-only infection, S. pneumoniae relative abundance was associated with driving the functional shift in all 9 pathways. By contrast, Moraxella catarrhalis was associated with driving 4 of 9 pathways and reducing the remaining 5 pathways. Among infants with RV-only infection, H. influenzae was associated with driving all 9 pathways, and Prevotella melaninogenica was associated with reducing all 9 pathways. Findings of the more granular module-level analysis were comparable to those of the broader pathway-level analysis, except that energy metabolism modules were reduced by S. pneumoniae and M. catarrhalis in RSV-only bronchiolitis and by H. influenzae in RV-only bronchiolitis (Supplementary Figure 1). Furthermore, in infants with RSV-only infection, S. pneumoniae was associated with driving carbohydrate and lipid metabolite modules, while Streptococcus mitis was associated with reducing these modules, demonstrating that species within the same genus may influence modules differently.
Multi-omic Analysis Showed That Virus and Dominant Bacteria Were Both Associated With Metabolomic Profiles
The 3 dominant bacterial operational taxonomic units from the 16S ribosomal RNA gene profiling (representing 80% of the total bacterial community) were characterized by metagenomic data as S. pneumoniae, H. influenzae, and M. catarrhalis species. Examining viral-bacterial associations in the current analytic cohort, we found that RSV-only infection was associated with a high relative abundance of S. pneumoniae and that RV-only infection was associated with high relative abundance of H. influenzae (Supplementary Figure 2). However, given the dominance of all 3 bacteria (ie, S. pneumoniae, H. influenzae, and M. catarrhalis) and previously published viral-microbial associations from the entire MARC-35 cohort [9], we included all 3 dominant species in the analysis.
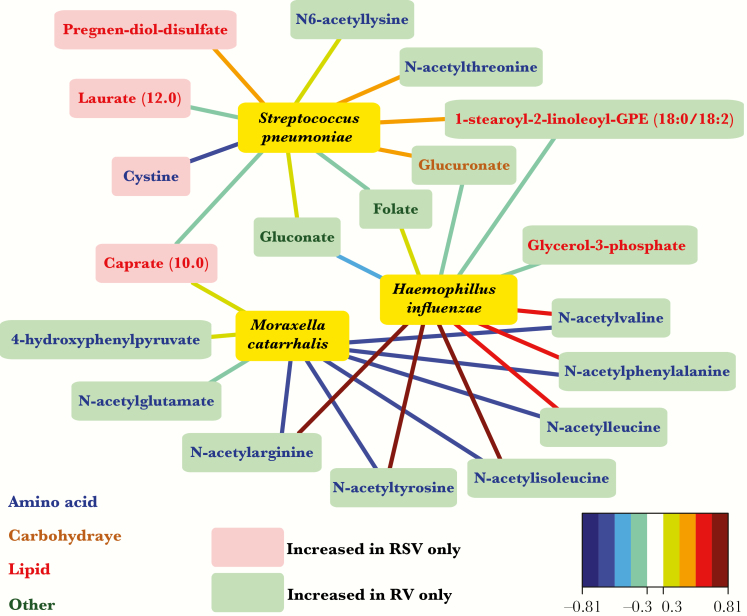
Among the 254 metabolites, the relative abundance of S. pneumoniae was positively correlated with metabolites from sphingolipid metabolism (Supplementary Figure 3). However, sphingolipid metabolites were not associated with RSV-only infection. Consistent with these results, the network analysis showed that the relative abundance of S. pneumoniae had only weak positive correlations with metabolites significantly increased in RSV-only infection (Figure 3). By contrast, H. influenzae was positively correlated with the N-acetyl metabolites from amino acid metabolism that are increased in RV-only infection and was negatively correlated with all other metabolites (Figure 3 and Supplementary Figure 3). The abundance of M. catarrhalis was positively correlated with metabolites from a range of pathways, but none of the metabolites were associated with either RSV-only or RV-only infections (Figure 3 and Supplementary Figure 3).
Figure 3.
Association of infants hospitalized with respiratory syncytial virus (RSV)–only and rhinovirus (RV)–only bronchiolitis with the nasopharyngeal microbiota. Analysis includes 106 infants. Network plot showing all correlations >0.5 between the relative abundance of the 3 dominant bacterial genera (yellow) and the 31 significant metabolites in RSV (red) or RV (green) infection. Nodes are colored red or green according to the significantly increased metabolite in RSV or RV infection. The metabolite names are colored according to the super-pathway. Edges are colored by canonical sparse partial least squares correlations. Relative abundance for the 3 dominant genera is based on 16S ribosomal RNA gene sequencing. Operational taxonomic units were identified to the species level, based on metagenomic data.
Permutational multivariate analysis of variance using distance matrices (adonis) was used to test whether the virus or the bacterial species had a greater association with the metabolomic profiles [22]. As the adonis function is based on sequential sum of squared deviations from a centroid, the order of the factors (ie, whether the virus was entered first or second in the model) was tested using both permutations. The virus (RSV only or RV only) was statistically significant when entered as the first factor (P = .04) but not when entered as the second factor (P = .14). Similarly, the bacterial species (dominance by S. pneumoniae, H. influenzae, M. catarrhalis, or a mixed community) was statistically significant when entered as the first factor (P = .049) but not when entered as the second factor (P = .10). These results suggest that both the viral etiology and the bacterial species are associated with the metabolomic profiles and that both are relevant in infants hospitalized with bronchiolitis.
Significant Metabolites Were Host Derived, Based on Computational Analysis
We used MIMOSA to ascertain which metabolites were either bacterially derived or host derived, based on metagenome orthology. Although there were significant metagenomic (metabolomic potential) differences between infants with RSV-only infection and those with RV-only infection, MIMOSA analysis of the 254 metabolites estimated that 19 metabolites (7%) were derived from bacteria (Supplementary Table 3). None of the 31 metabolites that significantly differed between infants with RSV-only infection and those with RV-only infection were bacterially derived (Table 2). Taken together, these results suggest that most metabolites that distinguish RSV-only from RV-only infections are host derived.
DISCUSSION
In this novel multi-omic analysis of nasopharyngeal airway samples collected from infants hospitalized with bronchiolitis, RSV-only and RV-only infections were associated with significantly different metabolic function. Infants with RSV-only bronchiolitis had significantly increased metabolites from a range of pathways, whereas infants with RV-only bronchiolitis had increased N-acetyl amino acids. Moreover, a higher viral genomic load positively correlated with a higher intensity of the metabolites associated with RSV and RV infections. RSV and RV co-occurred with bacterial species (eg, RSV with S. pneumoniae and RV with H. influenzae), and the associated metagenomes significantly differed between RSV-only and RV-only infections. Thus, these data are the first to demonstrate that RSV and RV are associated with different metabolic pathways and that the associated bacterial functional capacity is derived primarily from S. pneumoniae in RSV bronchiolitis and from H. influenzae in RV bronchiolitis.
Although the clinical phenotype of bronchiolitis due to different viruses is considered indistinguishable by some experts [23], comparison of the epidemiologic data of children with RV bronchiolitis to those with RSV bronchiolitis demonstrates that children with RV bronchiolitis have different demographic characteristics [2, 24], medical histories [25], short-term outcomes [6], and long-term outcomes [7, 25] and received different hospital treatment [25]. The present results provide the first multi-omic comparison demonstrating pathobiological differences between RSV and RV bronchiolitis. Higher genomic load of RSV and RV was positively correlated with a higher intensity of the metabolites that distinguished RSV infection from RV infection. This finding is intriguing because of the association between higher RSV genomic load and increased severity of illness [26], but the same association has not been found in RV infection [27]. While the relevance of genomic viral load requires additional validation owing to the epidemiological and now pathobiological differences between RSV and RV, future clinical trials for infants with bronchiolitis should consider stratifying infants on the basis of viral etiology.
Infants with RV-only infection hospitalized with bronchiolitis had altered amino acid metabolism, particularly related to N-acetyl metabolism. H. influenzae, which co-occurred with RV, was also associated with these metabolites. Nα-acetylation is the process of irreversibly transferring an acetyl group from acetyl coenzyme A to the α-amino group of an amino acid [28]. It is among the most abundant cotranslational and posttranslational protein modifications in eukaryotes but is less common in bacteria [29]. The functional implication of acetylation in RV bronchiolitis remains unclear. Notably, in asthma, there is an increase in histone acetyltransferase activity that is treatable with corticosteroids [30]. The association, although speculative, between RV wheezing illnesses during early childhood and an increased risk of asthma among school-aged children [31–33] may be explained in part by the altered host amino acid metabolism in RV bronchiolitis seen in the present results.
The bacterial (vs host) derivation of 7% of detected metabolites was comparable to findings from a previous report involving adult sputum, in which 4% of detected metabolites were microbial [34]. The KEGG orthologs from H. influenzae were associated with the generation of essential amino acids, but neither S. pneumoniae nor other microbes correlated with the generation of amino acids. Indeed, S. pneumoniae cannot synthesize amino acids [35], producing peptidases and proteases and using cell wall transporters to uptake amino acids [36]. Therefore, in addition to dietary intake, H. influenzae provides access to essential amino acids, which may have contributed to the observed increase in RV-associated N-acetyl metabolites. S. pneumoniae and H. influenzae were responsible for the same bacterial metabolic pathway functions in RSV and RV infections, respectively. Additionally, we found cases of S. pneumoniae driving and S. mitis reducing the same modules, suggesting important effects of different species within the same genera [20]. Taken together, these results suggest that the viruses and bacteria both have important influences on the metabolomic profiles, as supported by the adonis results. Thus, future bronchiolitis studies may need to account for both viral and bacterial communities, and clinicians may need to begin thinking of “viral” bronchiolitis as being due to a complex interplay among the infecting virus, microbiome, and host response.
The current results offer novel findings and additional evidence of microbial (virus and bacteria) and host interaction in the pathobiology of bronchiolitis as investigated within the MARC-35 cohort. We have previously found that infants with RV bronchiolitis have a shorter hospitalization duration [37, 38] and are associated with Haemophilus and Moraxella microbial dominance when compared to RSV, which is associated with microbial dominance by Streptococcus [9]. And although Haemophilus and Streptococcus are associated with an increased severity of illness [13, 39] and Moraxella with less severe bronchiolitis [13], the results from the MARC studies underscore the complex bacterial, viral, and host interaction in bronchiolitis pathobiology [40, 41].
The current study has several potential limitations. First, bronchiolitis is mostly a disease of the lower airways, and while nasopharyngeal samples may not provide a reliable representation of the lung microbiome [42], they are more easily obtained than lower airway samples [43]. Additionally, nasopharyngeal samples have further technical and ethical justification [43]. Second, the cross-sectional study design prevented temporal monitoring of viral and bacterial abundance and function; thus, subsequent studies would benefit from longitudinal sampling, including, ideally, the collection of samples prior to hospitalization and samples from noninfectious control infants. Third, antibiotic data were limited to use prior to enrollment, but neither the exact antibiotic used nor the duration of administration was analyzed. However, there were no significant differences in antibiotic use prior to enrollment between infants with RSV-only infection and those with RV-only infection, as shown in Table 1 (P = .75). Fourth, significant financial expense of the techniques used limited the number of samples that could be analyzed. Thus, the present results require validation not only in the full MARC-35 cohort, but also in an independent cohort. Indeed, such temporal and validation analyses would be necessary for biomarker identification. Finally, from an analytic standpoint, not all significant metabolites are present in the KEGG metabolic network model, meaning that MIMOSA would be unable to analyze them. Thus, it is possible that some metabolites considered to be host derived are products of novel microbial metabolism. Notwithstanding these limitations, the current study provides novel evidence to support the emerging hypothesis that RSV and RV induce bronchiolitis through distinct metabolic pathways.
Currently, most clinicians and researchers regard bronchiolitis as a relatively homogenous clinical entity. Although replication of our findings is needed, these data demonstrate the underlying heterogeneity of bronchiolitis by showing that RSV and RV are associated with different metabolic pathways and that both the virus and the co-occurring bacterial species play a role in the host metabolism. Bronchiolitis is also currently considered a viral condition, but these data provide further evidence that bronchiolitis involves a complex interplay among virus, microbiome, and host. Indeed, these results not only challenge long-standing ideas about bronchiolitis, but also, given the lack of effective pharmacotherapies for this very common condition [3–5], have important implications for the future development of novel treatments.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the patients of MARC-35, for study participation; Tulin Ayvaz, from the CMMR, for her work in sample processing; Ginger Metcalf, Donna Muzny, and Richard Gibbs, from the BCM Human Genome Sequencing Center, for their support in sequencing; and Dr Alkis Togias, from the National Institutes of Health (Bethesda, MD), for helpful comments about the study results.
Disclaimer. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (grants UG3 OD-023253, U01 AI-087881, R01 AI-114552, R01 AI-108588, and R21 HL-129909).
Potential conflicts of interest. J. M. M. has provided bronchiolitis-related consultation for Regeneron. N. J. A. and J. F. P. own shares of Diversigen, a microbiome research company. P. A. P. provided bronchiolitis-related consultation for Gilead, Novavax, and Regeneron. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hasegawa K, Mansbach JM, Camargo CA Jr. Infectious pathogens and bronchiolitis outcomes. Expert Rev Anti Infect Ther 2014; 12:817–28. [DOI] [PubMed] [Google Scholar]
- 2. Mansbach JM, Piedra PA, Teach SJ et al. ; MARC-30 Investigators Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med 2012; 166:700–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ralston SL, Lieberthal AS, Meissner HC et al. ; American Academy of Pediatrics Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014; 134:e1474–502. [DOI] [PubMed] [Google Scholar]
- 4. Corneli HM, Zorc JJ, Mahajan P et al. ; Bronchiolitis Study Group of the Pediatric Emergency Care Applied Research Network (PECARN) A multicenter, randomized, controlled trial of dexamethasone for bronchiolitis. N Engl J Med 2007; 357:331–9. [DOI] [PubMed] [Google Scholar]
- 5. Plint AC, Johnson DW, Patel H et al. ; Pediatric Emergency Research Canada (PERC) Epinephrine and dexamethasone in children with bronchiolitis. N Engl J Med 2009; 360:2079–89. [DOI] [PubMed] [Google Scholar]
- 6. Dumas O, Mansbach JM, Jartti T et al. . A clustering approach to identify severe bronchiolitis profiles in children. Thorax 2016; 71:712–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson DJ, Gangnon RE, Evans MD et al. . Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med 2008; 178:667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rubner FJ, Jackson DJ, Evans MD et al. . Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J Allergy Clin Immunol 2017; 139:501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mansbach JM, Hasegawa K, Henke DM et al. . Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol 2016; 137:1909–1913.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stewart CJ, Mansbach JM, Wong MC et al. . Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. a multiomic analysis. Am J Respir Crit Care Med 2017; 196:882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hasegawa K, Jartti T, Mansbach JM et al. . Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the United States and Finland. J Infect Dis 2015; 211:1550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009; 81:6656–67. [DOI] [PubMed] [Google Scholar]
- 13. Hasegawa K, Mansbach JM, Ajami NJ et al. ; the MARC-35 Investigators Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 2016; 48:1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Human T, Project M, Huttenhower C et al. . A framework for human microbiome research. Nature 2012; 486:207–214.22699609 [Google Scholar]
- 15. Caporaso JG, Lauber CL, Walters WA et al. . Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012; 6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res 2015; 43:W251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995; 57:289–300. [Google Scholar]
- 18. Lê Cao KA, González I, Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics 2009; 25:2855–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Team RC. R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing, 2014. http://www.r-project.org/. Accessed 16 January 2018. [Google Scholar]
- 20. Manor O, Borenstein E. Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe 2017; 21:254–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noecker C, Eng A, Srinivasan S et al. . Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. mSystems 2016; 1:e00013–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol 2001; 26:32–46. [Google Scholar]
- 23. Meissner HC. Viral bronchiolitis in children. N Engl J Med 2016; 374:62–72. [DOI] [PubMed] [Google Scholar]
- 24. Hasegawa K, Linnemann RW, Avadhanula V et al. . Detection of respiratory syncytial virus and rhinovirus in healthy infants. BMC Res Notes 2015; 8:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mansbach JM, Clark S, Teach SJ et al. . Children hospitalized with rhinovirus bronchiolitis have asthma-like characteristics. J Pediatr 2016; 172:202–204.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DeVincenzo JP, Wilkinson T, Vaishnaw A et al. . Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med 2010; 182:1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jartti T, Hasegawa K, Mansbach JM, Piedra PA, Camargo CA. Rhinovirus-induced bronchiolitis: lack of association between virus genomic load and short-term outcomes. J Allergy Clin Immunol 2015; 136:509–12.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta 2016; 1864:1372–401. [DOI] [PubMed] [Google Scholar]
- 29. Linda IH, Bruno PL, Alan JW. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol 2010; 77:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J 2005; 25:552–63. [DOI] [PubMed] [Google Scholar]
- 31. Mansbach JM, Camargo CA Jr. Respiratory viruses in bronchiolitis and their link to recurrent wheezing and asthma. Clin Lab Med 2009; 29:741–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Papadopoulos NG, Papi A, Psarras S, Johnston SL. Mechanisms of rhinovirus-induced asthma. Paediatr Respir Rev 2004; 5:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stone CA Jr, Miller EK. Understanding the association of human rhinovirus with asthma. Clin Vaccine Immunol 2015; 23:6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinn RA, Phelan VV, Whiteson KL et al. . Microbial, host and xenobiotic diversity in the cystic fibrosis sputum metabolome. ISME J 2016; 10:1483–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Härtel T, Eylert E, Schulz C et al. . Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J Biol Chem 2012; 287:4260–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ericsson AC, Personett AR, Grobman ME, Rindt H, Reinero CR. Composition and predicted metabolic capacity of upper and lower airway microbiota of healthy dogs in relation to the fecal microbiota. PLoS One 2016; 11:e0154646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mansbach JM, Piedra PA, Teach SJ et al. ; MARC-30 Investigators Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med 2012; 166:700–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jartti T, Aakula M, Mansbach JM et al. . Hospital length-of-stay is associated with rhinovirus etiology of bronchiolitis. Pediatr Infect Dis J 2014; 33:829–34. [DOI] [PubMed] [Google Scholar]
- 39. Stewart CJ, Mansbach JM, Wong MC et al. . Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. a multiomic analysis. Am J Respir Crit Care Med 2017; 196:882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mansbach JM, Hasegawa K, Ajami NJ et al. . Serum LL-37 levels associated with severity of bronchiolitis and viral etiology. Clin Infect Dis 2017; 65:967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hasegawa K, Mansbach JM, Ajami NJ et al. . The relationship between nasopharyngeal CCL5 and microbiota on disease severity among infants with bronchiolitis. Allergy 2017; 72:1796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Charlson ES, Bittinger K, Haas AR et al. . Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011; 184:957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Poole A, Urbanek C, Eng C et al. . Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol 2014; 133:670–8.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.